Abstract
A putative α-amylase gene of Thermotoga petrophila was cloned and expressed in Escherichia coli BL21 (DE3) using pET-21a (+), as an expression vector. The growth conditions were optimized for maximal expression of the α-amylase using various parameters, such as pH, temperature, time of induction and addition of an inducer. The optimum temperature and pH for the maximum expression of α-amylase were 22 °C and 7.0 pH units, respectively. Purification of the recombinant enzyme was carried out by heat treatment method, followed by ion exchange chromatography with 34.6-fold purification having specific activity of 126.31 U mg−1 and a recovery of 56.25 %. Molecular weight of the purified α-amylase, 70 kDa, was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The enzyme was stable at 100 °C temperature and at pH of 7.0. The enzyme activity was increased in the presence of metal ions especially Ca+2 and decreased in the presence of EDTA indicating that the α-amylase was a metalloenzyme. However, addition of 1 % Tween 20, Tween 80 and β-mercaptoethanol constrained the enzyme activity to 87, 96 and 89 %, respectively. No considerable effect of organic solvents (ethanol, methanol, isopropanol, acetone and n-butanol) was observed on enzyme activity. With soluble starch as a substrate, the enzyme activity under optimized conditions was 73.8 U mg−1. The α-amylase enzyme was active to hydrolyse starch forming maltose.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major carbohydrate storage product of terrestrial plants is starch which comprises an important part of the food consumed worldwide. In addition to its direct use as food, starch is also used in many industrial applications, such as manufacturing of high-fructose corn syrups, glues, sizing agent for the paper industry, ethanol production, etc. [1]. A very few of the numerous families of glycosidases and transglycosidases contain starch degrading enzymes which are termed as GH families. The α-amylase family (GH13) had been recognized very early [2–4] and includes enzymes that sometimes share only very limited sequential similarity.
Due to more predictable, controllable and reliable properties of the microbial enzymes, they are favoured than other sources including animals and plants [5]. Amylases are extensively dispersed in various sources including plants, animals, fungi and bacteria due to their role in the breakdown of polysaccharides [6–13]. In most industrial applications, amylases are used at high temperatures, such as 110 °C because starch becomes soluble only at 100 °C and higher temperatures [14]. Due to this reason in applied research, hyperthermophilic archaea are appealing attention because their enzymes show stability at very high temperatures [15]. To accomplish the demand of industries, the expedition for thermostable amylases with novel properties is being explored continuously [16, 17].
A variety of industrially important amylolytic enzymes have been produced by many hyperthermophiles [15]. From various hot environments, a large number of hyperthermophiles with more than 80 °C growth temperature have been isolated [18]. To study adaptation to higher temperatures, cellulosic metabolic pathways and microbial evolution, species of Thermotoga are considered as a model system [19]. Although there are many examples of amylase genes cloned and characterized from other Thermotoga sp. into Escherichia coli [20, 21], very little information is available regarding cloning of α-amylase genes from Thermotoga petrophila into mesophilic host. T. petrophila is a novel hyperthermophilic strain isolated from Kubiki oil reservoir [22]. The genome of this organism bears seven genes that code for α-amylases.
In the present study, cloning of one of the putative α-amylase gene from T. petrophila is reported. Expression of the cloned gene was studied in the mesophilic E. coli strain BL21 (DE3). Further, the recombinant α-amylase was purified and characterized. To the knowledge of the authors, this putative α-amylase gene from T. petrophila has been cloned, purified and characterized for the first time.
Methodology
Selection of Bacterial Strains and Plasmids
From German collection of microorganisms and cell cultures, DSMZ, the genomic DNA of T. petrophila was acquired. BL21 (+) of E. coli was used as host organism for expression of pET 21a (+) vector which was maintained in Lysogeny Broth (tryptone 1 %, yeast extract 0.5 %, NaCl 1 %).
Quantification of Genomic DNA and PCR Amplification
DNA quantification was carried out by spectrophotometric analysis using the spectrophotometer (Cecil-CE7200, Aquarius, UK). α-Amylase gene of T. petrophila was amplified by using the genomic DNA as template. By using the software DNASTAR, a pair of forward and reverse primers was designed on the basis of available DNA sequence information at NCBI database (Gene Bank accession no. KJ572116.1). The sequences of the forward and reverse primers are as follows. NdeI and HindIII restriction sites (shown in bold) were introduced at 5′ end of forward and reverse primers, respectively.
Forward: 5′ GCCATATGCTTTTGAGAGAGATAAACCGATACTGC 3′
Reverse: 5′ GCTCACTCCTGTACAACAAGAACAAAATCAAGGGGT 3′
The 50 μl master mixture for PCR reaction contained 5 μl 10× Taq polymerase buffer, 5 μl 200 μM dNTPs mixture, 5 μl 25 mM MgCl2, l μl 10 μM of each primer, l μl Taq DNA polymerase (10 units), template DNA 1 μl and 31 μl deionized water. Initial denaturation reaction was carried out for 5 min at 95 °C and then undergone 30 amplification rounds of 60, 90 and 60 s at 95, 54 and 72 °C, respectively. For final extension, this was followed by an incubation of 20 min at 72 °C. The amplified product was purified by using Thermo Scientific, GeneJET Gel Extraction Kit after running on agarose gel.
Cloning of α-Amylase Gene in pET-21a (+)
Amplified product of α-amylase gene from T. petrophila was purified by using Thermo Scientific GeneJET DNA purification kit. The purified PCR product was double restricted with Thermo Scientific HindIII and NdeI by using 10× Tango buffer. The double restricted amplified product of α-amylase gene was purified again by using Thermo Scientific GeneJET DNA purification kit after running the agarose gel electrophoresis.
The purified double restricted PCR product of T. petrophila α-amylase gene was ligated into the NdeI and HindIII sites of pET-21a(+) by using Thermo Scientific T4 DNA ligase in its specific buffer (T4 DNA ligase buffer). The reaction mixture for ligation contained the following: 1 μl 10× ligase buffer, 2 μl double restricted pET-21a(+), 4 μl double restricted α-amylase gene and 0.5 μl T4 ligase. Further, this was incubated at 22 °C overnight. The ligated product was then introduced into competent cells of E. coli BL21 (DE3) which were prepared according to the method of Cohen et al. [23]. According to this method, 1 % diluted culture of E. coli BL21 (DE3) was centrifuged at 4 °C and treated with ice cold 50 mM calcium chloride when its optical density reached at 0.6 at 600 nm, to make these cell permeable to external DNA. Validation of positive clones was carried out by colony PCR as well as by analysis of single restricted recombinant plasmid. The isolation of plasmid was carried out by following the method of Birnboim and Doly [24] in which overnight grown culture cells were harvested by centrifugation and treated with solution I (resuspension buffer), solution II (lysis buffer) and solution III (neutralization buffer). Plasmid DNA was precipitated by absolute ethanol and after washing with 70 % ethanol stored in TE buffer.
Expression of Recombinant α-Amylase Gene
For expression of the recombinant α-amylase enzyme, transformed E. coli BL21 cells were cultured in LB broth (100 ml) containing 100 μg/ml of ampicillin to an optical density of 0.5–0.7 at 37 °C. The cells were induced with 0.5 mM isopropyl-D-thiogalactopyranoside (IPTG) and were grown for an additional 4 h at 37 °C. After 4 h, these cells were pelleted down by centrifugation for 10 min at 4 °C. Supernatant was discarded and pellet was resuspended in Tris-HCl buffer (50 mM, pH 7.5 to 8). The cells were lysed by using sonication heat system for 10 min and centrifuged for 10 min at 4 °C. The pellet was again resuspended in Tris-HCl buffer (50 mM, pH 7.5). Expression of α-amylase enzyme was determined in both intracellular and extracellular fractions by using 12 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized after staining with Coomassie brilliant blue [25].
Quantification Assay
Enzyme quantification assay was performed by following dinitrosalicylic acid (DNS) method [26] for determination of reducing sugars. The liberated reducing sugars were measured by spectrophotometer (Cecil-CE7200, Aquarius, UK) at 550 nm. Maltose was used as a standard. Unit of enzyme activity was defined as “the amount of enzyme that liberates one μmole of reducing sugar per ml/min from substrate under assay conditions”.
Total Protein Determination
Total protein estimation was performed by following the method of Bradford [27]. In 100 μl enzyme sample, 5 ml of Bradford reagent and 900 μl phosphate buffer (0.05 M) were added. A control was run simultaneously which contained 5 ml Bradford reagent and 1 ml phosphate buffer (0.05 M). Absorbance was measured at 595 nm by using spectrophotometer (Cecil-CE7200, Aquarius, UK). Bovine serum albumin was used as a standard for the estimation of protein.
Optimization of Enzyme Expression
For maximum enzyme expression, different variables, such as temperature, pH, time of induction and different IPTG concentrations were optimized. Several pH values (4.0–9.0) of medium and different temperatures ranging from 16 to 42 °C were used and enzyme activity was evaluated. Enzyme activity was also calculated after incubation of recombinant E. coli BL21 (DE3) for different time periods between 1 and 7 h after induction with IPTG. Concentration of IPTG was also optimized ranging from 0.1 to 0.6 mM. Standard deviation values obtained by above-mentioned parameters were also calculated.
Effect of Time of Incubation on Enzyme Activity
To optimize the activity of recombinant enzyme, effect of time of incubation on enzyme activity was studied. Enzyme along with substrate was incubated for different time periods ranging from 5 to 30 min.
Enzyme Purification
Partial Purification by Heat Treatment
For partial purification of recombinant α-amylase, incubation was carried out at 70 °C for 1 h with 50 mM Tris buffer pH 8. After 1 h, sample was transferred on ice for 15 min to obtain maximum precipitation of host proteins which were later separated by centrifugation. The partially purified protein was analysed by running SDS-PAGE.
Ion Exchange Chromatography
The partially purified enzyme was filtered through 0.4 μm pore size filter and applied to anion exchange column (Bio-scale Mini Unosphere) which was equilibrated with 50 mM sodium phosphate buffer (pH 7.5). The flow rate was upheld at 1.5 ml min−1. The protein elution was carried out in 50 mM sodium phosphate buffer (pH 7.5) containing linear gradient of NaCl (0–1 M). Elution fractions were analysed by SDS-PAGE analysis.
Determination of Molecular Mass
The molecular mass determination of purified α-amylase was performed by SDS-PAGE analysis. SDS-PAGE electrophoresis (12 %) was accomplished by following Laemmli [25] using ColorPlus pre-stained protein ladder, broad range (10–230 kDa). Coomassie brilliant blue was used as staining dye for proteins visualization.
Characterization
Thermostability
Purified enzyme was characterized for its thermostability. The purified α-amylase was analysed for its activity at wide range of temperatures (60 to 100 °C). To determine the thermostability, the purified enzyme sample was incubated at 60, 70, 80, 90 and 100 °C for 1–4 h without substrate, and the relative activity was measured under standard conditions.
Effect of pH
The effect of pH on enzyme activity was determined by estimating α-amylase activity at various pH values ranging from 4 to 9 pH units. Three different buffers—citrate (0.05 M), phosphate (0.05 M) and glycine-NaOH (0.05 M)—were used. The stability of enzyme for pH was estimated by placing the purified enzyme in respective buffer for 1 to 4 h at room temperature. The remaining enzyme activity was calculated at standard enzyme assay conditions.
Metal Ions and EDTA Effect
The effect of EDTA and metal ions on activity of enzyme was measured by incubating the purified enzyme with 1 mM solution of EDTA and chloride salts of different metal ions (Mg+2, Cu+2, Ca+2, Co+2, Ni+2, Na+1, K+1 and NH4 +1) at room temperature for 1 h. The relative enzyme activity was determined using standard enzyme assay conditions.
Effect of Denaturing Agents
The influence of various inhibitors on activity of purified α-amylase was determined by incubating the enzyme with various concentrations (1–4 %) of different inhibitors, i.e. SDS, Tween 20, Triton X-100, β-mercaptoethanol, Tween 80 and urea, at standard assay conditions. The relative activity of enzyme was measured in comparison to the control, enzyme activity without inhibitor treatment.
Substrate Specificity
Substrate specificity of the α-amylase was measured by using 1.2 % (w/v) glycogen, soluble starch, amylopectin, starch and dextrin from corn and starch from rice as substrates.
Effect of Organic Solvents
The effect of organic solvents (ethanol, methanol, isopropanol, acetone and n-butanol) on the activity of purified α-amylase was calculated by incubating the purified enzyme with 10–40 % (v/v) of these solvents at room temperature for 1 h. The relative enzyme activity was calculated using standard enzyme assay conditions.
Analysis of Hydrolysed Product
Product hydrolysis was analysed by performing thin layer chromatography (TLC). Solvent mixture was prepared by dissolving ethyl acetate/water/methanol in ratio of 40:15:20. Maltose and starch standards (10 mg/ml) were utilized and 50 μl enzyme was prepared in solvent mixture and incubated with 1.2 % substrate at temperature with maximum enzyme activity: i.e. 90 °C for 10 min. Reaction was stopped by placing reaction mixture on ice after incubation. Standards (maltose and glucose) along with reaction mixtures were spotted on TLC plates (Kiesel gel 60 F254; Merck) using micro pipette at room temperature and were allowed to dry. It was then placed in solvent mixture to travel upward through capillary action. When the solvent reached near top of the plate, it was removed and silica plate was dried in oven for 15 min at 95 °C. Reduced sugars were estimated by staining with potassium permanganate stain.
Statistical Analysis
Protected least significant difference method was used for the determination of treatment effects [28]. Analysis of variance (ANOVA) was applied using software Costat.
Bioinformatics Studies
By using molecular evolutionary genetics analysis software (MEGA 5 Beta), phylogenetic analysis was performed. To infer evolutionary relationship among sequences, neighbor-joining method was used. Homology modelling for construction of tertiary structures of α-amylase was carried out by Swiss Model server version 8.05 [29]. Promod-II was used for model building which was based on the target template alignment.
Results
Cloning and Expression of α-Amylase Gene in pET-21a(+)
The concentration of genomic DNA acquired from DSMZ was estimated to be 90 ng/μl. Amplification of α-amylase gene was performed by polymerase chain reaction using specific primers. A 1914-bp band of α-amylase gene obtained after PCR (Fig. 1) was removed from the gel and purified by using Thermo Scientific, GeneJET Gel Extraction Kit. Purified PCR product obtained was double restricted with NdeI and HindIII and, after purification, was ligated into the respective sites of pET-21a(+) by using T4 DNA ligase. It was then transformed into the host bacterial cells of E. coli BL21 (DE3). Single restriction of recombinant plasmid with HindIII produced the single band of 7357 bp on agarose gel which confirmed the successful ligation of α-amylase gene with pET 21a (+) vector that was transformed into the E. coli BL21 (DE3) (Fig. 1). Ligation was also confirmed through colony PCR. The accession no. KT378295 for cloned α-amylase gene of T. petrophila was obtained from NCBI. α-Amylase activity was investigated in both intracellular and extracellular samples. For determination of α-amylase enzyme in intracellular samples, the bacterial cells were lysed by using the sonicator. Quantitative analysis was performed by using DNS method [26]. α-Amylase enzyme activity in intracellular samples was calculated to be 2.5 U/ml/min.
The agarose gel electrophoresis of amplified product of α-amylase and single restriction of recombinant pET-21a (+). Lanes 1 and 2 show the GeneRulerTM DNA ladder mix and amplified α-amylase gene (1914 bp), respectively, while lanes 6 and 7 show single restriction of recombinant expression vector (7357 bp), confirming the successful ligation of α-amylase gene in the expression vector
Optimization of α-Amylase Expression
To increase the yield of α-amylase, the following parameters were studied:
Effect of Incubation Temperatures and pH Values
Effect of incubation temperature on the expression of α-amylase was determined by incubating E. coli at different temperature ranging from 16 to 42 °C. Alpha amylase activity was maximum (2.2 U/ml/min) at 22 °C. Increase or decrease of temperature resulted in decrease of activity (Supplementary Fig. 1A). Similarly, effect of initial pH on the enzyme expression was determined by changing the pH values from 4 to 9 of culture medium for E. coli. Maximum α-amylase activity (2.23 U/ml/min) was exhibited at pH 7.0. Decline in enzyme activity was observed with further increase or decrease in pH value and enzyme activity was reduced to 0.521 U/ml/min at pH 9 (Supplementary Fig. 1B).
Effect of IPTG Concentrations and Induction Time
To improve the expression yield of α-amylase gene in E. coli, the effect of the concentration of the inducer (IPTG) was also studied. Following a 4-h induction period with 0.2–0.7 mM IPTG, the maximum enzyme activity (2.24 U/ml/min) was observed with 0.5 mM IPTG (Supplementary Fig. 1C). Similarly, the effect of IPTG induction (1 to 7 h, 37 °C) on expression of α-amylase was observed to be maximum (2.36 U/ml/min) after 4 h of incubation (Supplementary Fig. 1D).
Time of Incubation
The effect of time of incubation at 85 °C (5–30 min) was studied; maximum enzyme activity (3.562 U/ml/min) was achieved after 10 min of incubation (Supplementary Fig. 4).
Purification of Recombinant Enzyme
The crude α-amylase enzyme was partially purified by heat treatment. Enzyme was incubated at 70 °C for 1 h. All proteins of host cells (E. coli), denatured and precipitated at high temperature, were separated by centrifugation at 10,000×g for 10 min. α-Amylase was further purified by anion exchange chromatography by using Bioscale Mini Unospheres pre-packed column. The eluted fractions of α-amylase with maximum specific activity were pooled together and were purified to 34.60-fold with a specific activity of 126.31 U mg−1 (Table 1).
Molecular Mass Determination
The purified fraction of α-amylase enzyme was determined by SDS-PAGE analysis. The apparent single band at 70 kDa was observed in purified fractions but no band at this position was observed in the controls: non-induced vector only, vector induced with IPTG, cell lysate of wild E. coli BL21 (DE3) and vector containing α-amylase gene which was not induced with IPTG (Fig. 2).
SDS-PAGE analysis of the expression of cloned α-amylase gene. Lanes 1 to 8 represent ColorPlus prestained protein ladder, broad range (10–230 kDa), extract of wild E. coli, non-induced vector only, induced vector only, non-induced vector plus α-amylase gene, induced vector plus α-amylase gene, partially purified cloned α-amylase in E. coli and purified cloned α-amylase in E. coli. A distinct 70-kDa band present in lane 8 represents the purified recombinant α-amylase from T. petrophila
Characterization
Temperature and pH Stability
The optimum temperature for α-amylase activity was found to be 85 °C (3.20 U/ml/min). After pre-incubation (50–100 °C), relative activity of α-amylase enzyme was calculated [30]. The result showed that purified enzyme retained 100 % relative activity at 50, 60, 70 and 80 °C after 2 h of incubation (Fig. 3a, Table 2). The enzyme also showed reasonable stability at 90 °C (85 %) and at 100 °C (78 %). After 4 h of incubation, the relative activity of the enzyme was 80 % at 50 °C and 60 °C, 72 % at 70 °C, 63 % at 80 °C, 54 % at 90 °C and 45 % at 100 °C. Measurement of the pH stability (pH 4–9, 1–4 h) of purified α-amylase [31] indicated that the enzyme showed a good stability at a wide pH range (Table 2). Maximum relative activity (100 %) was obtained after 4 h at pH 7.0 (Fig. 3b). The enzyme exhibited 80 % relative activity at pH 6.0 after 4 h and 70 % after 3 h at pH 8.0. The enzyme activity was drastically decreased to 30 % at pH 9.0 after 1 h of incubation. At pH 4.0 and 5.0, the enzyme activity was 20 and 30 %, respectively, after 1 h of incubation.
Determination of α-amylase stability and substrate specificity. Effect of a temperatures (60–100 °C); b pH; c metal ions; d inhibitors; e organic solvents; f substrate specificity. Standard errors of the mean among three replicates are represented by error bars. The probability level of ≤0.05 is statistically significant
Effects of Different Metal Ions and EDTA
The effects of metal ions and EDTA were measured by incubating the purified enzyme with 1–10 mM solution of EDTA and various metal ions (Mg+2, Ca+2, Co+2, Cu+2, Na+1, K+1, NH4 +1, Hg+2 and Ni+2) at room temperature for 1 h [32]. α-Amylase activity was increased to 150 % in the presence of calcium chloride (Fig. 3c, Table 2). Other metal ions did not considerably affect the activity of purified recombinant α-amylase. However, the α-amylase activity was reduced to 86.33, 71.66, 62.66 and 98.8 % when incubated with 1 mM Mg+2, Ni+2, Co+2 and K+1 salts, respectively. The incubation of enzyme with 1 mM EDTA resulted in the decreased relative activity (58 %) and increased concentration (5–10 mM) of EDTA completely inhibited the enzyme activity.
Effect of Inhibitors and Organic Solvents
After incubating with various inhibitors [33], the results showed that relative activity was decreased to 53, 61 and 71 % when incubated with β-mercaptoethanol, Tween 80 and Tween 20, respectively, after 1 h incubation at the final concentration of 4 % (Fig. 3d). A complete retardation of the activity of enzyme was examined with 4 % SDS after 1 h incubation. Relative activity decreased up to 65 % in the presence of 2 % urea and 60 % in the presence of 2 % Triton X-100 after 1 h incubation. Similarly, results showed that the enzyme was highly stable in the presence of different organic solvents with concentration of 20 %, while higher concentrations slightly inhibited the enzyme activity [34] (Fig. 3e, Table 2). The relative activity was decreased to 87 % with acetone (40 %), 85 % with n-butanol (40 %) and 80 % with isopropanol (40 %).
Substrate Specificity
The purified enzyme showed maximum activity (3.25 U/ml/min) with 1.2 % soluble starch. However, less activity, such as 1.02, 1.021, 0.521, 0.5412 and 0.2145 U/ml/min was observed with starch from corn, rice, glycogen, amylopectin and dextrin from corn, respectively, as shown in Fig. 3f.
Analysis of Hydrolytic Product of α-Amylase
Thin layer chromatography technique was utilized for end product analysis of purified enzyme. Standards for glucose, maltose and starch prepared in at a concentration of 10 mg/ml were run against purified enzyme sample on silica gel plates. By analysis of TLC plates, it was observed that the end product of the purified recombinant α-amylase was maltose (Fig. 4).
Phylogenetic Analysis
The phylogenetic tree was constructed by using neighbor-joining method from recombinant α-amylase protein sequence from T. petrophila and reference strains (Fig. 5). On the basis of α-amylase protein sequence, it showed 97 % similarity with T. maritima and 99 % similarity Thermotoga sp.
Homology Modelling and Structural Analysis
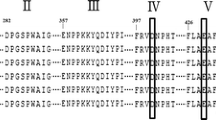
Homology modelling was carried out using SWISS Model version 8.05. Template was searched with Blast and HHBlits. The model was based on template 1gjuA (2.40 Å). The sequence identity with template was 96.38 % and E-value was 0.00e−1. The catalytic domain of the enzyme comprised of amino acids from 124 to 244. The sequence identity with template was 96.38 % and E-value was 0.00e−1 (Fig. 6a, b).
Discussion
More than 25 % of the industrially important enzymes belong to the class amylases [35]. The importance of amylases is due to their potential application in a number of industrial processes, such as paper, food, textiles industries [36], bread making [37], detergent, ethanol, fructose syrups production [38], fruit juices [39], alcoholic beverages [40], sweeteners [41], digestive aid and spot remover in dry cleaning [42]. The objective of the present research work was cloning, expression, purification as well as characterization of α-amylase gene obtained from strain of T. petrophila. An intracellular α-amylase gene, AmyB from T. maritima MSB8 [21], and an extracellular α-amylase gene, AmyE from Thermotoga neapolitana [43], were previously cloned and expressed into E. coli followed by characterization of the enzyme. After cloning and purification, the molecular weight of recombinant α-amylase was 70 kDa as determined by SDS-PAGE analysis. Galdino et al. [44] cloned and expressed amylase gene from Cryptococcus flavus and found the homogeneity on SDS-PAGE analysis with molecular mass 67 kDa. Purification of cloned α-amylase was carried out by heat treatment and anion exchange chromatography by using Bioscale Mini Unospheres pre-packed column which resulted in purification fold up to 159.41 and 78.57 % yield (Table 1). Various studies showed purification of amylases from different thermophiles such as Thermus filiformis, Geobacillus sp. and Pyrococcus sp. by using anion exchange chromatography [45–47]. Purification fold (34.6) and percentage yield (56.25 %) obtained in this study were much better than the previous reports. Chandra et al. [48] purified a thermostable α-amylase from Alicyclobacillus acidocaldarius with purification fold of 8.138, and Yang and Liu [34] reported α-amylase from Thermobifida fusca with percentage yield of 22 %.
A significant feature of this study was stability of purified α-amylase at a broad range of temperature (50 to 100 °C) and pH range (Table 2). These are the two important external physical factors which influence the enzyme activity. The temperature stability of α-amylase observed in this work was rationally higher than other reported thermostable α-amylases from T. maritima and Thermococcus profundus [30, 49].
Inflexible structures of temperature-tolerant enzymes are observed which are converted into flexible form by high temperatures [32]. This feature of the amylases makes them attractive for industrial applications. Enzymatic reaction of amylase is momentously dependent on pH value. Also, the pH tolerance of purified amylase was studied widely for its use in many industries. The purified α-amylase was moderately stable in acidic conditions (at pH 5 and 6) (Fig. 3b, Table 2). Gandhi et al. [50] cloned and expressed an α-amylase from Geobacillus stearothermophilus which retained its maximal activity at pH range of 6.0–8.0. However, Shaw et al. [6] reported pH stability of α-amylase from a Thermus sp. in a range of 5.0–8.0 while Gomes et al. [51] observed optimum pH 6.5–7.0 for α-amylase activity in Rhodothermus marinus.
The addition of metal ions at the concentration of 1, 5 and 10 mM, except for Hg+2 and Cu+2 did not inhibit the enzyme activity at the lower concentrations. α-Amylase activity was increased by 50 % with Ca+2 (Fig. 3c, Table 2). However, the addition of 1 mM metal chelating agent EDTA decreased the enzyme activity up to 50 % and the enzyme activity was totally inhibited addition with 10 mM EDTA. Addition of Hg+2 decreased α-amylase activity suggesting the thiol group existence at the active site of enzymes. Hg+2 react with protein sulfhydryl groups and residues of tryptophan and histidine as well as presence of cysteine residue at the active site of the enzyme [52]. These properties indicate the metaloenzyme nature of the enzyme which is later confirmed by the action of EDTA. Schumann et al. [53] reported a Ca+2-dependent amylase from T. maritima. However, Park et al. [54] cloned an amylase from Thermotoga neopolitana which was neither Ca+2-dependent nor showed any effect on addition of EDTA. Samie et al. [55] reported an enhancement in the amylase activity from Micrococcus sp. in the presence of metal ions while inhibition of the enzyme activity in the presence of EDTA.
The addition of 1 % (w/v) Tween 80, Tween 20 and β-mercaptoethanol did not considerably affect the enzyme activity while higher concentrations showed the inhibition of enzyme activity up to 60 % (Fig. 3d, Table 2). Increased concentration of β-mercaptoethanol caused changes in the conformation of the enzyme in the active site by breaking disulfide bonds that resulting in the loss of enzyme activity [56]. The addition of 1 % (w/v) SDS resulted in decrease of the enzyme activity by 65 % and higher concentrations of SDS completely inhibited the α-amylase. Total loss of enzyme activity with treatment of SDS indicates that hydrophobic interactions must be important in maintaining enzyme structure [57]. Samie et al. [55] reported that amylase from Micrococcus sp. was inhibited in the presence of various surfactants, such as SDS, urea and PMSF.
In all enzymatic reactions, organic solvents are being used for solubilizing of hydrophobic substrates [58]. The addition of the organic solvents at final concentration of 10 % showed a great stability for α-amylases. With higher concentrations, slight inhibition of the activity of the α-amylase was observed with relative activity of 90 %. The organic solvent-tolerant properties demonstrate potential ability of the recombinant α-amylase of T. petrophila for use in starch hydrolysing industries that produce ethanol, fructose, maltose and glucose syrups [59]. The recombinant α-amylase enzyme in this study showed enormous stability against the organic solvents even at high concentrations (40 %) (Fig. 3e, Table 2). Almazo et al. [60] reported that amylase activity from T. maritima was enhanced in the presence of methanol and butanol.
Among the tested substrates, α-amylase displayed highest activity with soluble starch, and moderate activity for starch from rice and corn. Very little enzymatic activity was observed with other substrates. Almost similar results for substrate specificity for α-amylase from T. maritima were obtained [61].
Hydrolysis of soluble starch by α-amylase activity was demonstrated by thin layer chromatography which resulted in the production of maltose as end product (Fig. 4). Classification of amylases is also based on the starch hydrolysis products [31]. As maltose is the major end product of the substrate catalysed by the recombinant α-amylase enzyme, it is therefore classified as saccharifying maltogenic type. Similarly, identification of the α-amylase enzyme was reported for Bacillus subtilis [33] and Clostridium acetobutylicum [62].
The catalytic site of α-amylase of T. petrophila contains two conserved amino acids, Asp468 and Glu414, that are involved in the enzyme catalytic activity (Fig. 6a, b). These conserved amino acids also play an important role in α-amylase catalytic function in T. maritima, Bacillus licheniformis, Bacillus amyloliquefaciens and Alteromonas haloplanctis [63–65]. In close vicinity to the catalytic site are present two conserved amino acids, Arg383 and Asn512, that perform a vital role in binding of Ca+2 that enhance the catalytic role of α-amylase (Fig. 6b) [66]. We have demonstrated that in the presence of Ca+2 ions, the activity of the α-amylase was increased by 50 % (Fig. 3c).
On the basis of all this study, it is suggested that the recombinant purified α-amylase enzyme from T. petrophila could be a potential candidate for use in many industrial processes; particularly, it could be helpful in starch saccharification due to the adaptive capability of the α-amylase to withstand unreceptive conditions used in industrial processes.
References
Van Der Maarel, M. J., Van Der Veen, B., Uitdehaag, J. C., Leemhuis, H., & Dijkhuizen, L. (2002). Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology, 94, 137–155.
Nakajima, R., Imanaka, T., & Aiba, S. (1986). Comparison of amino acid sequences of eleven different α-amylases. Applied Microbiology and Biotechnology, 23, 355–360.
MacGregor, E. A. (1988). α-Amylase structure and activity. Journal of Protein Chemistry, 7, 399–415.
Svensson, B. (1988). Regional distant sequence homology between amylases, α-glucosidases and transglucanosylases. FEBS Letters, 230, 72–76.
Burhan, A., Nisa, U., Gokhan, C., Omer, C., Ashabil, A., & Osman, G. (2003). Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochemistry, 38, 1397–1403.
Shaw, J. F., Lin, F. P., Chen, S. C., & Chen, H. C. (1995). Purification and properties of an extracellular a-amylase from Thermus sp. Botanical Bulletin of Academia Sinica, 36, 195–200.
Tomita, K., Nagata, K., Kondo, H., Shiraishi, T., Tsubota, H., Suzuki, H., & Ochi, H. (1990). Thermostable glucokinase from Bacillus stearothermophilus and its analytical application. Annals of the New York Academy of Sciences, 613, 421–425.
Ilori, M. O., Amund, O. O., & Omidiji, O. (1997). Purification and properties of an α-amylase produced by a cassava-fermenting strain of Micrococcus luteus. Folia Microbiologica, 42, 445–449.
Ribeiro, J. M., Rowton, E. D., & Charlab, R. (2000). Salivary amylase activity of the phlebotomine sand fly, Lutzomyia longipalpis. Insect Biochemistry and Molecular Biology, 30, 271–277.
Hagihara, H., Igarashi, K., Hayashi, Y., Endo, K., Ikawa-Kitayama, K., Ozaki, K., & Ito, S. (2001). Novel α-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Applied and Environmental Microbiology, 67, 1744–1750.
Zółtowska, K. (2000). Purification and characterization of alpha-amylases from the intestine and muscle of Ascaris suum (Nematoda). Acta Biochimica Polonica, 48, 763–774.
Bassinello, P. Z., Cordenunsi, B. R., & Lajolo, F. M. (2002). Amylolytic activity in fruits: comparison of different substrates and methods using banana as model. Journal of Agricultural and Food Chemistry, 50, 5781–5786.
Ikram-ul-Haq, Ashraf, H., Iqbal, J., & Qadeer, M. (2003). Production of alpha amylase by Bacillus licheniformis using an economical medium. Bioresource Technology, 87, 57–61.
Bentley, I., & Williams, E. (1996). Starch conversion. In Industrial enzymology (2nd ed., pp. 339–357). New York: Stockton.
Adams, M. W. (1993). Enzymes and proteins from organisms that grow near and above 100 degree C. Annual Reviews in Microbiology, 47, 627–658.
Karakaş, B., İnan, M., & Certel, M. (2010). Expression and characterization of Bacillus subtilis PY22 α-amylase in Pichia pastoris. Journal of Molecular Catalysis B: Enzymatic, 64, 129–134.
Sidkey, N., Abo-Shadi, M., Balahmar, R., Sabry, R., & Badrany, G. (2011). Purification and characterization of α-amylase from a newly isolated Aspergillus flavus F2Mbb. International Research Journal of Microbiology, 2, 96–103.
Stetter, K. O. (1996). Hyperthermophiles in the history of life. Paper presented at the Ciba Foundation Symposium 202-Evolution of Hydrothermal Ecosystems on Earth (And Mars?).
Frock, A. D., Notey, J. S., & Kelly, R. M. (2010). The genus Thermotoga: recent developments. Environmental Technology, 31, 1169–1181.
Kang, J., Kyung, M. P., Kyoung, H. C., Cheon, S. P., Go, E. K., Doman, K., & Jaeho, C. (2011). Molecular cloning and biochemical characterization of a heat-stable type I pullulanase from Thermotoga neapolitana. Enzyme and Microbial Technology, 48, 260–266.
Lim, W. J., Park, S. R., An, C. L., Lee, J. Y., Hong, S. Y., Shin, E. C., & Yun, H. D. (2003). Cloning and characterization of a thermostable intracellular α-amylase gene from the hyperthermophilic bacterium Thermotoga maritima MSB8. Research in Microbiology, 154, 681–687.
Takahata, Y., Nishijima, M., Hoaki, T., & Maruyama, T. (2001). Thermotoga petrophila sp. nov. and Thermotoga naphthophila sp. nov., two hyperthermophilic bacteria from the Kubiki oil reservoir in Niigata, Japan. International Journal of Systematic and Evolutionary Microbiology, 51(5), 1901–1909.
Cohen, S. N., Chang, A. C., & Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proceedings of the National Academy of Sciences, 69, 2110–2114.
Bimboim, H., & Doly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research, 7, 1513–1523.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Snedecor, G. W., & Cochrane, W. G. (1980). Statistical methods, 7th Ed. Ames, Iowa: Iowa State University Press. ISBN 0-81381560-6.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL Workspace: a web-based environment for protein structure homology modeling. Bioinformatics, 22, 195–201.
Chung, Y. C., Kobayashi, T., Kanai, H., Akiba, T., & Kudo, T. (1995). Purification and properties of extracellular amylase from the hyperthermophilic archaeon Thermococcus profundus DT5432. Applied and Environmental Microbiology, 61, 1502–1506.
Dheeran, P., Kumar, S., Jaiswal, Y. K., & Adhikari, D. K. (2010). Characterization of hyperthermostable α-amylase from Geobacillus sp. IIPTN. Applied Microbiology and Biotechnology, 86, 1857–1866.
Khajeh, M. (2011). Optimization of process variables for essential oil components from Satureja hortensis by supercritical fluid extraction using Box-Behnken experimental design. The Journal of Supercritical Fluids, 55, 944–948.
Matsuzaki, H., Yamane, K., Yamaguchi, K., Nagata, Y., & Maruo, B. (1974). Hybrid α-amylases produced by transformants of Bacillus subtilis. I. Purification and characterization of extracellular α-amylases produced by the parental strains and transformants. Biochimica et Biophysica Acta (BBA)-Protein Structure, 365, 235–247.
Yang, C. H., & Wen-Hsiung, L. (2003). Purification and properties of a maltotriose-producing α-amylase from Thermobifida fusca. Enzyme and Microbial Technology, 35, 254–260.
Sidhu, G., Sharma, P., Chakrabarti, T., & Gupta, J. (1997). Strain improvement for the production of a thermostable α-amylase. Enzyme and Microbial Technology, 21, 525–530.
Fogarty, W. M., & Kelly, C. T. (1990). Recent advances in microbial amylases. In Microbial enzymes and biotechnology (pp. 71–132). Springer.
Cheetham, P. S. J. (1980). Topics in enzyme and fermentation technology (p. 4). New York: Willey. chapter 6.
UpaDek, H., & Kottwitz, B. (1997). Application of amylases in detergents. In J. H. van Ee, O. Misset, & E. J Baas (Eds.). New York: Marcel Dekker.
Wiseman, A. (1980). Topics in enzyme and fermentation technology (Vol. 4). New York: Willey.
Macleod, A. M. (1979). In J. R. A. Pollock (Ed.), Brewing Science. London: Academic Press. R.J., 1(146–232).
Peppler, H. J., & Periman, D. (1978). Microbiological technology (2nd ed.). New York: Academic Press. Chapter 7–16.
Kathleen, T. A. A., T. (1996). Foundation in microbiology. USA: Brown Wm.c. 2nd edition (85).
Choi, K. H., Hwang, S., Lee, H. S., & Cha, J. (2011). Identification of an extracellular thermostable glycosyl hydrolase family 13 α-amylase from Thermotoga neapolitana. The Journal of Microbiology, 49, 628–634.
Galdino, A. S., Silva, R. N., Lottermann, M. T., Alvares, A. C. M., Moraes, L. M. P. D., Torres, F. A. G., & Ulhoa, C. J. (2011). Biochemical and structural characterization of amy1: an alpha-amylase from Cryptococcus flavus expressed in Saccharomyces cerevisiae. Enzyme Research, 2011, 1–7.
Egas, M. C., da Costa, M. S., Cowan, D. A., & Pires, E. M. (1998). Extracellular α-amylase from Thermus filiformis Ork A2: purification and biochemical characterization. Extremophiles, 2, 23–32.
Mollania, N., Khajeh, K., Hosseinkhani, S., & Dabirmanesh, B. (2010). Purification and characterization of a thermostable phytate resistant α-amylase from Geobacillus sp. LH8. International Journal of Biological Macromolecules, 46, 27–36.
Tachibana, Y., Leclere, M. M., Fujiwara, S., Takagi, M., & Imanaka, T. (1996). Cloning and expression of the α-amylase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. Journal of Fermentation and Bioengineering, 82, 224–232.
Chandra, M. S., Mallaiah, K., Sreenivasulu, P., & Choi, Y. L. (2010). Purification and characterization of highly thermostable α-amylase from thermophilic Alicyclobacillus acidocaldarius. Biotechnology and Bioprocess Engineering, 15, 435–440.
Liebl, W., Stemplinger, I., & Ruile, P. (1997). Properties and gene structure of the Thermotoga maritima alpha-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. Journal of Bacteriology, 179, 941–948.
Gandhi, S., Abu Bakar, S., Raja, N. Z. R. A. R., Thean, C. L., & Siti, N. O. (2015). Expression and characterization of Geobacillus stearothermophilus SR74 recombinant α-amylase in Pichia pastoris. Miomed Research International, 2015, 9.
Gomes, I., Gomes, J., & Steiner, W. (2003). Highly thermostable amylase and pullulanase of the extreme thermophilic eubacterium Rhodothermus marinus: production and partial characterization. Bioresource Technology, 90, 207–214.
Chivero, E. T., Mutukumira, A. N., & Zvauya, R. (2001). Partial purification and characterisation of a xylanase enzyme produced by a micro-organism isolated from selected indigenous fruits of Zimbabwe. Food Chemistry, 72, 179–185.
Schumann, J., Alexander, W., Rainer, J., & Karl, O. S. (1991). Topographical and enzymatic characterization of amylases from the extremely thermophilic eubacterium Thermotoga maritime. FEBS Letters, 282, 122–126.
Park, K. M., So-Young, J., Kyoung-Hwa, C., Kwan-Hwa, P., Cheon-Seok, P., & Jaeho, C. (2010). Characterization of an exo-acting intracellular α-amylase from the hyperthermophilic bacteriumThermotoga neapolitana. Applied Microbiology and Biotechnology, 86, 555–556.
Samie, N., Padma, R. M. R., & Masoumeh, A. (2011). Novel extracellular hyper acidophil and thermostable α-amylase from Micrococcus sp. NS 211. Starch-Stärke, 64, 136–144.
Anupama, A., & Jayaraman, G. (2011). Detergent stable, halotolerant α-amylase from Bacillus aquimaris vitp4 exhibits reversible unfolding. International Journal of Applied Biology and Pharmaceutical Technology, 2, 366–376.
Knob, A., Beitel, S. M., Fortkamp, D., Terrasan, C. R. F., & Almeida, A. F. D. (2013). Production, purification, and characterization of a major Penicillium glabrum xylanase using Brewer’s spent grain as substrate. BioMed Research International, 2013, 1–8.
Do, T. T., Quyen, T., & Dam, T. H. (2012). Purification and characterization of an acid-stable and organic solvent-tolerant xylanase from Aspergillus awamori VTCC-F312. ScienceAsia, 38, 157–165.
Shafiei, M., Ziaee, A.-A., & Amoozegar, M. A. (2012). Purification and characterization of a halophilic α-amylase with increased activity in the presence of organic solvents from the moderately halophilic Nesterenkonia sp. strain F. Extremophiles, 16, 627–635.
Almazo, J. Y. D., Alina, M., Agustin, L. M., Xavier, S., Fernando, G. M., & Gloria, S. R. (2008). Enhancement of the alcoholytic activity of α-amylase AmyA from Thermotoga maritima MSB8 (DSM 3109) by site-directed mutagenesis. Applied and Environmental Microbiology, 74, 165168–165177.
Ballschmiter, M., Fütterer, O., & Liebl, W. (2006). Identification and characterization of a novel intracellular alkaline α-amylase from the hyperthermophilic bacterium Thermotoga maritima MSB8. Applied and Environmental Microbiology, 72, 2206–2211.
Paquet, V., Croux, C., Goma, G., & Soucaille, P. (1991). Purification and characterization of the extracellular alpha-amylase from Clostridium acetobutylicum ATCC 824. Applied and Environmental Microbiology, 57, 212–218.
Brzozowski, A. M., David, M. L., Johan, P. T., Henrik, B. F., Allan, S., Torben, V. B., Zbigniew, D., Keith, S. W., & Gideon, J. D. (2000). Structural analysis of a chimeric bacterial α-amylase. High-resolution analysis of native and ligand complexes. Biochemistry, 39, 9099–9910.
Dickmanns, A., Meike, B., Wolfgang, L., & Ralf, F. (2005). Structure of the novel a-amylase AmyC from Thermotoga maritima. Acta Crystallographica, 2006, 262–270.
Aghajari, N., Georges, F., Charles, G., & Richard, H. (1997). Crystal structures of the psychrophilic a-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Science, 1998, 7564–7572.
Brayer, G. D., Yaoguang, L., & Stephen, G. W. (1995). The structure of human pancreatic a-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Science, 4, 1730–1742.
Author information
Authors and Affiliations
Corresponding author
Additional information
Asma Zafar and Muhammad Nauman Aftab contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary figure 1
(DOCX 111 kb)
Supplementary figure 2
(DOCX 1267 kb)
Supplementary figure 3
(DOCX 64 kb)
Supplementary figure 4
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Zafar, A., Aftab, M.N., ud Din, Z. et al. Cloning, Purification and Characterization of a Highly Thermostable Amylase Gene of Thermotoga petrophila into Escherichia coli . Appl Biochem Biotechnol 178, 831–848 (2016). https://doi.org/10.1007/s12010-015-1912-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1912-8