Abstract
A novel pH and thermo-tolerate halophilic alpha-amylase from moderately halophilic bacterium, Nesterenkonia sp.strain F was cloned and expressed in Escherichia coli. 16S rRNA sequence of the strain shared 99.46% similarities with closely related type species. Also, the genome sequence shared ANI values below 92% and dDDH values below 52% with the closely related type species. Consequently, it is proposed that strain F represents a novel species. The AmyF gene was 1390 bp long and encodes an alpha-amylase of 463 amino acid residues with pI of 4.62. The deduced AmyF shared very low sequence similarity (< 24%) with functionally characterized recombinant halophilic alpha-amylases. The recombinant alpha-amylase was successfully purified from Ni–NTA columns with a molecular mass of about 52 KDa on sodium dodecyl sulfate polyacrylamide gel electrophoresis. The enzyme was active over a wide range of temperature (25–75 °C) and pH (4–9) with optimum activity at 45 °C and 7.5, respectively. Also, although it was active over a various concentrations of NaCl and KCl (0–4 M), increasing activity of the enzyme was observed with increasing concentration of these salts. Low concentrations of Ca2+ ion had no activating effect, but high concentrations of the ion (40–200 mM) enhanced activity of AmyF. The enzyme activity was increased by increasing concentrations of Mg2+, Zn2+, Hg2+ and Fe3+. However, it was inhibited only at very high concentrations of these metal ions. Cu2+ did not decrease the amylase activity and the highest activity was observed at 100 mM of the ion. These properties indicate wide potential applications of this recombinant enzyme in starch processing industries. This is the first isolation, cloning and characterization of a gene encoding alpha-amylase from Nesternkonia genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alpha-amylases (E.C 3.2.1.1) are endo-acting enzymes that randomly hydrolyze internal α-1,4-glucosidic linkage of starch, glycogen and related polysaccharides to produce different size of oligosaccharides. These enzymes are classified as family 13 of the glycosyl hydrolases which have similar structure (β/α)8 barrel and the same catalytic mechanism (Pandey et al. 2000).They are among the most important commercial enzymes and are of great significance for biotechnology. Amylases covering about 25–30% of the world enzyme market and have a wide application in starch processing, brewing, alcohol production, textile and other industries (Nigam 2013; Gupta et al. 2003). Although alpha-amylases can be derived from several sources, including plants, animals and microorganisms, microbial amylases are widely used in industrial processes due to large productivity, consistency, environmental protection and cost effectiveness (Mobini-Dehkordi and Javan 2012). Despite benefits, most of these microbial enzymes cannot be used in harsh and diverse conditions of industries. Therefore, there is ongoing interest in finding and developing novel amylases with properties suitable for use in various industries.
Extremophiles are a group of microorganisms that can grow in diverse conditions including extremes of pH, pressure, temperature and salinity. These microorganisms can produce enzymes that function in these extreme conditions, where other microbial enzymes cannot tolerate these conditions (Van Den Burg 2003). Halophilic microorganism, one of the extremophiles, is classified in two categories, extremely halophilic archea and moderately halohilic bacteria. Extremely halophilic archea require high salt concentration (15 to 30% NaCl) for their survival and growth. On the other hand, moderately halophilic bacteria can grow over a wide range of salinities (3 to 15% NaCl) (Ventosa et al. 1998). Halobacteria produce a number of enzymes that have optimal activity at high salinities and could, therefore, be used in many harsh industrial processes where the concentrated salt solution used would otherwise inhibit many enzymatic conversions. Due to the adaptation and activity at different salt concentrations, these groups of enzymes have various potential biotechnological applications in environmental bioremediation, food processing and pharmaceuticals (Ventosa and Nieto 1995; Oren 2010). Considering the functional importance of halophilic enzymes, to date several halophilic amylases have been purified and their biochemical properties reported (Kumar et al. 2016). Earlier, a novel halophilic, SDS and surfactant stable, raw starch digesting and two organic solvent tolerant amylases from a moderately halophilic bacterium Nesterenkonia sp. strain F were purified and characterized (Shafiei et al. 2010, 2011, 2012).
At present, the fermentation and enzyme production technology for extremophiles is not considered to be standard procedure. Therefore, various molecular techniques have been applied to improve the quality and performance of the microbial enzymes for their applications in many industries (Van Den Burg 2003). Gene analysis and prediction of the protein's three-dimensional structure can reveal the molecular properties of the protein. As a result, DNA manipulation and targeted protein engineering provide enzymes that have more biotechnological applications. With this perspective, studies on the molecular cloning and structure prediction of halophilic enzymes are extremely important for understanding the molecular basis of halophilic enzyme properties. However, little molecular studies on halophilic amylases have been reported yet (Wang et al. 2019).
In this paper, we described analysis of a novel halophilic alpha amylase gene and 3D structure prediction of the enzyme from Nesterenkonia sp. strain F. Furthermore, the gene was cloned and expressed with N-terminal histidine tag in E. coli. Then, the recombinant enzyme was purified and biochemical characterization of the novel enzyme was made. To the best of our knowledge this is the first report of cloning and characterization of an alpha-amylase from Nesterenkonia genus.
Materials and methods
Bacterial strain and vectors
The bacteria used in this study were Nesterenkonia sp.strain F and Escherichia coli. Nesterenkonia sp.strain F (NCBI:txid795955), which was isolated from Aran-Bidgol Lake located in the center of Iran (Sarikhan et al. 2011), was identified physiologically, morphologically and molecularly by Amoozegar et al (2013) and was deposited in Iranian Biological Resource Center (IBRC) (IBRC-M 10,223). E. coli DH5α as host strain and T/A vector pTG-19 T from Vivantis (Vivantis Technologies, Malaysia) were used for gene cloning. E. coli BL21(DE3) and vector pET-28b from Novagen were used for gene expression.
Genome analysis
The genome sequence of Nesterenkonia sp.strain F was available in NCBI GeneBank (AFRW00000000.1) (Sarikhan et al. 2011). Analysis of the genome sequence was performed using bioinformatics tools. 16S rRNA gene sequence of strain F (GU5_RS0113080) was analyzed by performing BLASTn program of NCBI to search similar sequences in the database.
To compare strain F genome with other Nesterenkonia species member genomes, average nucleotide identity (ANI) values were determined using two online software tools: (1) EZGenome (Yoon et al. 2017) (https://www.ezbiocloud.net/tools/ani), which calculates ANI by the OrthoANIu algorithm (Lee et al. 2016) and uses USEARCH (Edgar 2010) instead of BLAST (Altschul et al. 1990) and (2) JSpeciesWS (Richter et al. 2015) (http://jspecies.ribohost.com/jspeciesws/), which estimates ANI based on BLAST + (Camacho et al. 2009) (ANIb).
Also, Digital DNA–DNA hybridization (dDDH) values were calculated using the Genome-to-Genome Distance Calculator (GGDC) version 2.1 (Auch et al. 2010; Meier-Koltho et al. 2013) (http://ggdc.dsmz.de/). For determining high-scoring segments pairs (HSP), BLAST + (the recommended default) (Camacho et al. 2009) was used. Genome-to-genome distance values were calculated according to three formulas: Formula 1—the length of all HSPs divided by total genome length; Formula 2—sum of all identities found in HSPs divided by overall HSP length; Formula 3—sum of all identities found in HSPs divided by total genome length.
Genomic DNA extraction
For extraction of genomic DNA, the Nesterenkonia sp.strain F was grown in starch medium containing 1% (w/v) peptone, 0.5% (w/v) yeast extract, 1% (w/v) soluble starch and 7.5% (w/v) NaCl (pH 7) at 33 °C for 72 h, then cells were harvested. Approximately 0.5 g of cells was suspended in 1 ml of 10 mM Tris–HCl (pH 8). Cell lysis was performed by two rapid freeze/thaw cycles in liquid nitrogen and a 37 °C water bath. Then, incubation of the mixture for 1 h at 56 °C was followed by addition 1 ml of lysis cell solution (10 mM Tris, 0.3 M NaCl, 20 mM EDTA, 2% SDS, 100 µg/ml proteinase K). The DNA solution was extracted with chloroform/Isoamyl alcohol (24 V/1 V), and the DNA was precipitated with 0.6 volume of cold isopropanol. After 2 h at room temperature, DNA was pelleted by centrifugation (16,000 × g for 30 min) and re-suspended in TE (1 ml).
Construction of expression vector
Halophilic alpha-amylase gene was amplified from Nesterenkonia sp.strain F genome by PCR. The primers were designed according to the genome sequence of Nesterenkonia sp.strain F. Following oligo-deoxyribonucleotides were used as primers: F 5ʹ-GCGGAATTCATGGTCGCCGCGCT-3ʹ and R 5ʹ-CCAAGCTTGCCGCGCAGCCAG-3ʹ. Forward and reverse primers contained restriction endonuclease sites for EcoRI and HindIII, respectively.
The PCR product was cloned in to T/A vector pTG-19 T by using the procedure described in the instructions for the Vivantis. After transformation in competent cell of E.coli DH5α, the plasmids were isolated and diagnostic restriction analysis was carried out using the restriction endonucleases to select a recombinant plasmid with appropriate orientation of the amylase gene fragment.
The selected recombinant pTG-19 T-amylase plasmid was digested with EcoRI and HindIII. Then, the fragment was gel purified and ligated into the fusion protein expression vector, pET28b previously digested with the same restriction enzymes. The plasmid contains an IPTG or lactose inducible T7 promoter and a hexa-histidine sequence that encodes histidine-tag and facilitates purification of the recombinant enzyme with Ni-chelate columns when fused at N-terminal of the enzyme. The resulting pET28b-amyl plasmid was transformed into competent E.coli BL21(DE3) for protein expression. Colonies were screened and the recombinant expression plasmids were analyzed by the restriction enzyme digestion and sequencing.
AmyF Sequence analysis
After sequencing of the alpha-amylase gene, BLASTn program of NCBI was used to search for similar sequences in the database. Also, similarity of the corresponding alpha-amylase amino acid sequence with protein sequences registered in the protein databases was determined by BLASTp program (http://www.ncbi.nlm.nih.gov/BLAST/). Also, information on family and conserved domains of the amylase was obtained from structure program of NCBI (https://www.ncbi.nlm.nih.gov/Structure). Amino-acid sequence alignments were carried out using ESPript 3.0 program (http://espript.ibcp.fr/ESPript/ESPript/). Theoretical pI and molecular mass corresponding to the amino acid sequence were predicted by ProtParam tool (https://web.expasy.org/protparam/).
Protein structure analysis
Since the three-dimensional structures, crystallography information of ligand–protein complexes and the function of many relevant proteins have been recorded on PDB database (https://www.rcsb.org/), structural information of the novel halophilic amylase should be predicted. Therefore, three-dimensional structure of the enzyme was predicted with SWISS-MODEL (https://swissmodel.expasy.org/) (Arnold et al. 2006), Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) (Kelley et al. 2015) and I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Yang et al. 2015). The softwares predict the 3D model using the crystallographic templates in the PDB database. In the next step, the proSA-web software (https://prosa.services.came.sbg.ac.at/prosa.php) was used to validate the proposed structures. This software evaluates the predicted model based on z-score that indicates overall model quality and measures the deviation of the total energy of the structure with respect to an energy distribution derived from random conformations (Wiederstein and Sippl 2007).
Expression and purification of the recombinant alpha-amylase
E.coli BL21(DE3) cells harboring pET28b-amyl were grown in 50 ml of LB medium (1% (w/v) peptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl) containing 30 µg/ml of kanamycin at 37 °C with shaking until the absorbance at 600 nm was approximately 0.6. The Lac promoter and protein expression was subsequently induced by adding Lactose to a final concentration of 10 mM and incubating at 25 °C for further 8 h. Cells were harvested by centrifugation at 5000 × g for 20 min at 4 °C, and the pellets were suspended in 630 µl of lysis buffer (10 mM Tris pH 7.5, 0.5 mM EDTA, 1 mM PMSF). Then, sonication of the cell suspension was performed for 2 min at 20 °C using ultrasonic sonicator. The supernatant was incubated for 20 min on ice and centrifuged at 12,000 × g for 20 min at 4 °C. The supernatant was used as cellular extract. The recombinant enzyme was purified from the cellular extract by using Ni–NTA columns according to the manufacturerʼs instructions.
Electrophoresis and activity staining
Purity of the protein samples was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) with a 12% separating gel and a 5% stacking gel (Laemmli 1970). Electrophoresis was carried out at 100 V constant voltage on vertical slabs. Marker protein with molecular weight ranging from 9 to 170 KDa was used for estimation of the molecular weight of purified recombinant alpha-amylase. The gel was stained with Coomassie Brilliant Blue R-250 and destained in methanol–acetate–water (1 V:1 V:12 V). Native PAGE was performed under the same conditions as described above except for the absence of SDS in the gel and buffer system. After electrophoresis, the native polyacrylamide gel was soaked for 1 h in 20 mM Tris–HCl buffer (pH 7.0) containing 1% soluble starch at 45 °C with slow shaking. Enzyme activity was visualized by staining the gel with iodine solution (1% I2 and 5% KI) until a clear band appeared.
Amylase activity assay
Recombinant alpha-amylase activity was determined by measuring the amount of reducing sugar released during enzymatic hydrolysis of 1% soluble starch using dinitrosalicylic acid (DNS) method (Miller 1959). 50 µl of purified enzyme was mixed with 450 µl of 1% soluble starch in 20 mM Tris–HCl buffer (pH 7.0), and the mixture was incubated for 10 min at 45 °C. Then the reaction was stopped by the addition of 500 µl of DNS and the mixture was boiled for 5 min. After incubation at room temperature, absorbance of the solution was measured at 540 nm. As a control, a reaction mixture without enzyme was performed. One unit of enzyme activity was defined as the amount of enzyme that releases 1 µmol of reducing sugar as maltose per minute under assay conditions.
Biochemical characterization of the recombinant alpha-amylase
To determine optimum pH of the recombinant enzyme, the following buffers were used in the reaction mixture to investigate the amylase activity at various pH values (4 to 10): 0.1 mM KH2PO4/K2HPO4 (pH 4 to 6), 0.1 mM NaH2PO4/ Na2HPO4 (pH 6 to 7), 0.1 mM Tris–HCl (pH 7.5 to 9) and 0.1 mM glycin/NaOH ( pH 9 to 11). The effect of temperature on activity of the recombinant alpha-amylase was determined by incubating the reaction mixture at various temperatures ranging from 15 to 75 °C. The effect of NaCl and KCl concentrations on the purified enzyme activity was determined in the presence of 0–4 M of each salt in enzyme reaction mixture. The enzyme activities were measured under standard assay conditions. The effect of metal ions on the amylase activity was determined at 45 °C in 20 mM Tris–HCl buffer (pH 7.0) containing 1% soluble starch and various concentrations (0.1 mM to 200 mM) of Ca2+, Fe2+, Mg2+, Cu2+, Hg2+ and Zn2+metal ions.
Results
Genome sequence analysis
16S rRNA gene analysis indicated that strain F fell into the genus Nesterenkonia and shared the highest identity of 99.46% with Nesterenkonia halophila and about 98–95% identity with other species within Nesterenkonia genus members.
Whole genome comparisons were performed by calculating average nucleotide identity (ANI) and in silico DNA–DNA hybridization (dDDH) between strain F and closest members. Genome sequence analysis of the strain F in EZGenome indicated that OrthoANIu average identity to the closest relative Nesterenkonia halophila was 91.72%, while the average nucleotide identity (ANIb) calculated by JSpeciesWS was 91.11%. ANI values between strain F and other members were below than 77.8% (Table 1). All ANI values were below the 95% cut-off generally accepted for differentiating two species (Richter and Rosselló-Móra 2009). Digital DNA–DNA hybridization (dDDH) values were estimated according to three mathematical formulas. dDDH values showed that the strain F shared only about 52% similarity with Nesterenkonia halophila. However, the results indicated that strain F showed less than 23.9% similarity with the other Nesterenkonia species (Table1). The dDDH relatedness values between strain F and other Nesterenkonia species were below the threshold value (70%) for species delineation (Goris et al. 2007).
Nucleotide sequence of the AmyF gene
The novel alpha-amylase gene was successfully cloned from Nesterenkonia sp.strain F in T/A vector pTG-19 T and the recombinant plasmid pTG-19 T-amyF was made. Analysis of nucleotide sequence of the alpha-amylase revealed the presence of a unique ORF of 1389 bp, starting with an ATG codon and terminating with TGA stop codon. The full-length sequence of the halophilic alpha-amylase gene was termed AmyF (amylase from Nesterenkonia sp.F) and deposited in GenBank database with accession number MF523655.1. The GC content of AmyF was 70%. Investigation of data bases revealed that no amylase of Nasterenkonia genus has ever been sequenced and cloned.
Amino acid sequence analysis and comparison
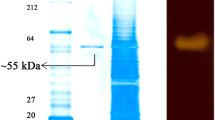
The amino acid sequence deduced from AmyF contained 462 amino acids and encodes a protein with a molecular mass of about 50.8 kDa. The molecular mass was in agreement with the value also estimated by SDS-PAGE of about 52 kDa (Fig. 1a). The amino acid sequence was analyzed with the NCBI database. Blastp homology search indicated that the recombinant alpha-amylase (AmyF) shows the highest level of similarity (74%) to putative enzymes such as α-amylase from Nesterenkonia sp. AN1 (GeneBank ID: EXF24087.1), glycoside hydrolase family 13 protein from Nesterenkonia sp. GY239 (TLP74224.1) and alpha-glucosidase from Nesterenkonia Aurantiaca (TDS83030.1). Furthermore, AmyF has 73.6% similarity to putative α-amylase from Nesterenkonia jeotgali (KUG60193.1) and 72.6% to putative alpha-glucosidases from Promicromonospora thailandica (RIA13948.1) and Haloactinobacterium album (SEE90629.1). The putative enzymes were reported only in NCBI database and no further research has been done on their cloning and properties. However, AmyF shared very low sequence similarity with cloned and functionally characterized halophilic alpha-amylases from Halothermothrix orenii (AAN52525.1) (24.3%) and Pseudoalteromonas sp. M175 (AGC81924.3) (21.5%).
PAGE analysis of AmyF: a SDS-PAGE analysis with 12% denaturating acrylamide separating gel. Lanes: M, the molecular-weight protein marker; 1, the cell lysate of an induced E.coli BL21; 2, the cell lysate of a non-induced recombinant BL21 harboring pET28b-amyF; 3, the after 8 h induction; 4, extracted enzymatic solution of the induced BL21 harboring pET28b-amyF cell lysate before purification; 5, the purified AmyF Ni–NTA chromatography. b Native PAGE analysis using 12% acrylamide gel. Alpha-amylase activity was visualized by activity staining. The band related to the recombinant alpha-amylase is indicated by an arrow
The amino acid distribution and theoretical isoelectric point of AmyF were analyzed (Table 2). The amino acid composition of the halophilic alpha-amylase revealed the presence of hydrophobic amino acids (Ala, Ile, Leu, Phe, Trp, and Val) (36%), acidic amino acids (Asp and Glu) (17.4%) and Arg (8%). Lysine content of the enzyme was very low (0.9%). The isoelectric point of AmyF was calculated to be 4.62. The low isoelectric point of AmyF is mainly due to the fewer number of basic amino acids as has been observed for most halophilic proteins (Madern et al. 2000). Family of the protein and the conserved domains were determined from structure program of NCBI using the gene and amino acid sequences. The obtained results showed that this protein is a member of the glycosyl hydrolyses superfamily that posses a (β/α)8 TIM barrel fold and it has the conserved domain of GH family13 (GH13).
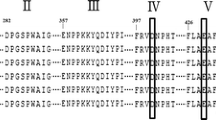
Amino acid sequence alignments were done with ESPript 3.0 program to determine conserved regions. As AmyF showed homology to the alpha-amylase family enzymes belonging to GH13, multiple alignment of AmyF (AWO12117.1) was performed with other alpha-amylases from Halorhabdus tiamatea SARL4B (ZP_08560505), Natronococcus sp. Ah-36 (BAA05516), Haloterrigena turkmenica DSM 5511 (YP_003403666) and Nitrosococcus halophilus Nc4 (YP_003528298) of GH family13 (Supplementary Fig. S1). Four highly conserved regions commonly found in alpha-amylases (MacGregor et al. 2001) were also identified in the AmyF amino acid sequence. The four conserved regions, designated I, II, III and IV, were found as Val14 to His20, Gly120 to Gly128, Ala180 to Val186, and Val239 to Asp244, respectively. Three of the regions are involved in formation of the three catalytic residues (catalytic triad) of AmyF and are conserved among amylases. These residues are catalytic nucleophile Asp124 in Region II, catalytic proton donor Glu183 in region III and Asp244 in region IV which involved in substrate binding and substrate distortion (Henrissat 1991). Furthermore, the amino acid sequence of AmyF contain two conserved histidine residues (His20 and His243) in region I and IV which are involved in the stabilization of the transition state (Santorelli et al. 2016).
Comparisons of AmyF amino acid sequence with other alpha-amylase indicated that similar to the structure of other alpha-amylases, AmyF contains three domains: Domain A (amino acids Met1-Phe46 and Glu101-Glu396), Domain B (Arg47-Pro100) and Domain C (Leu397-Gly462). Domain A folds as a centeral (β/α)8 TIM-barrel and contains the catalytic triad. A large loop between the third β-strand and third helix of the barrel forms Domain B. Domain C which made up of β-strands (Greek key structure), is located at C-terminal end of the protein following of the catalytic (β/α)8 TIM-barrel (MacGregor et al. 2001).
Structure modeling
To model the third structure of the enzyme, the SWISS-MODEL, Phyre2 and I-TASSER were used. The softwares used PDB protein database to predict three-dimentional (3D) structure of the protein. The Phyre2 server automatically selects templates using PDB database and performs modeling based on crystallographic proteins contained in the PDB database. The phyre2 software delivers a 3D model with the help of structural comparison information. I-TASSER modeling starts from the structure templates identified by multiple template alignments programs from the PDB library. I-TASSER only uses the templates of the highest significance in the threading alignments, the significance of which is measured by the Z-score. SWISS-MODEL generates 3D protein model of a target sequence by extrapolating experimental information from an evolutionary related protein structure of SWISS-MODEL template library SMTL that serves as a template. The best template identified by Phyre2 and I-TASSER was crystal structure of isomaltase enzyme from Saccharomyces cerevisiae (PDB code: 3a47A). The alignment z-score by HHSEARCH2 on I-TASSER server was 1.34, which indicated a good alignment. The best structure predicted by the SWISS-MODEL was provided based on crystal structure of alpha-glucosidase from the Halomonas sp.H11 bacterium (PDB code: 3wy4). The GMQE score of the predicted structure was 0.66. The 3D structures of AmyF predicted by the sofwares are shown in Fig. 2. The predicted structures demonstrated that the alpha-amylase from Nesterenkonia sp. F contains three domains with a (α/β)8 motif. ProSA-web program was used to qualify the predicted 3D structures provided by the servers. The results indicated that Phyre2 and SWISS-MODEL model z-scores were − 8.35 and − 7.77, respectively. Therefore, both the structures were reliable structures.
Predicted three-dimensional structures and electrostatic potential surface map of AmyF. a 3D structure of AmyF constructed by Swiss-Model based on the crystal structure of alpha-glucosidase from the Halomonas sp.H11 bacterium (PDB code: 3wy4). b 3D structure of AmyF constructed by Phyre2 based on crystal structure of isomaltase enzyme from Saccharomyces cerevisiae (PDB code: 3a47A). c is the 180° rotated view of a. d Position of the catalytic triad (Asp124, Glu183 and Asp244) is indicated by green arrows in structure of a. f and e show surface electrostatic potentials for a and c, respectively. Red and blue indicate the negative and positive electrostatic potentials, respectively
Expression, purification and activity staining of the recombinant alpha-amylase
The novel alpha-amylase gene (AmyF) was sub-cloned from pTG-19 T-amyF in pET28b expression vector. The resulting pET28b-amyF was transformed into competent E.coli BL21(DE3) for protein overexpression. AmyF expression was induced with 10 mM Lactose and the amylase activity of cellular extract was measured at various hours after induction. The alpha-amylase activity was increased with the increase of cell growth and reached a maximum level at 8 h. Purification of the recombinant protein from the bacterial lysate was achieved by Ni–NTA chromatography column and the purified AmyF migrated as a single band on SDS-PAGE (Fig. 1a). The specific activity of recombinant alpha-amylase in the cellular extract was found to be 0.61 U/mg. After purification by Ni–NTA chromatography, specific activity of the purified enzyme was measured at 3.85 U/mg. Activity staining of the Native-PAGE of the purified alpha-amylase showed one protein band which confirmed the enzyme activity (Fig. 1b).
Effect of pH, temperature and salt concentration on enzyme activity
Activity of the recombinant alpha-amylase was assayed in the presence of various range of pH (3 to 10) and the relative activities are shown in Fig. 3a. Optimum pH of the recombinant alpha-amylase was measured at pH of 7 to 7.5 and the enzyme shown high activity at a pH range of 6.5 to 8. However, the enzyme was active at acidic and alkaline values, as 58 and 50% of the maximum activity remained at pH of 4 and 9, respectively. Also, the effect of temperature on the activity of AmyF was measured in the range of 15 °C to 75 °C (Fig. 3b). The optimum temperature for the recombinant enzyme was 45 °C and it was active at in both low-temperature and high-temperature, as 50 and 60% of the maximum activity was measured at 25 °C and 75 °C, respectively. The effect of NaCl and KCl on the recombinant alpha-amylase activity was determined at various concentrations of the salts. As shown in Fig. 4, it was observed that the amylase was active in various concentrations of NaCl and KCl and activity of the enzyme was increased with increasing concentration of the salts. AmyF showed maximum activity in the presence of 4 M NaCl and 3.5 M KCl and retained about 31% of the maximal activity even in the absence of the salts.
the Effects of pH and temperature on the activity of AmyF. a The effect of pH was determined at different pH (4–10) at 45 °C. b The effect of temperature was determined at the range of 15–75 °C at pH of 7. The relative activities were defined as the percentage of the maximum activity detected in the assay. Values are mean ± SD of three independent experiments
The effect of NaCl (a) and KCl (b) concentrations on AmyF. The activities were determined in the presence of 0–4 M of each salt in enzyme reaction mixture under standard assay conditions. The relative activities were defined as the percentage of the maximum activity detected in the assay. Values are mean ± SD of three independent experiments
Effect of metal ions on enzyme activity
The effect of metal ions on recombinant alpha-amylase activity was examined by measuring the activity in the presence of various concentrations of the metals. All of the metal ions tested (Ca2+, Fe3+, Mg2+, Cu2+, Hg2+ and Zn2+) stimulated the activity of AmyF at different concentrations. The results indicated that the recombinant alpha-amylase was active in the absence of metal ions and did not require any specific metal ions for catalytic activity. The amylase activity increased as the concentration of Ca2+ (40 mM and above) and Cu2+ (0.1 mM and above) ions increased (Fig. 5a). As shown in Fig. 5b activity of the amylase was increased by increasing concentrations of Fe3+ and Mg2+ as maximum activity was observed at 100 mM and 1 mM of the ions, respectively. But higher concentrations of these ions decrease the enzymatic activity. In the presence of Zn2+, the optimum activity was observed at concentrations of 0.1 to 0.5 mM, and high enzyme activity (above 75%) was at 1 to 40 mM range of Zn2+, although higher concentrations decreased the enzyme activity Similarly, maximum activity was measured at 0.1 to 2 mM of Hg2+ and enzyme activity was inhibited at above concentrations (Fig. 5c).
The effect of various concentrations of metal ions on AmyF. a The effect of CaCl2 (filled circle) and CuSO4 (open circle) on the amylase activity of AmyF. b The effect of FeCl3 (filled circle) and MgSO4 (open circle) on the amylase activity of AmyF. c The effect of ZnSO4 (filled circle) and HgCl3 (open circle) on the amylase activity of AmyF. The amylase activity in the presence of the ions was determined at 45 °C in 20 mM Tris–HCl buffer (pH 7.0). The relative activities were defined as the percentage of the maximum activity detected in the assay. Values are mean ± SD of three independent experiments
Discussion
Halophiles are attracting growing attention because of the hyperhalophilicity their enzymes. These enzymes can function under high ionic strengths where most enzymes cannot function at these conditions. Despite that many enzymes have been identified and many of them have opened their way to the industry and biotechnology, the present enzymes cannot supply the entire needs of the industry (Van Den Burg 2003). Due to the importance of alpha-amylase in the industrial sector and assigning of 15 to 20 % of the world market in saccharification sector to this enzyme (Mojsov 2012), today, many halophilic amylases from various sources have been purified and characterized. In our previous studies, we purified and characterized three halophilic alpha-amylases from Nesterenkonia sp.strain F (Shafiei et al. 2010, 2011, 2012). However, genetic studies and understanding the molecular adaptation to extreme salinity of the halophilic enzymes can reveal the new biotechnological potential and uses of these enzymes in industry. Also, the determination of the 3-D structure of some proteins from halophiles has allowed the performance of comparative analysis between halophilic proteins and their non-halophilic homologs in order to understand the relationships between structure and particular biochemical and biophysical properties (Ventosa et al. 2005). In comparison to the extensive use of ‘extremozymes’ from thermophilic and alkaliphilic Bacteria and Archaea, very few halophilic enzymes have thus far found applications in industry and biotechnology. In part, this is due to the limited demand for salt-tolerantenzymes in current manufacturing or related processes (Oren 2010). With the aim of industrial production and expanding the industrial application of useful halophilic alpha-amylases, here we report cloning, expression and characterization of AmyF gene encoding a novel pH and thermo-tolerant halophilic α-amylase from the halophilic bacterium Nesterenkonia sp.strain F. This is the first report about the cloning, sequencing and characterization of a gene encoding moderately halophilic alpha-amylase from Nesternkonia genus. We recorded the sequence of the gene in the NCBI with the registration number MF523655.
Analysis of Nesterenkonia sp.strain F genome sequence indicated that the strain shared 99.46% 16S rRNA sequence similarities with closely related type species. Also, it shared ANI values below 92% and dDDH values below 52% with the closely related type species. Consequently, it is proposed that strain F represents a novel species.
In the present study, the transformation and expression of a halophilic α-amylase gene in E. coli BL21(DE3) was successfully carried out. The results of this study showed that the produced recombinant enzyme had a specific activity of 3.85 U/mg. Most of functionally characterized recombinant halophilic alpha-amylase was cloned and expressed in E. coli BL21(DE3) (Wang et al. 2019, 2018; Yamaguchi et al. 2011; Qin et al. 2014). Mijts and Patel (2002) and Onodera et al. (2013) expressed the cloned halophilic alpha-amylase genes from Halotermothrixorenii bacteria and halophilic Archaeon Haloarcula japonica in the E.coli Top10 and JM109 strains, respectively. However, Jabbour et al. (2012) expressed a recombinant halophilic alpha-amylase in the E. coli M15.
In the present study the AmyF gene was cloned and expressed in one of the well-known expression vector, pET28b. This vector has a sequence related to His-tag at both ends of carbonyl and amine and the polyhistidine tag is suitable for easily purification of the gene product with a Ni2+-chelating column. Also, the vector contained the T7 promoter for large-scale expression of the recombinant gene and induced whit IPTG (isopropyl β-d-1-thiogalactopyranoside) and lactose. IPTG is used to induce low-level recombinant proteins. However, because of cost constraints, for larger scale microbiological processes, the use of Lactose is preferred as an inducer of the expression system (Hoffman et al. 1995). Also, optimizing the induction of gene expression by these inducible materials is very important for reducing costs in industrial applications. Therefore, in this research attempts were made to optimize the type of inducer and induction time in order to prevent the formation of inclusion body. Induction of the recombinant gene expression by various concentrations of IPTG and lactose inducers was analyzed (data not shown). The results indicated that the highest expression was achieved at concentrations of higher than 0.1 mM of IPTG. But, due to the high expression and production of recombinant protein, a large amount of inclusion bodies were formed, and the produced enzyme showed little activity as compared to Lactose induction. Finally, the highest amount of active recombinant enzyme production was obtained with induced by lactose 10 mM for 8 h. Among recombinant halophilic alpha-amylases, the alpha amylases from Alkalibacterium sp.SL3 and Pseudoalteromonas sp.M17 were cloned in pET28a and expressed with 0.3 mM of IPTG (Wang et al. 2018, 2019). In the studies of Yamaguchi et al. (2011), Mijts and Patel (2002) and Jabbour et al. (2012) pTAF, pTrcHis and pJET were used as expression vector, respectively.
All efforts were made to reduce purification process of the recombinant enzyme. For this purpose, the His-tag sequence was linked to the α-amylase gene and purification of the recombinant protein using the affinity chromatography was performed. After affinity chromatography, single protein band with a molecular weight of approximately 52 kDa on the SDS-PAGE gel was observed which indicate successful purification and homogeneity of the purified enzyme. The value of molecular mass of the recombinant alpha-amylase was comparable with that obtained for the organic-solvent-tolerant halophilic alpha-amylase previously purified from Nesterenkonia sp.F (57 kDa) (Shafiei et al. 2011), but different from those which were obtained for the other purified alpha-amylases of Nesterenkonia sp. F (110 and 100 kDa) (Shafiei et al. 2010, 2012). The molecular mass was lower than those of recombinant halophilic amylases from Haloarcula japonica (83 kDa) (Onodera et al. 2013), Zunongwangia profunda (66 kDa) (Qin et al. 2014) and Halothermothrix orenii (62 kDa) (Mijts and Patel 2002). However, similar molecular masses have been calculated for other recombinant alpha-amylases such as those from Natranococcus sp. Ah-36, with relative molecular mass of 52 kDa (Kobayashi et al. 1994); from Haloterrigena turkmenica, 54 kDa (Santorelli et al. 2016); and from Alteromonas haloplanctis A23, 50 kDa (Feller et al. 1992). It should be noted that the His-tag sequences have no effect on the function and activity of the recombinant enzyme and also did not interfere with salt tolerance and other biochemical properties of AmyF.
Alignment of the protein sequence showed that the highest sequence similarity of AmyF (74%) was with putative amylases and the sequence had very little similarity to other sequenced halophilic amylases. The data confirmed that the recombinant halophilic alpha-amylase AmyF is a novel alpha-amylase. Also, AmyF contains 17.4% acidic amino acids. Halophilic proteins typically have an excess of acidic amino acids on their surface. The high negative charges on the surface are neutralized mainly by tightly bound water dipoles and make the enzyme more soluble and more flexible at high salt concentrations, conditions in which non-halophilic proteins tend to aggregate and become rigid (Gomes and Steiner 2004).
By comparing the amino acid composition of the halophilic alpha-amylases with the other halophilic amylases, it was revealed that AmyF was more acidic than Amy13 (16.7%) (Jabbour et al. 2012) and halophilic amylases from Haloarcula hispanica (16.6%) (Hutcheon et al. 2005), Halomonas meridian (12.4%) (Coronado et al. 2000) and Zunongwangia profunda (15.4%) (Qin et al. 2014), but it was less acidic than the halophilic amylases from Alkalibacterium sp. SL3 (20.7%) (Wang et al. 2019) and Natronococcus sp. Ah-36 (24%) (Kobayashi et al. 1994). Moreover, the hydrophobic content of this enzyme is highest among other halophilic amylases (36%) (Table 2). The amino acid content of this enzyme was most similar to that of E. coli JM109 hydrophobic amino acids (35.5%), acidic amino acids (17%) and basic amino acids (8%)) (Wei et al. 2013).The high content (17.4%) of aspartic and glutamic acid residues, low lysine content (0.9%), increased number of small hydrophobic amino acids (alanine, glycine and valine) (24.4%) and decreased number of large hydrophobic residues (isolucine, leusine, phenylalanine and methionine) (16.9%) are the main features of halophilic enzymes (Bolhuis et al. 2008). In addition, high hydrophilic amino acids are displayed on the surface of AmyF which enhances hydration and solubility of the enzyme in the presence of high salt concentrations (Supplementary Fig. S1).
In the CAZy database, the alpha-amylases are grouped with different kinds of glycosyl hydrolases in Family 13 and alpha-amylases sequences contain at least four conserved regions (Janeček and Zámocká 2020). Amino acid sequence analysis of AmyF indicated that the protein has four highly conserved regions and catalytic triad. Ragion I of AmyF contained VDIVPNH and none of the structure templates of the highest significance in the threading alignments have the same sequence. Among the highly conserved amino acids in region I (Asp 15, Asn 19 and His 20), Asp 15 is important for active site integrity. The closest sequence with the conserved sequence of region II of AmyF (GFRVDVAHG) was found in alpha-glucosidase from Bacillus sp. AHU2216 (PDB: 5ZCB) (Auiewiriyanukul et al. 2018) (GFRVDAISHI). The highest variations are found in the amino acid sequences of the conserved regions of III and IV of alpha-amylase family. However, Glu183 in region III, and Asp244 in region IV which have a major role in active site, have the highest conservation (Nielsen and Borchert 2000).
Homology modeling and 3D-structure prediction data indicated that the best structure templates of AmyF were isomaltase enzymes from Saccharomyces cerevisiae (PDB: 3a47A) and Halomonas sp.H11 bacterium (PDB: 3wy4) (Shen et al. 2015). Both of these templates were described as monomer that is correlated with the SDS-PAGE results of AmyF. However, sequence identities of 3a47A and 3wy4 with AmyF were 36% and 32.8%, respectively. Also, ProSA-web program results indicated that the predicted 3D-structures were a reliable structure and AmyF has a similar structure with the structure of GH13 superfamily members.
Salt affects the second or third structure of a protein, and the presence of salt in an enzyme solution can reduce the activity of the enzyme. So, enzymes that can maintain their activity in the presence of different salt concentrations will have many applications in industries that have processes under these salt conditions. The recombinant alpha-amylase retained its activity over a wide range of NaCl and KCl salts. In the presence of KCl, this enzyme showed the highest activity at 3.5 M. But in the case of NaCl, increasing activity of the enzyme was observed with increasing concentration of this salt. The high activity of this enzyme at high salt concentrations is due to the high amount of acidic amino acids on the surface of this enzyme. Negative charges on the halophilic protein binds significant amounts of hydrated ions and produce a hydrated ion networks, thus reducing its surface hydrophobicity, enhancing protein solubility and decreasing the tendency to aggregate at high salt concentration (Gomes and Steiner 2004; Hough and Danson 1999).
Most halophilic enzymes lose their activity at a salt concentration below 1–2 M (Madern et al. 2000). At low salt concentrations, the high amount of acidic amino acids increase electrostatic repulsions, and eventually the protein structure becomes unstable and the enzyme activity is lost (Hutcheon et al. 2005). Halophilic amylases from Natronococcus sp. Ah-36 (Kobayashi et al. 1994) and Alkalibacterium sp. SL3 (Wang et al. 2019), which, respectively, have 24 and 20.7% acidic amino acids, have high activity at high salt concentrations, whereas they show no activity in the absence of salt. However, AmyF also maintained about 31% of its activity even in the absence of salts, which could be due to the medium amount of acidic amino acids. Similar to AmyF (17.4% acidic amino acids), halophlic alpha-amylases from Haloarcula hispanica (16.6% acidic amino acids) (Hutcheon et al. 2005), Haloarcula japonica (18.3% acidic amino acids) (Onodera et al. 2013) and Amy13 (16.7% acidic amino acids) (Jabbour et al. 2012) have activity even in the absence of NaCl. These properties can increase the application potential of AmyF and its use in various industries whose processes are performed under different salt conditions. All three alpha-amylases previously purified from Nesterenkonia sp. F showed the highest activity at NaCl concentrations of about 0.25–0.75 M and their activity decreased by about 50% in 4 M of the salt (Shafiei et al. 2010, 2011, 2012). Whereas the recombinant halophilic amylase from this bacterium showed increased activity with increasing salt concentration and reached its maximum activity in 4 M of NaCl.
Effect of temperature and pH on the activity of recombinant halophilic amylase was also investigated. It was determined that although the optimal activity of this enzyme was in pH of 7–7.5, it was active at a wide range of pH, showing 58 and 50% activity in pH of 4 and 9, respectively. Therefore, this enzyme has the potential to be used in both acidic and alkaline environments and will have significant future applications in industries. Most of the halophilic amylases that have been sequenced so far have optimum activity in pH 7–7.5 except for the amylases from Natonococcus sp. Ah-36 (Kobayashi et al. 1994) and Haloterrigena turkmenica (Santorelli et al. 2016) which has shown optimal activity in pH of 8.7 and 8.5, respectively. However, neither of them was capable to had high activity under both acidic and alkaline conditions. Halophilic alpha-amylase from Pseudoalteromonas sp. M175 was relatively similar to AmyF in terms of pH activity-profile, but in terms of activity in the presence of NaCl, this enzyme showed maximum activity in 1 mM of the salt and its activity were decreased gradually with increasing NaCl concentration (Wang et al. 2018). The alpha-amylases previously purified from Nesterenkonia sp.F did not show any activity at pH 4 and at pH 9 they showed only about 15% activity (Shafiei et al. 2010, 2011, 2012).
Optimal temperature for AmyF activity (45 °C) was similar to those of AmySL3 and AmyHj from Alkalibacterium sp. SL3 (Wang et al. 2019) and Haloarcula japonica (Onodera et al. 2013), respectively. Higher optimum temperature was reported for Amy13 (80 °C) (Jabbour et al. 2012), Haloterrigena turkmenica (55 °C) (Santorelli et al. 2016) and Natranococcus sp. Ah-36 (55 °C) (Kobayashi et al. 1994) halophilic amylases. The maximum activity of recombinant enzyme from Halothermotrix orenii was reported at 65 °C which the maximal activity was depended on the presence of NaCl and CaCl2 (Mijts and Patel 2002). Some other recombinant halophilic alpha-amylases such as those from pseudoalteromonas sp. M175 (25 °C) (Wang et al. 2018) and Zunongwangia profunda (35 °C) (Qin et al. 2014) showed lower optimal activity. However, AmyF exhibited the highest range of thermal activity among all recombinant halophilic alpha-amylases, it maintained 50 and 60% of its activity at 25 °C and 75 °C, respectively. Therefore, this enzyme can be used in both low-temperature and high-temperature processes in a wide range of industries.
Studies of the effect of ions on the recombinant enzyme have shown that this enzyme does not require any ions to function, as the purified enzyme in 20 mM Tris–HCl buffer (pH 7.0) showed a specific activity of 3.85 U/mg. However, different ions increased the activity of this enzyme.
Many alpha-amylases are metalloenzymes containing at least one calcium ion and require the ion for activity (Van der Maarel et al. 2002). Although low concentrations of Ca2+ ion had no activating effect, high concentrations of the ion (40–200 mM) enhanced activity of AmyF. Except for Haloarcula hispanica (Hutcheon et al. 2005) and Zunongwangia profunda (Qin et al. 2014) amylases that increase their activity at 1–8 mM calcium concentration, Ca2+ had no significant effect on activity of most of recombinant halophilic amylases. The activity of either of these enzymes was not measured at high concentrations of calcium ion. Given that the activity of AmyF was calculated in the absence of NaCl, the increase in activity in the presence of high concentrations of Ca2+ could be due to offsetting the effect of Na+ on the stabilization of the enzyme structure by Ca2+.
The effect of various metal ions on AmyF activity indicated that the enzyme activity was increased by increasing concentrations of Zn2+, Hg2+ and Fe3+ metal ions and the activity was reduced only at very high concentrations of these metal ions. However, Cu2+ ions did not decrease the amylase activity and the highest amylase activity was observed at 100 mM concentration of the ion and at a concentration of 200 mM showed only 8% reduction in activity. About other recombinant halophilic alpha amylase, Cu2+, Hg2+, Fe3+and Mn2+ strongly decreased activity of Amy175 from Pseudoalteromonas sp. M175 at 10 mM concentration (Wang et al. 2018) and Cu2+, Zn2+, Fe3+ Ni2+, Co2+ and Al3+ completely inhibited Amy13 (Jabbour et al. 2012). Also, the purified alpha-amylases from Nesterenkonia sp. F were completely inhibited by Zn2+, Cu2+ and Fe3+ ions at 10 mM (Shafiei et al. 2010, 2011, 2012). These results indicating that the recombinant alpha amylase has completely different and more prominent properties for industrial applications than the purified amylases from this bacterium and can be used in a wide range of industries, particularly in the detergent production for use in various areas where their water includes various metals.
Conclusion
In the present study, a novel pH and thermo-tolerant halophilic alpha-amylase gene was isolated from the halophilic bacterium Nesterenkonia sp.F and cloned in E.coli. The homology analaysis of the protein sequence confirmed that AmyF is a novel alpha-amylase. For understanding the biochemical properties of the enzyme, expression in an E. coli expression system and purification with His-tag was performed. To the best of our knowledge, this is the first isolation, cloning and characterization of a gene encoding alpha-amylase from Nesternkonia genus. Biochemical characterization of the amylase indicated that the enzyme is halophilic, pH and thermo-tolerant alpha-amylase. Also, although different metal ions make this enzyme more active, the recombinant alpha-amylase has activity in the absence of metal ions. Therefore, it can be concluded that this enzyme is not dependent on metal ions for activity. These properties indicate that AmyF has potential applications in a variety of industries that whose processes are performed under different conditions of pH, temperature, metal ions or salts. Therefore, AmyF is a good candidate for using in food, textile and waste treatment industries.
References
Altschul SF, Gish W, Miller W, Ew M, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amoozegar MA, Samareh-Abolhasani B, Shafiei M, Didari M, Hamedi J (2013) Production of halothermotolerant α-amylase from a moderately halophilic bacterium, Nesterenkonia Strain F. Prog Biol Sci 2:85–97
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISSMODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Auch AF, Jan M, Klenk HP, Göker M (2010) Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom Sci 2:117
Auiewiriyanukul W, Saburi W, Kato K, Yao M, Mori H (2018) Function and structure of GH13_31 α-glucosidase with high α-(1→4)-glucosidic linkage specificity and transglucosylation activity. FEBS Lett 592:2268–2281
Bolhuis A, Kwan D, Thomas JR (2008) Halophilic adaptations of proteins. In: Siddiqui KS, Thomas T, Uversky V (eds) Protein adaptation in extremophiles: design, selection and applications. Nova Science Publishers, New York, pp 71–104
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Coronado MA, Vargas C, Mellado E, Tegos G, Drainas C, Nieto JNJ, Ventosa A (2000) The alpha-amylase gene amyH of the moderate halophile Halomonas meridiana: cloning and molecular characterization. Microbiology 146:861–868
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Feller G, Lonhienne T, Deroanne C, Libioulle C, Van Beeumen J, Gerday C (1992) Purification, characterization, and nucleotide sequence of the thermolabile alpha-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. J Biol Chem 267:5217–5221
Gomes J, Steiner W (2004) The biocatalytic potential of extremophiles and extremozymes: review. Food Tech Biotech 42:223–235
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B (2003) Microbial α-amylases: a biotechnological perspective. Process Biochem 38:1599–1616
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR (1995) Lactose fed-batch overexpression of recombinant metalloproteins in Escherichia coli BL21 (DE3): process control yielding high levels of metal-incorporated, soluble protein. Protein Expr Purif 6:646–654
Hough DW, Danson MJ (1999) Extremozymes. Curr Opin Chem Biol 3:39–46
Hutcheon GW, Vasisht N, Bolhuis A (2005) Characterization of a highly stable alpha-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 9:487–495
Jabbour D, Sorger A, Sahm K, Antranikian G (2012) A highly thermoactive and salt-tolerant a-amylase isolated from a pilotplant biogas reactor. Appl Microbiol Biotechnol 97:2971–2978
Janeček Š, Zámocká B (2020) A new GH13 subfamily represented by the α-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 24:207–217
Kelley L, Mezulis S, Yates C et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Kobayashi T, Kanai H, Aono R, Horikoshi K, Kudo T (1994) Cloning, expression, and nucleotide sequence of the alpha-amylase gene from the haloalkaliphilic archaeon Natronococcus sp. strain Ah-36. J Bacteriol 176:5131–5134
Kumar S, Grewal J, Sadaf A, Hemamalini R, Khare SK (2016) Halophiles as a source of polyextremophilic alpha-amylase for industrial applications. AIMS Microbiol 2:1–26
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
MacGregor EA, Janecek S, Svensson B (2001) Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim Biophys Acta 1546:1–20
Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4:91–98
Meier-Koltho JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Mijts BN, Patel BK (2002) Cloning, sequencing and expression of an α-amylase gene, amyA, from the thermophilic halophile Halothermothrix orenii and purification and biochemical characterization of the recombinant enzymea. Microbiology 148:2343–2349
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Mobini-Dehkordi M, Javan FA (2012) Application of alpha-amylase in biotechnology. J Biol Today’s World 1:39–50
Mojsov K (2012) Microbial alpha-amylases and their industrial applications: a review. IntJ Manag IT Eng 2:583–609
Nielsen JE, Borchert TV (2000) Protein engineering of bacterial alpha-amylases. Biochim Biophys Acta 1543:253–274
Nigam PS (2013) Microbial enzymes with special characteristics for biotechnological applications. Biomolecules 3:597–611
Onodera M, Yatsunami R, Tsukimura W, Fukui T, Nakasone K, Takashina T, Nakamura S (2013) Gene analysis, expression, and characterization of an intracellular α-amylase from the extremely halophilic archaeon Haloarcula japonica. Biosci Biotechnol Biochem 77:281–288
Oren A (2010) Industrial and environmental applications of halophilic microorganisms. Environ Technol 31:825–834
Pandey A, Nigam P, Soccol CR, Soccol VT, Singh D, Mohan R (2000) Advances in microbial amylases. Biotechnol Appl Biochem 31:135–152
Qin Y, Huang Z, Liu Z (2014) A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: molecular cloning, heterologous expression and biochemical characterization. Extremophiles 18:271–281
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Santorelli M, Maurelli L, Pocsfalvi G, Fiume I, Squillaci G, La Cara F, Del Monaco G, Morana A (2016) Isolation and characterisation of a novel alpha-amylase from the extreme haloarchaeon Haloterrigena turkmenica. Int J Biol Macromol 92:174–184
Sarikhan S, Azarbaijani R, Yeganeh LP, Fazeli AS, Amoozegar MA, Salekdeh GH (2011) Draft genome sequence of Nesterenkonia sp. strain F, isolated from Aran-Bidgol Salt Lake in Iran. J Bacteriol 193:5580–5580
Shafiei M, Ziaee AA, Amoozegar MA (2010) Purification and biochemical characterization of a novel SDS and surfactant stable, raw starch digesting, and halophilic α-amylase from a moderately halophilic bacterium, Nesterenkonia sp. strain F. Process Biochem 45:694–699
Shafiei M, Ziaee AA, Amoozegar MA (2011) Purification and characterization of an organic-solvent-tolerant halophilic α-amylase from the moderately halophilic Nesterenkonia sp. strain F. J Ind Microbiol Biotechnol 38:275–281
Shafiei M, Ziaee AA, Amoozegar MA (2012) Purification and characterization of a halophilic alpha-amylase with increased activity in the presence of organic solvents from the moderately halophilic Nesterenkonia sp. strain F. Extremophiles 16:627–635
Shen X, Saburi W et al (2015) Structural analysis of the α-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism. Acta Crystallogr D Biol Crystallogr 71:1382–1391
Van Den Burg B (2003) Extremophiles as a source for novel enzymes. Curr Opin Microbiol 6:213–218
Van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155
Ventosa A, Nieto JJ (1995) Biotechnological applications and potentialities of halophilic microorganisms. World J Microbiol Biotechnol 11:85–94
Ventosa A, Nieto JJ, Oren A (1998) Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544
Ventosa A, Sanches-Porro C, Martin S, Mellado E (2005) Halophilic archaea and bacteria as a source of extracellular hydrolytic enzymes. In: Gunde-Cimerman N, Oren A, Plemenitaš A (eds) Adaptation to life at high salt concentrations in Archaea, Bacteria, and Eukarya. Springer, Dordrecht, pp 339–354
Wang X, Kan G, Ren X, Yu G, Shi C, Xie Q, Wen H, Betenbaugh M (2018) Molecular cloning and characterization of a novel α-amylase from antarctic sea ice bacterium Pseudoalteromonas sp. M175 and Its primary application in Detergent. Biomed Res Int 1:1–15. https://doi.org/10.1155/2018/3258383
Wang G, Luo M, Lin J, Lin Y, Yan R, Streit WR, Ye X (2019) A new extremely halophilic, calcium-independent and surfactant-resistant alpha-amylase from Alkalibacterium sp. SL3. J Microbiol Biotechnol 29:765–775
Wei Y, Wang X, Liang J, Li X, Du L, Huang R (2013) Identification of a halophilic α-amylase gene from Escherichia coli JM109 and characterization of the recombinant enzyme. Biotechnol Lett 35:1061–1065
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
Yamaguchi R, Tokunaga H, Ishibashi M, Arakawa T, Tokunaga M (2011) Salt-dependent thermo-reversible α-amylase: cloning and characterization of halophilic α-amylase from moderately halophilic bacterium, Kocuria varians. Appl Microbiol Biotechnol 89:673–684
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8
Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Acknowledgements
Financial support for this work was provided by the Research grant from Shahid Chamran University of Ahvaz.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Solat, N., Shafiei, M. A novel pH and thermo-tolerant halophilic alpha-amylase from moderate halophile Nesterenkonia sp. strain F: gene analysis, molecular cloning, heterologous expression and biochemical characterization. Arch Microbiol 203, 3641–3655 (2021). https://doi.org/10.1007/s00203-021-02359-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02359-7