Abstract
Purpose
Knowledge of soil N cycling and the associated functional microbial groups of N2O production under different management measures could provide clues for the restoration of degraded meadows in alpine ecosystems.
Materials and methods
We investigated soil N2O emissions, the genes related to N2O production and reduction (AOA-amoA, AOB-amoA, nirK, nirS, and nosZ), and associated microbial communities in four meadows (continuous grazing, grazing exclusion by fencing, grazing exclusion by combined fencing and reseeding, and undisturbed meadow) in the Tibetan Plateau to reveal the mechanism underlying potential N2O emissions in alpine meadows.
Results and discussion
Compared to the grazing meadow, fencing and fencing + reseeding meadows had lower N2O emissions and lower abundances of AOA-amoA, AOB-amoA, nirK, nirS, and nosZ genes, suggesting that grazing exclusion could decrease the soil N-turnover potential. However, the higher N2O emissions compared to those of undisturbed meadows indicated that longer restoration periods were necessary. Seeding the fenced meadow did not alter soil N2O emission or the abundance of AOA-amoA, AOB-amoA, nirK, nirS, and nosZ genes, possibly owing to the similar soil nutrient status compared to that of the fencing meadow. Grazing exclusion also resulted in significant changes in the community diversity and composition of microbes harboring these functional genes, especially nirK and nosZ communities. N2O emissions were significantly associated with microbial communities involved in N2O production and reduction but not with the gene abundance of AOA-amoA, AOB-amoA, nirK, nirS, and nosZ. Soil dissolved organic nutrients, including C and N, and soil moisture were the controlling factors for N2O production by altering the community composition of nirK- and nosZ-type denitrifiers, such as Bradyrhizobiaceae, Rhizobiaceae, Brucellaceae, Ochrobactrum, and Proteobacteria.
Conclusions
Our results indicated that grazing-induced elevation of potential N2O emissions from meadow soil could be alleviated by grazing exclusion, including sole fencing and a combination of fencing and reseeding, by changing soil dissolved organic nutrients and moisture thus regulating the microbial communities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
As the “third pole” of the Earth, the Tibetan Plateau has the largest alpine meadow ecosystem in the world, and provides huge ecological and social benefits to the local population (Chen et al. 2013). However, intensive human activities (e.g., continuous cultivation and grazing) have caused several negative consequences to alpine meadows, such as rapid degeneration of the structure and function of vegetation and microbial communities, resulting in the decline of soil quality and loss of ecological function (Kooch et al. 2020). Most importantly, overgrazing is considered a major cause of widespread meadow degradation, and it has resulted in severe environmental problems, such as accelerating greenhouse gas (GHG) emissions (Liu et al. 2018).
N2O contributes 8% to the global GHG emissions, and its 100-year global warming potential is 310 times higher than that of CO2 (Harter et al. 2014). N2O is stable over 100 years under natural conditions and results in stratospheric ozone depletion (Fu et al. 2020). Soil is the primary N2O source, producing more than 60% of the atmospheric N2O (Seitzinger and Kroeze 1998). Particularly, pasture is one of the most important sources of GHG, as grazing accelerates N cycling through nutrient recycling via animal manure and animal trampling, which compacts soil and decreases hydraulic conductivity and air permeability (Zhong et al. 2017).
In the Tibetan Plateau, grazing exclusion was adopted to maintain ecosystem functions, which can reduce GHG emissions and greatly contribute to the mitigation of global climate change (Zhong et al. 2017). Fencing is a commonly used method for grassland restoration, which can increase N storage potential and thus reduce soil N2O emissions (Song et al. 2019; Kooch et al. 2020). However, the underlying mechanisms of N2O emission regulation under grazing exclusion remain unclear, such as the association between soil variables and N2O emissions, functional microbes involved in N cycling, and direct or indirect pathways of environmental forces driving N2O emissions. These topics are the basis of guidelines on restoration measures in fragile ecosystems. N2O emissions are primarily produced from two microbial pathways central to N biogeochemical cycling: nitrification and denitrification pathways, which are associated with the supply, leaching, and transformation of N in soil systems (Levy-Booth et al. 2014; Song et al. 2019). Thus, a better understanding of the structure and functioning of microbial communities involved in N transformation is required to control N2O emissions and increase N storage. Certain functional microbial groups determine N availability and losses from the ecosystem, and the abundance of N functional genes (NFGs) can be used to assess the strength of distinct N-cycling pathways (Sun and Badgley 2019; Zhong et al. 2018). The most commonly used N-cycling marker genes in the process of N2O production include amoA of ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA), nirS/K for nitrite reduction, and nosZ for N2O reduction (Stone et al. 2015). Previous studies have reported that nitrification is the major pathway of soil N2O emission in forest and grassland ecosystems (Song et al. 2019; Tzanakakis et al. 2019). Ammonia oxidation, catalyzed by an ammonia monooxygenase encoded by the amoA gene, is the first and rate-limiting step in nitrification mediated by AOA- and AOB-type microbes (Fan et al. 2019). Furthermore, denitrification is also fundamental in N cycling, which contributes to N loss and N2O emissions, in which nitrate (NO3−) is finally reduced to N2 (NO3−-N → NO2−-N → NO → N2O → N2) by denitrifiers that harbor denitrifying genes, such as nirK/S, nosZ, napA, narG, and norB (Levy-Booth et al. 2014). Although the N-cycling process is well documented, how microbial communities that harbor these genes respond to grazing exclusion remains unknown. Moreover, the biotic and abiotic regulation pathways for N2O emissions are unclear, especially in the alpine meadow ecosystem.
Soil N2O emissions are highly variable and can largely be attributed to the coupling of soil properties, vegetation characteristics, and microbial interaction pathways (Glenn et al. 2017). Soil moisture (SM) is a sensitive factor regulating soil N2O emissions because it directly determines oxygen availability in soil pores and regulates the activity of nitrifiers and denitrifiers within the soil profile (Wu et al. 2017). However, the effect of SM on N2O emissions has been previously reported to be positive (Qin et al. 2020), negative (Cai et al. 2016), or neutral (Xie et al. 2015). Thus, the mechanisms underlying soil N2O emission feedback in response to variation in moisture remain unclear. In addition, pH is a vital factor affecting microbial community composition and structure in several ecosystems (Fierer and Jackson 2006; Delgado-Baquerizo et al. 2018) and can regulate soil N2O emissions by shaping nitrifier and denitrifier communities (Brenzinger et al. 2015). Moreover, soil N2O emissions are likely to be positively related to available N, including NH4+-N and NO3−-N, which provide N substrate for nitrification or denitrification processes producing N2O and together with C availability can directly reshape the composition and diversity of functional microbial communities (Zhang et al. 2019c; Yang et al. 2020a, b). In addition, by providing available sources and energy for nitrifiers and denitrifiers and regulating the living environment of microorganisms, dissolved organic matter is also very important to regulate soil N2O emissions (Cai et al. 2014; Qin et al. 2020). The soil N2O emissions are also mitigated by plant properties. Zhang et al. (2019b) found that plant diversity and biomass were the direct drivers of N-cycling potential and related microbial diversity and community composition, while Gu et al. (2019) found that soil N2O emissions significantly correlated with aboveground plant biomass/belowground plant biomass (AGB/BGB). However, the lack of understanding of quantitative links or empirical relationships between microbial functional groups and environmental factors driving N2O emissions limits our ability to effectively reduce GHG emissions and maintain ecosystem services and functions in the Tibetan Plateau.
To determine the underlying mechanism of N2O emission in the meadow ecosystem, we performed experiments in four managed meadows in the Tibetan Plateau, namely, grazing, fencing, fenced + reseeding, and undisturbed meadows. We aimed to assess the effects of grazing exclusion on the potential soil N2O emissions and the abundance and diversity of nitrifying and denitrifying communities. We hypothesized that (i) fenced and fenced + reseeded management can reduce potential N2O emissions, but longer restoration periods will be required to achieve the lowest emissions in the undisturbed meadow, and (ii) soil organic nutrients and moisture are the main controlling factors of N2O emissions by regulating the diversity and community composition of nitrifiers and denitrifiers.
2 Materials and methods
2.1 Study area
The field experiment was carried out at the Bangjietang experimental site of the Alpine Meadow Ecosystem Research Station, Institute Geographic Science and Natural Resources Research, Chinese Academy of Sciences. The study site was located in the northwest part of the Tibetan Plateau in Tibet Province, PR China (32°21′N, 91°40′E; 4672 m a.s.l.). This study area has a typical plateau continental climate with a mean annual precipitation of 409.0 mm, with most of it occurring during the summer season (July to August) (Wang et al. 2020b). The mean annual temperature is about −3.0 °C, annually ranging from −15.0 to 7.9 °C in January and July, respectively, and the annual accumulated temperature (> 0 °C) is 846 °C. The soil is mainly composed of Mattic-Cryic Cambisol (Alpine meadow soil, Cambisols in FAO/UNESCO taxonomy). The vegetation comprises a typical alpine meadow, with Kobresia pygmaea, Stipa purpurea, Kobresia robusta, Kengyilia thoroldiana, and Elymus nutans being the main species.
2.2 Experimental design
The experimental area covered an area of 200 hm2 and was freely grazed by yaks until the establishment of the experiment in 2009 (Wang et al. 2020b). Four meadow types were established: grazing meadow (GM), grazing exclusion by fencing meadow (FM), grazing exclusion by fencing + reseeding meadow (FRM), and undisturbed meadow (UM). Twenty plots (4 treatments × 5 replications) were established in a randomized complete block design, and each plot covered 1 hm2. The sites had similar soil type, altitude, and slope gradient and aspect. The four meadows were characterized as follows. (a) GM was grazed by four yaks per hectare. The grazing intensity was determined according to annual plant biomass that can feed the yaks and was allowed at intervals after accounting for the time required for plant re-growth instead of continuous grazing throughout the year. (b) FM was enclosed by iron fencing in 2009 to restore vegetation without grazing livestock. FM had similar grazing management to that of GM before fencing. (c) FRM was fenced and seeded in 2009. FRM also had similar grazing management to that of GM before fencing. Elymus nutans Griseb., Poa cymophila Keng., and Kengyilia thoroldiana (Oliver) J. L. were seeded at a depth of 3 cm, spaced 20 cm apart, at seeding densities of 3, 5, and 8 g m−2, respectively. (d) UM was an original alpine meadow that had not been disturbed by human activities or livestock.
2.3 Vegetation surveys and soil physicochemical analysis
Soil sampling and vegetation surveys were conducted in September 2018, when the AGB was the highest. Nine 10 × 10 m blocks were established in each plot and separated by 10–20 m along a diagonal line. Then, in each block, three 1 × 1 m quadrats were set up for plant community and soil sampling. After litter, stones, and debris were removed, soil samples were collected from each block with a soil auger from the top 15 cm of the soil profile along with an S-shaped pattern. Finally, nine composite samples from each block were combined thoroughly to form one sample. Combining multiple soil samples can reduce the errors caused by soil heterogeneity at sampling points. Each collected soil sample was divided into two subsamples. One subsample was immediately stored at −80 °C for DNA extraction, and the other was air-dried to determine the soil physicochemical properties. Furthermore, a vegetation survey was conducted in each quadrat. Plants were dug up to determine the following vegetation characteristic parameters: coverage, AGB/BGB, and richness. AGB was obtained by harvesting and air-drying aboveground parts, and BGB was obtained by washing the roots under a tap and drying at 70 °C for 48 h. Plant diversity was estimated using the Shannon–Wiener index.
Soil pH was determined using a combination pH meter with a soil-to-water ratio of 2.5:1 (v/w). Soil water content was measured by oven-drying the soil to a constant mass at 105 °C. Soil total nitrogen (TN) was detected using Kjeldahl digestion. Soil NH4+-N and NO3−-N were extracted with 2 M KCl at a 1:5 ratio and measured using a continuous flow auto analyzer (AutAnalyel, Bran+Luebbe GmbH, Norderstedt, Germany), as described by Zhang et al. (2016). Soil organic carbon (SOC) was determined by the potassium dichromate oxidation method. Total phosphorus (TP) was measured using melt-molybdenum, antimony, and scandium colorimetry. Dissolved organic carbon (DOC) and dissolved organic nitrogen (DON) were analyzed using a TOC analyzer (LiquiTOC II, Elementar Analysensysteme GmbH, Langenselbold, Germany) after extraction with distilled water. Soil available phosphorus (AP) was extracted using 0.5 M NaHCO3 at pH 8.5 and measured using the molybdate ascorbic acid method with a UV spectrophotometer (Camspec, Cambridge, UK).
2.4 N2O flux measurements
The static closed-chamber method was used to collect in situ fluxes of N2O. In brief, five static chambers consisted of a PVC pipe (height 25 cm, radius 10 cm). A movable cap was randomly inserted into each plot and separated by about 50 m to collect N2O gas, and the soil depth was 10 cm (Hall et al. 2014). Soil N2O gases were sampled every 2 weeks from July to September in 2018, that is, on July 2, July 16, July 30, September 13, and September 27. Insulating cotton was used to wrap the outer walls of the chambers to prevent temperature fluctuation caused by external radiation and wind interference. N2O sampling was conducted between 10:00 and 16:00 every sampling day to minimize the diurnal temperature variations (Rowlings et al. 2015; Mafa-Attoye et al. 2020; Zhang et al. 2021). Gas samples were collected from the headspace of each chamber at 0, 10, 20, and 30 min using a 30-mL airtight syringe and transferred to gas sampling bags (30 mL) sealed with a butyl rubber stopper. After each gas collection, the cap was removed and ventilation for 5 min was allowed to avoid the influence of pressure changes on N2O emissions. N2O fluxes were measured simultaneously in the laboratory after the gas sampling. The concentration of N2O was determined using a gas chromatography (Agilent Technologies Inc., Santa Clara, CA, USA) equipped with an electron capture detector with N2O electron capture detectors. The average hourly N2O emission was calculated using the following equation (Zhong et al. 2013; Li et al. 2021):
where \({\text{R}}_{{\text{N}}{2}{\text{O}}}\) is the N2O emission rate (μg·m−2·h−1), \({\text{h}}\) is the height of the static chamber (m), \(\frac{{\Delta}{\text{C}}}{{\Delta}{\text{t}}}\) is the rate of N2O concentration increase in sealed chamber per unit time (× 10−9 min−1), \({\text{M}}\) is the N molecular mass in a mole of N2O (28 g·mol−1), \(\frac{273}{\text{273} + \frac{{\text{T}}{1}+{\text{T}}{2}}{2}}\) is the temperature correction coefficient, and T1 and T2 are the temperatures inside the static chamber at the first and last sampling, respectively, (°C).
2.5 Microbial analysis
2.5.1 Microbial DNA extraction
DNA was extracted from 0.5 g of frozen soil using a FastDNA SPIN kit (MP Biomedicals, Cleveland, OH, USA), according to the manufacturer’s protocols. The concentration and purity of extracted DNA were checked with a NanoDrop® ND-2000 spectrometer (Thermo Fisher Scientific, Wilmington, DE, USA) at 260/280 nm: concentration ≥ 20 ng/μL, OD 260/280 = 1.8–2.0. All purified DNA samples were stored at − 80 °C until analysis.
2.5.2 Quantitative PCR analysis
Quantitative PCR (qPCR) with ABI 7500 real-time quantitative PCR system (Applied Biosystems, Foster City, CA, USA) was used to determine the absolute abundance of NFGs with selected primer pairs and thermal cycling conditions (Table S1). Tenfold serially diluted plasmids of each gene were subjected to real-time PCR assays in triplicate to generate standard curves (Liu et al. 2018). Negative controls, samples, and standard series were run in triplicate in 96-well plates. The qPCR reaction mixture (20 μL) used for amplification comprised 10 μL of SYBR® Premix ExTaq™ (TaKaRa), 1 μL template DNA, 0.8 μL of each forward and reverse primer (20 μM), and 7.4 μL ddH2O. Furthermore, a melting curve analysis was performed following each assay to ensure the specificity of amplification.
2.5.3 Amplicon sequencing and phylogenetic classification
After DNA extraction, a portion of DNA was used as a template for PCR with primer pairs and thermal cycling conditions chosen to amplify the AOA-amoA, AOB-amoA, nirS, nirK, and nosZ genes and to investigate the microbial community associated with N cycling by amplicon sequencing. Fragments of nirK and nosZ were amplified using the primer pairs nirK876 (5′-ATYGGCGGVAYGGCGA-3′)/nirK1040 (5′-GCCTCGATCAGRTTRTGGTT-3′) and nosZ2F (5′-CGCRACGGCAASAAGGTSMSSGT-3′)/nosZ2R (5′-CAKRTGCAKSGCRTGGCAGAA-3′) (Henry et al. 2004), respectively. The primer pairs for AOA, AOB, and nirS were the same as those for qPCR (Table S1). Mothur software v. 1.30.1 was used to process the high-throughput sequence, and quality-controlled sequences were obtained after filtering out low-quality reads, adaptors, and barcodes (Schloss et al. 2009). All high-quality effective sequences were clustered into operational taxonomic units (OTUs) using the software Usearch, with a 97% sequence identity threshold, and the most abundant representative sequence for each OTU was chosen for phylogenetic classification. Taxonomic assignment was conducted with RDP classifier version 2.2 (http://rdp.cme.msu.edu/) against the FunGene database (http://fungene.cme.msu.edu/) and a confidence threshold of 0.7 (Fish et al. 2013). The sequences have been deposited into the NCBI Sequence Read Archive (SRA) with accession number SRP300131.
2.6 Statistical analysis
One-way analysis of variance (ANOVA) followed by Duncan’s tests for multiple comparisons was performed to assess the effect of grazing exclusion on plant characteristics, soil properties, N-cycling functional genes, and N2O emissions at a significance level of 0.05. Principal coordinates analysis (PCoA) and pairwise comparisons with a permutational multivariate ANOVA were performed to examine the overall differences between the NFG assemblages and related microbial community structures, based on Bray–Curtis distances. Spearman’s correlation was used to examine the relationships of soil physicochemical and vegetation properties with NFG abundances and N2O emissions, while Mantel’s test was used to evaluate the relationships of the structure of functional microbial communities with soil physicochemical and vegetation properties and N2O emissions. Multiple linear regression (MLR) was used to identify the most important drivers of N2O emissions, including soil properties and vegetation characteristics (Pahlavan-Rad et al. 2020). Partial least squares path modeling (PLS-PM) was used to reveal the direct and indirect pathways to N2O emissions with the index of goodness of fit (GOF) representing the statistical accuracy of estimates. Random forest modeling (RFM) and Spearman’s correlation were also used to identify the critical microbial taxa (relative abundance > 0.01) affecting soil N2O emissions (Wang et al. 2021). All statistical analyses were conducted in R v.3.6.1 software (R core team) using the “vegan,” “ggplot2,” “relaimpo,” “plspm,” and “rfPermute” packages.
3 Results
3.1 Soil and vegetation properties and N2O emissions
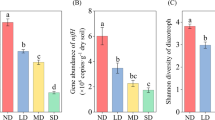
Soil physical properties and nutrients differed significantly among the four meadows (Table 1). In general, SM, SOC, TP, DOC, DON, and AP showed an increasing trend after grazing exclusion, and the highest values were found in UM. Contrastingly, NO3−-N and NH4+-N were the lowest in FRM, while the FM site showed the lowest C/N ratio. pH showed a narrow variation ranging from 8.32 to 8.90, and the lowest value was found in UM. Moreover, the highest and lowest TN occurred in UM and FRM, respectively. Vegetation characteristics also varied among the four meadows (Table 2). AGB, BGB, and coverage in FM, FRM, and UM were higher than those in GM. The highest and lowest Shannon diversity was found in FM and UM, respectively, and no significant differences in species richness existed among the four meadows (P > 0.05). Soil N2O emissions ranged from 7.3 to 38.22 μg·m−2·h−1 (Fig. 1) and were significantly lower in FM and FRM compared to those in GM, while being higher than that in UM (P < 0.05). No significant difference was observed in the N2O emissions between FM and FRM (P > 0.05).
Soil N2O emissions under different meadows. Means and standard errors are presented per treatment (n = 5). Different letters indicate significant differences between different treatments (Duncan’s test, P < 0.05). GM, grazing meadow; FM, fencing meadow; FRM, fencing + reseeding meadow; UM, undisturbed meadow
3.2 Abundances of NFGs
NFG abundances varied significantly among the four meadows (P < 0.05; Fig. 2). Considering nitrification, the highest abundance of AOA-amoA among the meadows was found in UM. AOA-amoA and AOB-amoA abundances in FM and FRM were significantly lower than those in GM, and no significant difference in AOB-amoA was found among FM, FRM, and UM. For denitrification, similar trends were found in nirS, nirK, and nosZ. The highest gene abundances of nirS, nirK, and nosZ were obtained in UM, followed by those in GM, FM, and FRM, and the NFG abundances in UM were significantly higher than those in GM, FM, and FRM (P < 0.05). PCoA indicated a clear separation in the functional genes among different meadows (Fig. 3), which suggested that the N-cycling function varied significantly among the four treatments. The first two principal coordinate axes explained 56.23% of the total variation.
Abundance of N-cycling functional genes under different meadows. Means and standard errors are presented per treatment and measurement date (n = 5). Different letters indicate significant differences between different treatments (Duncan’s test, P < 0.05). GM, grazing meadow; FM, fencing meadow; FRM, fencing + reseeding meadow; UM, undisturbed meadow
3.3 Diversity and composition of N-cycling microbial communities
The diversity and richness of N-cycling microbial communities were characterized by Shannon and Chao1 indexes, respectively (Table 3). Overall, the diversity and richness of functional microbial communities significantly differed among the four meadows (P < 0.05) and showed different fluctuation trends. Regarding nitrifiers, the lowest diversity and richness of the AOA community were observed in UM, whereas no significant difference was found among those of GM, FM, and FRM (P > 0.05). Contrastingly, AOB showed no significant difference among the four meadows (P > 0.05). Regarding denitrifiers, Chao1 richness of nirS and nosZ showed a similar trend, with higher values found in GM and FRM, whereas Shannon diversity showed no significant differences. The diversity and richness of nirK in GM, FM, and FRM were not significantly different (P > 0.05). However, the highest Shannon index and lowest richness of nirK were found in UM.
The changes in the composition of five functional communities at the class and genus levels are shown in Fig. S1 and Fig. 4, respectively. AOA community composition was primarily ascribed to Crenarchaeota (Fig. 4A). Besides, unclassified archaea were also the main components of the AOA community. Crenarchaeota abundance in FM and FRM was higher than that in UM and GM. In the AOB community, Nitrosospira was the dominant genus detected in all samples (Fig. 4B). Meanwhile, unnamed ammonia-oxidizing bacteria ensemble and unclassified Nitrosomonadales belonging to the class Betaproteobacteria accounted for a large proportion of the AOB community (Fig. 4B; Fig. S1B). Regarding denitrifiers, the major community contributions to nirK-type denitrifying bacterial communities were unclassified Bacteria and Proteobacteria, while the nirS-type denitrifying bacterial community was composed of Bradyrhizobium, unclassified Brucellaceae, unclassified Bradyrhizobiaceae, and unclassified Rhizobiaceae (Fig. 4C and D). The OTUs of nosZ-type microbes of each site were mainly annotated to 10 genera (relative abundance > 0.01), such as Bradyrhizobium, Ochrobactrum, Pseudomonas, and Paracoccus (Fig. 4E). At the class level, unclassified Proteobacteria was the major component of the nirS community (Fig. S1C). Meanwhile, both nirK- and nosZ-denitrifying bacteria mainly comprised Alphaproteobacteria (Fig. S1D and E). Although large proportions of the N-cycling microbial communities were not identified to the finer levels of classification, PCoA at the OTU level significantly (P < 0.05) separated the N-cycling microbial community among treatments (Fig. 5), indicating large shifts in functional microbial communities after grazing exclusion. The first two principal coordinate axes explained 72.51%, 44.13%, 45.55%, 45.78%, and 35.38% of the total variation for AOA, AOB, nirS, nirK, and nosZ communities, respectively.
3.4 Regulatory pathways of multiple drivers on N2O emissions
The correlations among soil properties, vegetation characteristics, and N2O emissions are shown in Table 4. Soil N2O emissions were negatively correlated with SM, SOC, TN, TP, DOC, DON, AP, and NO3−-N, while showing a positive correlation with soil pH (P < 0.05). AGB, BGB, and coverage were also significantly and positively correlated with soil N2O emission (P < 0.05). Contrastingly, the abundance of none of the NFGs was significantly correlated with the N2O emissions (P > 0.05; Fig. 6A). Mantel’s test showed that, except that of AOB, microbial community compositions were significantly correlated with soil N2O emissions (P < 0.05). MLR indicated that the most important variables were DOC, DON, and SM, with a total explained variance of over 55% (Fig. 6B). Finally, PLS-PM was performed to further explore the complex biotic and abiotic pathways for regulating N2O emission (Fig. 7). SM had direct positive effects on AOB and nirK communities and negative effects on the nirS community. Furthermore, DOC had a negative direct effect on the nosZ community. Although DOC had a negative direct effect on N2O emissions, it had larger effects on the AOB and nosZ communities. The direct negative contributors to N2O emissions were nirK and nosZ communities, while the AOB community was a positive contributor. Overall, the effect of all driving factors explained 84.6% of the total variance in N2O emissions. RFM indicated that 56.94% of the N2O emission variance was explained by 36 taxa, and only eight taxa (~ 22.2%) were identified as significant predictors of N2O emissions (Fig. 8; P < 0.05), including nirK, nosZ-harboring microbes. Brucellaceae of nirK was the main predictor of soil N2O emissions. Spearman’s rank correlation showed that N2O emissions were significantly correlated with most of the identified taxa (Table 5). The correlation analysis results indicated that Proteobacteria of nosZ-type bacteria and Bradyrhizobiaceae and Rhizobiaceae of nirK-type bacteria were the main drivers that were suppressing N2O emissions, whereas Brucellaceae of nirK-type bacteria was positively related to N2O emissions.
The results of Spearman’s correlation analysis and Mantel’s test. A Spearman’s rank correlation analysis of individual gene abundance with soil properties, vegetation characteristics, and N2O emissions showed above in A. The blue and red colors show, respectively, positive and negative relationships between two variables. The deeper the color and the larger the square, the stronger correlation relationships. Significant correlations are indicated by *P < 0.05, **P < 0.01, ***P < 0.001. Functional microbial composition (based on OTU) was related to each environmental factor and N2O emissions by partial Mantel tests using Bray–Curtis distance shown in an interaction network in the bottom of A. Edge color corresponds to the Mantel’s R statistic for the corresponding distance correlations. The nodes’ size of environmental factor is proportional to the number of connections with significant correlation to microbial communities through partial Mantel tests. Insignificant correlations (P > 0.05) are not shown in mantel test. B Relative variable importance in multiple linear regression. GM, grazing meadow; FM, fencing meadow; FRM, fencing + reseeding meadow; UM, undisturbed meadow; SM, soil moisture; SOC, soil organic C; TN. total N; TP, total P; DOC, dissolved organic C; DON, dissolved organic N; AP, soil available P; C/N, the ratio of SOC and TN; AGB, aboveground biomass; BGB, belowground biomass
Partial least squares path modeling (PLS-PM) shows the standardized effect of abiotic and biotic factors on potential soil N2O emissions. Each box represents an observed latent variable. The loading for the microbial community that creates the latent variables are shown in the dashed rectangle. Larger path coefficients are reflected in the width of the arrow; blue indicates a positive effect and red, a negative effect. Path coefficients are calculated after 1000 bootstraps. Coefficients of inner model differ significantly from 0 are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001. The diversity and richness of N-cycling microbial communities were characterized by Shannon and Chao1 indexes. The model is assessed using the goodness of fit statistic. Path coefficients that were not significantly different from 0 were shown as dashed lines. SM, soil moisture; DOC, dissolved organic C; DON, dissolved organic N; abund., gene abundance
Random forest modeling analysis identified the main taxa predicting the changes in N2O emissions (showing only the significant results). Significance levels of each predictor are as follows: *P < 0.05 and **P < 0.01. GM, grazing meadow; FM, fencing meadow; FRM, fencing + reseeding meadow; UM, undisturbed meadow
4 Discussion
4.1 Effects of grazing exclusion on soil N2O emissions
Elucidating the N2O emission mechanisms is important to develop effective management strategies for meadows that are sensitive to climate change. In this study, we aimed to determine the underlying mechanisms of grazing exclusion on soil N2O emissions and to recognize the nitrifying and denitrifying communities associated with N2O emission. As expected, our results showed that soil N2O emissions dropped significantly after grazing exclusion in the grazing meadow (P < 0.05), but longer restoration periods were required to reach the lowest emissions recorded in the undisturbed meadow. This was consistent with the findings of Wang et al. (2020a) and Yin et al. (2020), reporting that soil N2O emissions decreased during revegetation and increased after grazing. The high N2O gas emissions in grazing meadows can be largely attributed to urine and dung deposited in the meadows by the grazing yaks, which are an important source of N2O (Zhu et al. 2020; Chen et al. 2020). Furthermore, animal trampling can increase soil compaction and create an anaerobic environment conducive to denitrification, the main process behind N2O emissions in grazing soil (Brucek et al. 2009). Besides, aggregate destruction and suppression of plant growth caused by animal trampling are also the main factors facilitating rapid N turnover (Uchida et al. 2008). In this study, N2O emissions were negatively correlated to soil nutrients (e.g., DOC, DON, and SOC) and plant characteristics (Table 4). Contrastingly, previous studies suggested that the decrease in N2O emissions during the natural restoration of abandoned farmlands was caused by decreasing soil C and N availability, which suppressed denitrifying activities responsible for N gas production (Zhang et al. 2019c). This difference may be attributable to the composition and diversity of functional microorganisms and complex soil–microbe interactions in those studies. Thus, it is important to identify the driving path of environmental factors and specific functional microorganisms involved in N2O emissions.
4.2 Effects of grazing exclusion on NFGs and related microbial communities
Compared to those with grazing, the abundance of all NFGs involving nitrification and denitrification showed decreasing trends after grazing exclusion (Fig. 2), suggesting that fencing and reseeding could decrease the soil N-turnover potential. This conclusion is contrary to several studies, which indicated that restoration increased the abundance of NFGs for nitrification and denitrification (Song et al. 2019). In a semiarid area, 10-year fencing and vegetation restoration may not increase NFG abundances because microorganisms are likely to suffer nutrient limitation in the early stages of vegetation restoration with insufficient water (Cui et al. 2020; Wang et al. 2020b). However, vegetation restoration over decades or a century in UM can significantly increase NFG abundances, which is consistent with our previous finding in grasslands of a semiarid area under long-term grazing prohibition (Song et al. 2019). In this study, seeding the fenced meadow did not alter the soil N2O emission or N-cycling potential, possibly owing to the soil nutrient status being similar to that of FM (Table 1). Correlation analysis showed that all NFG abundances except that of AOB-amoA showed positive correlations with soil nutrients (e.g., SOC, TN, TP, NO3−-N) (Fig. 6A). These results were consistent with those of Wang et al. (2020b) and Song et al. (2019), which indicated that a favorable environment is necessary to facilitate microorganism growth.
This study showed the drastic variations in the composition and structure of functional microbial communities (Figs. 4 and 5; Fig. S2). At the genus level, significant changes in the relative abundance of N-cycling microorganisms were observed in the four treatments, particularly in nirK and nosZ communities. Similar changes in microbial composition occurred during the restoration of abandoned farmland, as reported by Wang et al. (2020a). Several prominent explanatory variables were responsible for reshaping N-cycling microbial communities. Mantel’s test showed that N-cycling functional microbial composition, except that of AOB, was significantly related to SM, pH, soil nutrients (e.g., SOC, TN, TP, NO3−-N), and vegetation characteristics, i.e., BGB and coverage (Fig. 6A), agreeing with previous studies (Zhong et al. 2017). Several studies have indicated that SM increases lead to increasing gene abundance and changes in the composition and diversity of nitrifying and denitrifying microbial communities (Prosser and Nicol 2012; Fan et al. 2019), and soil nutrients may be the main factors inducing changes in microbial community composition and diversity, as microorganisms have different resource acquisition strategies and niche specialization and differentiation (Henry et al. 2008; Fan et al. 2014; Zhong et al. 2017). Moreover, plant community structure can also act as a major driver for soil N cycling by providing different compositions and amounts of litter and root exudates, further improving soil quality (Trivedi et al. 2020). Plant species also influence the structure and distribution of microbial communities observed by selecting the microbial communities they host, and further regulate the ecological process of N cycling. Yang et al. (2020a, b) and Li et al. (2020) reported that pH was one of the most important factors shaping communities of nitrifiers and denitrifiers; however, the obvious effect of pH commonly occurred in acid soil where pH ranged from 4.5 to 6.8 (Brenzinger et al. 2015). In our study, the roles of pH on microbial communities could not be confirmed in alkaline soil with a narrow variation (8.32–8.90).
4.3 Possible drivers of N2O emissions
The copy number of functional genes is widely used to characterize the potential of specific N-cycling pathways. However, no gene abundances had a significant correlation with N2O emissions in our study (Fig. 6A). Similarly, Fan et al. (2019) reported that the correlation between N2O emissions and the abundance of related functional genes was not significant in a fertilizing field experiment. Moreover, Yin et al. (2020) found that enhancement of N2O emissions by grazing is related to soil physicochemical characteristics rather than to the gene abundance of nitrifiers and denitrifiers in alpine grassland; however, their statistical methods were limited because correlation analysis may not identify the interaction effects of environmental factors and microbes on N2O emissions. The linkage between microbial functional genes and ecosystem processes may be too complex to simply predict the biogeochemical processes using the copy number of functional genes (Zhang et al. 2019c). Thus, gene abundance will unlikely explain the simultaneous N2O emission because such rates are dominated by real-time environmental variables, especially dissolved organic nutrients and specific microbial taxa (Yin et al. 2020). Furthermore, the activity of microbial enzymes and related functional gene expression should also be considered to fully understand the evolution of the N-cycling and N2O emission processes during long-term restoration of meadow ecosystems. Indeed, the soil microbial community structure showed a strong relationship with soil N2O emissions in our study (Fig. 6A). Similarly, Wang et al. (2020a) found that restoration in abandoned farmland has the potential to mitigate N2O emissions by changing N cycle-related microbial communities. Thus, the composition and structure of the N-cycling microbial community may be more capable of predicting N2O emissions than the single indicator of gene abundance.
Plant–soil–microbe regulation mechanisms to suppress the emissions of N2O occur during grazing exclusion and restoration (Kravchenko et al. 2017). However, we found that vegetation characteristics had a minor effect on N2O emissions by MLR (Fig. 6B). Plants can change the microbial community by secreting different matters into the soil, such as carbon compounds, or by changing the nutrient status through litter decomposition (Zhang et al. 2019a). This suggests that the influence of plants on microbial processes is mainly indirect, while soil has a greater direct impact on the N2O production process by providing effective nutrients to microorganisms and reaction substrates for the N cycle. SM, DOC, and DON were the primary environmental variables determining the efficacy of fencing and restoration in reducing N2O emissions (Figs. 6 and 7). Similarly, Cai et al. (2016) found that watering increased DOC concentrations by 72–234% but decreased N2O emissions by 33–60% in an experiment with increased precipitation from a mixed grassland site in southern Alberta, Canada. Grazing exclusion directly suppressed potential soil N2O emissions by changing AOB, nirK, and nosZ communities and indirectly by altering SM, DOC, and DON (Fig. 7). Similarly, Mehnaz et al. (2019) found an interaction effect of P availability and denitrifiers on N2O emissions and NO3− leaching. Dissolved organic matter, which can supply these microbes with additional available organic nutrients, was the major controlling factor for nitrification and denitrification processes in soils (Levy-Booth et al. 2014). A meta-analysis showed that increasing C and N bioavailability can improve microbial N immobilization. (Cao et al. 2021). In addition to dissolved organic nutrients, soil moisture content is an important factor for soil N2O emission, which affects the activity of soil microbes and determines the processes for N2O production (Feng et al. 2018; Qin et al. 2020; Zuo et al. 2020).
Regarding microbial properties, nirK and nosZ microbial communities were the main inhibitors of N2O emissions (Fig. 7), which is in line with several studies (Jones et al. 2013; Aamer et al. 2020). Wang et al. (2020a) reported that nirK and nosZ communities may play predominant functional roles in denitrification during restoration in abandoned farmland. Mitigation of N2O emissions from soil has been widely attributed to denitrifiers harboring nosZ, as the only known biotic sink for N2O catalyzing N2O reduction to N2 (Shaaban et al. 2018). In contrast, AOB were the main promoters of N2O emissions. Although AOB gene abundance was lower than that of AOA in our study, AOB contributed to the vast majority of nitrification, as proven by several reports (Tzanakakis et al. 2019). Therefore, the inhibition of N2O emissions by grazing exclusion mainly affects denitrification, whereas the nitrification process still plays a positive role in N2O emissions. Moreover, using meta-transcriptomes and dual-label 15 N-18O isotope analysis, Fang et al. (2019) revealed a shift in the N2O emissions pathway from nitrification to nitrifier denitrification after fumigation, supporting further research in natural ecosystems. Surprisingly, we found that the nirK community has a significant inhibitory effect on soil N2O emissions, in contrast to their codified gene functions for promoting N2O production (Qin et al. 2020). Several studies have also reported a negative relationship between the nirK community and N2O emissions (Fan et al. 2019; Huang et al. 2019). Hallin et al. (2018) indicated that nirK genomes are more likely to carry nosZ than nirS genomes. Thus, nirK microbes usually co-occur with nosZ microbes. nirK-type denitrifiers may be more likely to harbor potential for complete denitrification, suggesting that lower N2O emissions do not reflect lower denitrification, but could be the result of N2O reduction to N2 by nosZ (Fan et al. 2019). This explanation might fit the results because labile C and N are generally considered to enhance the strength of N cycling by providing substrates and energy sources for microorganisms (Stow et al. 2005), which is also consistent with the high N-cycling potential and low N2O emissions in UM (Figs. 1 and 2). Furthermore, Miller et al. (2008) and Morley and Baggs (2010) found simple C sources (e.g., glucose and sucrose) to be more likely to induce complete denitrification than complex C sources. Previous studies have also reported that nosZ communities are sensitive to oxygen concentration (Feng et al. 2018). In our study, high SM in UM supports that denitrification attained completion with the reduction of N2O to N2, suggesting that N2O production may be lower than N2O reduction by grazing exclusion, leading to a decrease in N2O emission. RFM and Spearman’s correlation also supported the above arguments (Fig. 8; Table 5), with N2O emissions being mainly suppressed by Proteobacteria of nosZ-type bacteria and Bradyrhizobiaceae and Rhizobiaceae of nirK-type bacteria. The inhibitory effect of Bradyrhizobium on N2O emission has been verified by laboratory culture experiments (Akiyama et al. 2016). Although we identified several taxa strongly associated with N2O emissions, only a few taxa (~ 22.2%) were identified as significant contributors to N2O emissions (Fig. 8). This suggested that widespread functional redundancy also existed in the N-cycling process of the alpine meadow ecosystem, which may be an important factor influencing soil GHG emissions (Wang et al. 2021).
5 Conclusions
The results of this study supported our hypothesis that grazing-induced increase in N2O emissions in meadow soil could be alleviated by grazing exclusion, including fencing and a combination of fencing and reseeding. Soil DOC, DON, and moisture were the main environmental drivers of N2O emissions, as they changed the community abundance and structure of nirK- and nosZ-type denitrifiers rather than N-cycling gene abundance. Only 22.2% of the microbial taxa were identified as predictors of N2O emissions, suggesting a functional redundancy for N cycling in meadows. Our results revealed the regulatory mechanisms for N2O emissions during grazing-to-fencing conversion and provided insights for the management of alpine meadow ecosystems.
References
Aamer MS, M H, M U, Huang GQ, Liu Y, Tang HY, Rasul F, Ma QY, Li ZL, Rasheed A, Peng Z, (2020) Biochar mitigates the N2O emissions from acidic soil by increasing the nosZ and nirK gene abundance and soil pH. J Environ Manage 255:7
Akiyama H, Hoshino YT, Itakura M, Shimomura Y, Wang Y, Yamamoto A, Tago K, Nakajima Y, Minamisawa K, Hayatsu M (2016) Mitigation of soil N2O emission by inoculation with a mixed culture of indigenous Bradyrhizobium diazoefficiens. Sci Rep 6:8
Brenzinger K, Dorsch P, Braker G (2015) pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front Microbiol 6:11
Brucek P, Simek M, Hynst J (2009) Long-term animal impact modifies potential production of N2O from pasture soil. Biol Fertil Soils 46:27–36
Cai Y, Wang X, Tian L, Zhao H, Lu X, Yan Y (2014) The impact of excretal returns from yak and Tibetan sheep dung on nitrous oxide emissions in an alpine steppe on the Qinghai-Tibetan Plateau. Soil Biol Biochem 76:90–99
Cai YJ, Chang SX, Ma B, Bork EW (2016) Watering increased DOC concentration but decreased N2O emission from a mixed grassland soil under different defoliation regimes. Biol Fertil Soils 52:987–996
Cao Y, He Z, Zhu T, Zhao F (2021) Organic-C quality as a key driver of microbial nitrogen immobilization in soil: a meta-analysis. Geoderma 383:114784
Chen H, Zhu Q, Peng C, Wu N, Wang Y, Fang X, Gao Y, Zhu D, Yang G, Tian J, Kang X, Piao S, Ouyang H, Xiang W, Luo Z, Jiang H, Song X, Zhang Y, Yu G, Zhao X, Gong P, Yao T, Wu J (2013) The impacts of climate change and human activities on biogeochemical cycles on the Qinghai-Tibetan Plateau. Glob Chang Biol 19:2940–2955
Chen Z, Wang Q, Zhao J, Chen Y, Wang H, Ma J, Zou P, Bao L (2020) Restricted nitrous oxide emissions by ammonia oxidizers in two agricultural soils following excessive urea fertilization. J Soils Sediments 20:1502–1512
Cui Y, Wang X, Zhang X, Ju W, Duan C, Guo X, Wang Y, Fang L (2020) Soil moisture mediates microbial carbon and phosphorus metabolism during vegetation succession in a semiarid region. Soil Biol Biochem 147:107814
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-Gonzalez A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N (2018) A global atlas of the dominant bacteria found in soil. Science 359:320–325
Fan FL, Yin C, Tang YJ, Li ZJ, Song A, Wakelin SA, Zou J, Liang YC (2014) Probing potential microbial coupling of carbon and nitrogen cycling during decomposition of maize residue by C-13-DNA-SIP. Soil Biol Biochem 70:12–21
Fan X, Yin C, Chen H, Ye M, Zhao Y, Li T, Wakelin SA, Liang Y (2019) The efficacy of 3,4-dimethylpyrazole phosphate on N2O emissions is linked to niche differentiation of ammonia oxidizing archaea and bacteria across four arable soils. Soil Biol Biochem 130:82–93
Fang WS, Yan DD, Huang B, Ren ZJ, Wang XL, Liu XM, Li Y, Ouyang CB, Migheli Q, Cao AC, Wang QX (2019) Biochemical pathways used by microorganisms to produce nitrous oxide emissions from soils fumigated with dimethyl disulfide or allyl isothiocyanate. Soil Biol Biochem 132:1–13
Feng ZJ, Sheng YQ, Cai F, Wang W, Zhu LZ (2018) Separated pathways for biochar to affect soil N2O emission under different moisture contents. Sci Total Environ 645:887–894
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631
Fish JA, Chai BL, Wang Q, Sun YN, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:14
Fu QL, Xi RZ, Zhu J, Hu HQ, Xing ZQ, Zuo JC (2020) The relative contribution of ammonia oxidizing bacteria and archaea to N2O emission from two paddy soils with different fertilizer N sources: a microcosm study. Geoderma 375:12
Glenn EP, Jordan F, Waugh WJ (2017) Phytoremediation of a nitrogen-contaminated desert soil by native shrubs and microbial processes. Land Degrad Dev 28:361–369
Gu X, Wang Y, Laanbroek HJ, Xu X, Song B, Huo Y, Chen S, Li L, Zhang L (2019) Saturated N2O emission rates occur above the nitrogen deposition level predicted for the semi-arid grasslands of Inner Mongolia, China. Geoderma 341:18–25
Hall MKD, Winters AJ, Rogers GS (2014) Variations in the diurnal flux of greenhouse gases from soil and optimizing the sampling protocol for closed static chambers. Commun Soil Sci Plant Anal 45:2970–2978
Hallin S, Philippot L, Loffler FE, Sanford RA, Jones CM (2018) Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol 26:43–55
Harter J, Krause HM, Schuettler S, Ruser R, Fromme M, Scholten T, Kappler A, Behrens S (2014) Linking N2O emissions from biochar-amended soil to the structure and function of the N-cycling microbial community. ISME J 8:660–674
Henry S, Baudoin E, Lopez-Gutierrez JC, Martin-Laurent F, Brauman A, Philippot L (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods 59:327–335
Henry S, Texier S, Hallet S, Bru D, Dambreville C, Cheneby D, Bizouard F, Germon JC, Philippot L (2008) Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ Microbiol 10:3082–3092
Huang R, Wang Y, Liu J, Li J, Xu G, Luo M, Xu C, Ci E, Gao M (2019) Variation in N2O emission and N2O related microbial functional genes in straw- and biochar-amended and non-amended soils. Appl Soil Ecol 137:57–68
Jones CM, Graf DRH, Bru D, Philippot L, Hallin S (2013) The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J 7:417–426
Kooch Y, Moghimian N, Wirth S, Noghre N (2020) Effects of grazing management on leaf litter decomposition and soil microbial activities in northern Iranian rangeland. Geoderma 361
Kravchenko AN, Toosi ER, Guber AK, Ostrom NE, Yu J, Azeem K, Rivers ML, Robertson GP (2017) Hotspots of soil N2O emission enhanced through water absorption by plant residue. Nat Geosci 10:496–500
Levy-Booth DJ, Prescott CE, Grayston SJ (2014) Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol Biochem 75:11–25
Li PP, Zhang SQ, Li F, Zhang YT, Han YL (2020) Long term combined fertilization and soil aggregate size on the denitrification and community of denitrifiers. Appl Soil Ecol 156:103718
Li Z, Li L, Xia S, Zhang R, Zhang R, Chen P, Pan J, Liu Y (2021) K fertilizer alleviates N2O emissions by regulating the abundance of nitrifying and denitrifying microbial communities in the soil-plant system. J Environ Manage 291:112579
Liu SB, Zamanian K, Schleuss PM, Zarebanadkouki M, Kuzyakov Y (2018) Degradation of Tibetan grasslands: consequences for carbon and nutrient cycles. Agric Ecosyst Environ 252:93–104
Mafa-Attoye TG, Baskerville, MA Ofosu E, Oelbermann M, Thevathasan NV, Dunfield, KE (2020) Riparian land-use systems impact soil microbial communities and nitrous oxide emissions in an agro-ecosystem. Sci Total Environ 724:138148
Mehnaz KR, Keitel C, Dijkstra FA (2019) Phosphorus availability and plants alter soil nitrogen retention and loss. Sci Total Environ 671:786–794
Miller MN, Zebarth BJ, Dandie CE, Burton DL, Goyer C, Trevors JT (2008) Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol Biochem 40:2553–2562
Morley N, Baggs EM (2010) Carbon and oxygen controls on N2O and N2 production during nitrate reduction. Soil Biol Biochem 42:1864–1871
Pahlavan-Rad MR, Dahmardeh K, Hadizadeh M, Keykha G, Mohammadnia N, Gangali M, Keikha M, Davatgar N, Brungard C (2020) Prediction of soil water infiltration using multiple linear regression and random forest in a dry flood plain, eastern Iran. Catena 194:104715
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531
Qin H, Xing X, Tang Y, Zhu B, Wei X, Chen X, Liu Y (2020) Soil moisture and activity of nitrite- and nitrous oxide-reducing microbes enhanced nitrous oxide emissions in fallow paddy soils. Biol Fertil Soils 56:53–67
Rowlings DW, Grace PR, Scheer C, Liu S (2015) Rainfall variability drives interannual variation in N2O emissions from a humid, subtropical pasture. Sci Total Environ 512–513:8–18
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Seitzinger SP, Kroeze C (1998) Global distribution of nitrous oxide production and N inputs in freshwater and coastal marine ecosystems. Global Biogeochem Cy 12:93–113
Shaaban M, Wu YP, Khalid MS, Peng QA, Xu XY, Wu L, Younas A, Bashir S, Mo YL, Lin S, Zafar-ul-Hye M, Abid M, Hu RG (2018) Reduction in soil N2O emissions by pH manipulation and enhanced nosZ gene transcription under different water regimes. Environ Pollut 235:625–631
Song ZL, Wang J, Liu GB, Zhang C (2019) Changes in nitrogen functional genes in soil profiles of grassland under long-term grazing prohibition in a semiarid area. Sci Total Environ 673:92–101
Stone MM, Kan JJ, Plante AF (2015) Parent material and vegetation influence bacterial community structure and nitrogen functional genes along deep tropical soil profiles at the Luquillo Critical Zone Observatory. Soil Biol Biochem 80:273–282
Stow CA, Walker JT, Cardoch L, Spence P, Stow CA (2005) N2O emissions from streams in the Neuse River watershed, North Carolina. Environ Sci Technol 39:6999–7004
Sun S, Badgley BD (2019) Changes in microbial functional genes within the soil metagenome during forest ecosystem restoration. Soil Biol Biochem 135:163–172
Trivedi P, Leach JE, Tringe SG, Sa TM, Singh BK (2020) Plant-microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18:607–621
Tzanakakis VA, Taylor AE, Bakken LR, Bottomley PJ, Myrold DD, Dörsch P (2019) Relative activity of ammonia oxidizing archaea and bacteria determine nitrification-dependent N2O emissions in Oregon forest soils. Soil Biol Biochem 139:107612
Uchida Y, Clough TJ, Kelliher FM, Sherlock RR (2008) Effects of aggregate size, soil compaction, and bovine urine on N2O emissions from a pasture soil. Soil Biol Biochem 40:924–931
Wang H, Shu D, Liu D, Liu S, Deng N, An S (2020a) Passive and active ecological restoration strategies for abandoned farmland leads to shifts in potential soil nitrogen loss by denitrification and soil denitrifying microbes. Land Degrad Dev 31:1086–1098
Wang J, Wang X, Liu G, Zhang C, Wang G (2021) Bacterial richness is negatively related to potential soil multifunctionality in a degraded alpine meadow. Ecol Indic 12:106996
Wang J, Wang XT, Liu GB, Wang GL, Wu Y, Zhang C (2020b) Fencing as an effective approach for restoration of alpine meadows: evidence from nutrient limitation of soil microbes. Geoderma 363:10
Xie Y, Zhang J, Meng L, Muller C, Cai Z (2015) Variations of soil N transformation and N2O emissions in tropical secondary forests along an aridity gradient. J Soils Sediments 15:1538–1548
Wu D, Cárdenas LM, Calvet S, Brüggemann N, Loick N, Liu S, Bol R (2017) The effect of nitrification inhibitor on N2O, NO and N2 emissions under different soil moisture levels in a permanent grassland soil. Soil Biol Biochem 113:153–160
Yang D, Xiao X, He N, Zhu W, Liu M, Xie G (2020a) Effects of reducing chemical fertilizer combined with organic amendments on ammonia-oxidizing bacteria and archaea communities in a low-fertility red paddy field. Environ Sci Pollut Res 27:29422–29432
Yang LQ, Liu R, Ju XT (2020b) Effect of carbon rate and type amended with ammonium or nitrate on nitrous oxide emissions in a strong ammonia oxidation soil. J Soils Sediments 20:1253–1263
Yin M, Gao X, Tenuta M, Li L, Gui D, Li X, Zeng F (2020) Enhancement of N2O emissions by grazing is related to soil physicochemical characteristics rather than nitrifier and denitrifier abundances in alpine grassland. Geoderma 375
Zhang C, Li J, Wang J, Liu G, Wang G, Guo L, Peng S (2019a) Decreased temporary turnover of bacterial communities along soil depth gradient during a 35-year grazing exclusion period in a semiarid grassland. Geoderma 351:49–58
Zhang C, Song ZL, Zhuang DH, Wang J, Xie SS, Liu GB (2019b) Urea fertilization decreases soil bacterial diversity, but improves microbial biomass, respiration, and N-cycling potential in a semiarid grassland. Biol Fertil Soils 55:229–242
Zhang J, Fan Z, Chen H, Ju S (2016) The design of a double layer microwave absorber based on studying microwave absorbing characteristics, 2016 IEEE International Conference on Microwave and Millimeter Wave Technology (ICMMT), pp. 1032–1034
Zhang K, Li X, Cheng X, Zhang Z, Zhang Q (2019c) Changes in soil properties rather than functional gene abundance control carbon and nitrogen mineralization rates during long-term natural revegetation. Plant Soil 443:293–306
Zhang Y, Liu D, Jia Z, Zhang P (2021) Ridge and furrow rainfall harvesting can significantly reduce N2O emissions from spring maize fields in semiarid regions of China. Soil Tillage Res 209:104971
Zhong J, Wei Y, Wan H, Wu Y, Zheng J, Han S, Zheng B (2013) Greenhouse gas emission from the total process of swine manure composting and land application of compost. Atmospheric Environ 81:348–355
Zhong L, Li FY, Wang Y, Zhou X, Zhou S, Gong X, Bai Y (2018) Mowing and topography effects on microorganisms and nitrogen transformation processes responsible for nitrous oxide emissions in semi-arid grassland of Inner Mongolia. J Soils Sediments 18:929–935
Zhong L, Zhou X, Wang Y, Li FY, Zhou S, Bai Y, Rui Y (2017) Mixed grazing and clipping is beneficial to ecosystem recovery but may increase potential N2O emissions in a semi-arid grassland. Soil Biol Biochem 114:42–51
Zhu Y, Merbold L, Leitner S, Xia L, Pelster DE, Diaz-Pines E, Abwanda S, Mutuo PM, Butterbach-Bahl K (2020) Influence of soil properties on N2O and CO2 emissions from excreta deposited on tropical pastures in Kenya. Soil Biol Biochem 140:107636
Zuo J, Hu H, Fu Q, Zhu J, Xing Z (2020) Biological-chemical comprehensive effects of goethite addition on nitrous oxide emissions in paddy soils. J Soils Sediments 20:3580–3590
Funding
This work was financially supported by the National Natural Sciences Foundation of China (41761008), the Natural Science Basic Research Program of Shaanxi Province (2019KJXX-081; 2021JM-605), the Under the auspices of Strategic Priority Program of the Chinese Academy of Sciences (CAS) (XDA20040200), and the Chinese Universities Scientific Fund (2452018336).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Huaiying Yao
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Wang, X., Wang, J. et al. Grazing exclusion reduces soil N2O emissions by regulating nirK- and nosZ-type denitrifiers in alpine meadows. J Soils Sediments 21, 3753–3769 (2021). https://doi.org/10.1007/s11368-021-03035-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-021-03035-5