Abstract
Background and aims
Agricultural abandonment has been taken as one of the most widely used strategies for ecological restoration. Abandoned croplands expanded worldwide since the middle of the twentieth century and will increase considerably in the future. Whether agricultural abandonment alters the abundances of soil carbon (C) and nitrogen (N) mineralization genes, and whether these functional genes are linked to alterations in C and N mineralization rates remain unknown.
Methods
We took advantage of a well-established chronosequence of nature restoration sites on abandoned croplands which represent a century of vegetation succession in China’s Qinling Mountains, to determine soil C and N mineralization rates, and their relationships with vegetation succession, edaphic characteristics, soil microbial communities, and functional genes based on GeoChip 5.0.
Results
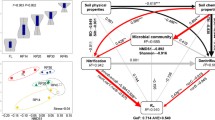
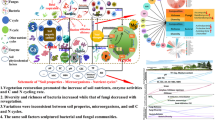
We found that soil C and N mineralization rates, microbial biomass and composition, and the number and abundances of functional genes were significantly affected by vegetation succession following agricultural abandonment. Soil C and N mineralization rates increased by sixfold and threefold, respectively, along the cropland to forest successional gradient. The NO3-N, NH4-N and soil available phosphorus (P) concentrations were important factors associated with the C and N mineralization genes. The shifts in edaphic environments (i.e., soil moisture, cation exchange capacity, and C/P ratio), vegetation-derived substrates (i.e., soil organic C and N), and soil microbial composition (i.e., Fungi/Bacteria ratio, amounts of Gram-negative bacteria, Actinomycetes, and arbuscular mycorrhizal fungi), rather than the abundances of functional genes involved in mineralization processes, controlled soil C and N mineralization rates along the long-term vegetation successional gradient.
Conclusions
Abandoning cropland for natural succession could facilitate the recuperation of soil C and N processes and alter soil microbial composition and functional genes. The abundances of microbial functional genes are less important than previously expected in regulating soil C and N mineralization rates.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Abandoned croplands have been expanding worldwide since the middle of the twentieth century due to environment conservation programs, policy incentives, urbanization, and depopulation in villages (Rey Benayas et al. 2007; Campbell et al. 2008; Cramera et al. 2008; Zhang et al. 2013). For instance, the U.S. Conservation Reserve Program established in 1985 and China’s ‘Grain-for-Green’ Program initiated in 1999 both converted massive croplands into abandoned agricultural lands (Zhang et al. 2013). The collapse of the Soviet Union in the early 1990s drove the expansion of abandoned croplands in Eastern Europe (Song et al. 2018). The area of global abandoned croplands is estimated to be 385–472 million hectares (Campbell et al. 2008). Moreover, abandoned croplands will increase considerably as agricultural abandonment is now one of the most widely used strategies for ecological restoration (Rey Benayas et al. 2007; Morriën et al. 2017). When agricultural lands are abandoned, the land cover changes from crops to secondary vegetation while the biotic community composition, ecological processes, and functions may evolve accordingly (Awiti et al. 2008; Cramera et al. 2008; Zhang et al. 2013; Zhang et al. 2018). Thus, knowledge of key ecological processes in abandoned croplands is imperative for better understanding and managing these ecosystems (Stanturf et al. 2014; Morriën et al. 2017).
Soil carbon (C) and nitrogen (N) mineralization are key processes regulating nutrient cycling and soil fertility in terrestrial ecosystems (Reich et al. 1997; Pathak and Rao 1998; Miller et al. 2003; Whitman et al. 2016; Clivot et al. 2017). It has been well documented that over 90% of N in most surface soils occurs in organic forms (Kelley and Stevenson 1995). Driven by microbes, mineralization processes release N from organic compounds into inorganic forms which are available to plants and often limit vegetation productivity (Reich et al. 1997). Organic C compounds are converted into CO2 during mineralization which produces a major flux of CO2 from soil pools into the atmosphere (Tian et al. 2017). Thereby, soil C and N mineralization have received increasing attention in ecology, agriculture, and global change research in recent years (Davidson and Janssens 2006; Tardy et al. 2015; Whitman et al. 2016; Meyer et al. 2018), in particular for ecosystems undergoing land-use changes.
Previous studies have investigated soil C and N mineralization rates and their driving factors in forests, pastures, wetlands, and some other ecosystems (Reich et al. 1997; Bedard-Haughn et al. 2006; Strickland et al. 2010; Don et al. 2017). Vegetation types, land-use regimes, climate, edaphic factors, and soil microorganisms have been identified as important parameters affecting soil C and N mineralization (Bedard-Haughn et al. 2006; Chatterjee et al. 2008; Strickland et al. 2010; Sun et al. 2013; Xiao et al. 2017). Among these factors, microbes are known as the engines of soil biogeochemical cycles and play vital roles in mediating soil C and N mineralization (Wang et al. 2014; Graham et al. 2016; Don et al. 2017; Chen et al. 2018). However, the linkages between microbial functional genes and ecosystem processes are not well understood (Graham et al. 2016; Trivedi et al. 2016).
The GeoChip, a comprehensive functional gene array targeting different gene families, provides a powerful technique for researchers to comprehensively analyze the relationships between microbial functional genes and microbial-mediated biogeochemical processes including soil C and N cycling (Tu et al. 2014; Van Nostrand et al. 2016). For instance, the GeoChip 5.0 (60 K) covers 60 gene families which contain important genes involved in C-degradation pathways including glucose, lactose, starch, hyaluronic acid, sucrose, pectin, terpenes, protein, lipids, phospholipids, tannins, vanillin, hemicellulose, and lignin decomposition. Previous studies have used the GeoChip to investigate how the functional genes respond to environmental changes (Paula et al. 2014; Xue et al. 2016; Zhang et al. 2017). Scientists have found that forest-to-pasture conversion alters the abundances of some C and N cycling genes (Paula et al. 2014), and degenerative succession affects C degradation gene composition (Zhang et al. 2017). However, whether vegetation restoration following agricultural abandonment alters the abundances of soil C and N mineralization genes and whether these functional genes are linked to the alterations in soil C and N mineralization rates remain unknown.
To understand how the mineralization rates of soil C and N, microbial composition, and functional genes respond to agricultural abandonment, we used a well-established chronosequence of nature restoration sites on abandoned croplands which represent a century of vegetation succession in China’s Qinling Mountains (Zhang et al. 2018). We determined soil C and N mineralization rates in laboratory incubation, and explored their relationships to vegetation succession, edaphic characteristics, microbial community, and functional genes based on GeoChip 5.0. Changes in land-use or vegetation may alter soil physico-chemical properties and microbial processes which may also alter the abundance of related functional genes (Hallin et al. 2009; Zhang et al. 2017). Therefore, we hypothesized that the soil C and N mineralization rates, soil microbial composition, and functional genes would display significant differences among the vegetation successional stages. We further hypothesized a positive correlation between the abundances of C and N mineralization genes and rates of C and N mineralization, respectively.
Materials and methods
Site description and soil sampling
This study was performed at south aspect of Qinling Mountains, a famous international biodiversity hotspot in central China. All the sampling plots were located in the Foping National Nature Reserve (FNNR), a member of UNESCO/MAB World Network of Biosphere Reserves (Zhang et al. 2018). This region belongs to transitional climatic zones between subtropical humid zone and warm temperate zone, with a mean annual average temperature of 11. 8 °C, mean average temperature in growing season of 20.5 °C, and 220 frost free days per year. Long-term average annual precipitation is 938 mm, with 66% falling from June to September. Soil type is yellow-brown soil developed from granite, and natural vegetation is dominated by deciduous broad-leaved trees (i.e., Quercus serrata var. brevipetiolata, Castanea mollissima, Dendrobenthamia japonica var. chinensis) (Zhang et al. 2018).
A total of 16 plots representing cropland-to-forest conversion chronosequence (successional gradient: abandoned cropland→ grassland→shrubland→secondary forest), including 4 croplands, 4 grasslands, 4 shrublands, and 4 secondary forests, were selected in a small catchment (107.81°-107.86°E, 33.53°-33.62°N) (Fig. 1). All stands were located within 5 km of the Yueba Conservation Station of the FNNR with similar soil parent material and topography. Main crop in this area was maize (Zea mays L.), thus, the history of agricultural use is comparable. All plots were selected from ex-arable lands identified by irrigation ditches and cropland ridges (Fig. 1). Since abandonment, natural vegetation in the ex-arable lands was allowed to establish with no fertilization, clipping, pest control, and other human disturbances. The stand age of each plot, namely the years since abandonment, was inquired from landowners or estimated from the tree-rings of the oldest pioneer trees. The selected grasslands were 1, 1, 2, and 15 years since abandonment respectively, which were dominated by Artemisia lavandulaefolia, Erigeron annuus, and Aster ageratoides. The stand age of shrubland sites was 12, 15, 16, and 21 years respectively, and the dominant plants were Broussonetia papyrifera, Rubus flosculosus, Pueraria lobata, and Artemisia lavandulaefolia. The stand age of secondary forest stands was 22, 60, 80, and 110 years, respectively, and the dominant plants were Castanea mollissima, Dendrobenthamia japonica var. chinensis, Quercus serrata var. brevipetiolata, and Bashania fargesii. The croplands were planted with maize (Z. mays L.) when soils were sampled.
Three quadrats were selected randomly in each plot for soil sampling. Ten surface soil cores were sampled randomly from the top 10 cm by sterilized shovels in each sampling quadrat. The soil samples were pooled by sampling quadrat, thus, three mixed soil samples were collected within each plot and a total of 48 soil samples were collected. In field, the fresh soil samples were kept in sterilized plastic bags and immediately placed in carry-on ice box. Three additional soil cores were sampled randomly (0–10 cm depth) using a 100-cm3 stainless steel cutting rings in each plot for measuring soil bulk density. All the soil samples were collected at the same day in vegetation growing season (i.e., August 13th, 2016). After transporting the carry-on ice box to lab, we subdivided each fresh soil samples into two subsamples. One subsample was kept at −80 °C for soil genomic DNA extraction, and the other was used to determine phospholipid fatty acids (PLFAs), C and N mineralization rates, and physico-chemical properties.
Soil physico-chemical properties and C and N mineralization rates measurement
The concentration of NO3-N in fresh soils was extracted with pure water and measured by phenol disulfonic acid spectriphotometric method (Lu 2000). The NH4-N was extracted from fresh soil samples with a 2 M KCl solution and the concentration was determined by indophenol blue colorimetry (Lu 2000). Fresh soils were dried to constant weight (at 105 °C) for calculating moisture. The intact soil cores sampled by cutting rings were dried (at 105 °C) to constant weight for measuring soil bulk density. Air-dried subsamples were ground to pass through 1.0 mm sieves for measuring the available phosphorus (P) (by 0.5 M NaHCO3 extraction (1:20), Mo-Sb colorimetric method), available K (by 1 M NH4OAC extraction (1:20), atomic absorption spectrophotometer), cation exchange capacity (CEC) (the ammonium saturation method), total P (molybdenum blue colorimetry), total potassium (K) (atomic absorption spectrophotometer) (Lu 2000). Soil pH was measured in soil-water (1:2.5 w/v) suspension using a pH meter. Soil organic C (SOC), total N, and their δ13C and δ15N values were determined by an isotope ratio mass spectrometer (Delta-V Advantage, Germany). The concentration of soil organic N (SON) was estimated by subtracting the inorganic N (NO3-N plus NH4-N) from total N. The stable isotopic compositions of soil are expressed in δ notation (‰):
where Rsample is the ratio of the heavy to the light isotope (13C/12C or 15N/14N) of a sample; Rstd is ratio of a standard, i.e., Pee Dee Belemnite (δ13C = 0.0‰) for C or air (δ15N =0.0‰) for N (Marin-Spiotta et al. 2009; Cheng et al. 2013).
Rates of soil C and N mineralization were measured in a 26-day laboratory incubation experiment with the modification of the methods of Tian et al. (2017) and Wang et al. (2014). Briefly, fresh soil equivalent to 30 g dry weight for each sample were placed in a 500 ml glass jar. The soil moisture was kept as the natural state (i.e., field moisture) and the loss of water during incubation was supplemented by adding deionized water. All samples were pre-incubated at 20.5 °C (mean average temperature in growing season in the study sites) for 3 days. A glass cup containing 10 mL of 0.1 M NaOH solution was placed in each glass jar, which was collected at 1, 4, 7, 12, 19, and 26 d after incubation and was titrated against 0.1 M HCl to determine CO2 production from soils (Wang et al. 2014). All the glass jars with soil were sealed and incubated at 20.5 °C in the dark in an incubator. Three additional glass jars only with a cup containing 10 mL of 0.1 M NaOH were sealed. These bottles were used for accounting the CO2 trapped from the air. Soil C mineralization rate was calculated based on the cumulative amount of C mineralized during 26-day incubation and expressed as the mean mineralized C (mg) per kg dry soil per day (Supplementary Fig. S1). The concentrations of NO3-N and NH4-N in soils at the start and end of incubation period were measured to estimate the soil N mineralization rate which was calculated based on the change in inorganic N (NO3-N plus NH4-N) content in the soil before and after incubation and expressed as the mean mineralized N (mg) per kg dry soil per day (Tian et al. 2017).
Soil microbes and functional genes
The PLFAs were applied to assess the soil microbial biomass and community composition (Frostegård and Bååth 1996; Kaiser et al. 2015). Fresh soil equivalent to 4 g dry weight for each sample was used to prepare the PLFAs following the standard protocol (Prasanna et al. 2016). Briefly, lipids in the fresh soil samples were extracted by chloroform-methanol-phosphate buffer mixture (1:2:0.8). The lipid fractions were separated on a solid-phase extraction column (CNWBOND Si SPE Cartridge) by successively using 10 ml chloroform, 10 ml acetone, and 10 ml methanol. The separated phospholipids were evaporated under N2 gas and then trans-esterified to fatty acid methyl esters. The obtained fatty acid methyl esters were extracted with hexane, evaporated, analyzed by gas chromatography in Agilent 6890 N with Agilent 19091B-102 columns (25 m × 200 μm × 0.33 μm)), and identified based on the MIDI Sherlock system (MIDI Inc., Newark, DE, USA). Similar to Frostegård and Bååth (1996) and Prasanna et al. (2016), the fatty acids were summed notionally into different ‘microbial groups’, i.e., Gram positive bacteria, Gram negative bacteria, Anaerobe, Actinomycetes, Eukaryote, Fungi, and Arbuscular mycorrhiza Fungi (AM fungi), and expressed as nmol g−1 soil. We used all of the PLFAs to represent the total soil microbial biomass (Kaiser et al. 2015).
The soil genomic DNA was extracted from 0.4 g soil samples by kit (PowerSoil® DNA Isolation Kit DNA), and the purity, concentration, and integrity of DNA were assessed. The genomic DNA samples were detected by agarose gel electrophoresis, and the purity of DNA was assessed by Nanodrop One. The concentration of DNA was determined by Qubit 3.0. The amount of each DNA sample was more than 2 μg. All the DNA samples passed quality control, and the three DNA samples from the same plot were pooled into a DNA sample. Thus, a total of 16 DNA samples were used in the downstream experiments.
The microbial functional genes were determined by GeoChip 5.0 according to standard protocols (Bracho et al. 2016) and were conducted by Guangdong Magigene Biotechnology Co., Ltd. China. The GeoChip 5.0 (60 K arrays) used our study contains 57 603 gene probes (oligonucleotide) from 393 gene families. Here, gene refers to each unique sequence targeted by microarrays. A gene family includes all gene sequences assigned with the same name (e.g., named amoA), which codes for the same class of proteins (Paula et al. 2014). More information about the GeoChip 5.0 could be obtained from the website (http://glomics.com/gch-tech.html). Briefly, 800 ng of soil community DNA was labeled with the fluorescent dye Cy-3, hybridized to GeoChip 5.0 60 K microarrays, and scanned with a NimbleGen MS200 Microarray Scanner using techniques described previously (Cong et al. 2015; Bracho et al. 2016). The scanned images of hybridized GeoChips were converted and extracted using Agilent Feature Extraction 11.5 software (Agilent Technologies, Inc., CA, USA); the extracted data were then analyzed using the GeoChip data analysis pipeline (http://ieg.ou.edu/microarray/) according to Paula et al. (2014) and Bracho et al. (2016). These GeoChip data represent a broad picture of the microbial functional communities in each sample. Since our objective in the present study was to examine C and N mineralization, we mainly focused on the gene families involved in C degradation (60 gene families), ammonification (i.e., gdh, ureC) and nitrification (i.e., amoA, hao).
Statistical analyses
One-way ANOVA, followed by Least Significant Difference (LSD) test, was employed to verify the effect of vegetation successional stage on soil physico-chemical properties, rates of C and N mineralization, soil microbial biomass and community identified by PLFA biomarkers, the number and abundances (signal intensities) of functional genes involved in C and N mineralization (SPSS 17.0 for Windows, SPSS, Inc.). The p-values of multiple comparisons were corrected by Bonferroni’s correction.
The dissimilarities in gene composition between successional stages were tested by Analysis of Similarities (ANOSIM) in the package of ‘vegan’ in R (R Development Core Team 2013). We conducted redundancy analysis (RDA) to analyze the relationships among functional genes and soil physico-chemical variables. First, we used ‘Forward Selection of Explanatory Variables’ to select the first three environmental variables (Canoco 5.0). Then, the selected environmental variables were analyzed by RDA in Canoco (Version 5.0 for Windows, Ter Braak and Smilauer 2012).
Pearson correlation analysis was used to explore relationships of C (or N) mineralization rates with the abundances of functional genes, soil physico-chemical properties, and the soil microbial community composition (total PLFAs, microbial groups PLFAs, Gram positive/Gram negative bacteria, and Fungi/Bacteria ratio). Pearson correlation analysis was conducted to test the relationships between PLFAs and the studied soil physico-chemical variables, and stepwise regression analysis was performed to pick the main soil variables influencing microbial groups.
Results
Soil C and N mineralization rates
Soil C mineralization rates were significantly affected by vegetation successional stage (P < 0.05, Fig. 2), with highest mean values (36.60 mg C kg−1 dry soil d−1) in forests and lowest mean values (5.06 mg C kg−1 dry soil d−1) in croplands. Soil C mineralization rates in the shrublands (with a mean of 16.32 mg C kg−1 dry soil d−1), grasslands (9.93 mg C kg−1 dry soil d−1), and croplands displayed large variations and were significantly lower than that in forest stands. Vegetation successional stage significantly affected soil N mineralization rates with significantly higher level in forests (with a mean of 2.25 mg N kg−1 dry soil d−1) compared to croplands (with a mean of 0.53 mg N kg−1 dry soil d−1) (Fig.2).
Soil microbial functional genes
Vegetation successional stage significantly affected the number (51 out of 60 gene families) and abundances (52 out of 60 gene families) of most of the gene families involved in C-degradation (Supplementary Tables S1, S2). The number of genes (800 ng DNA) involved in C-degradation ranged from 5154 to 5726 with a mean of 5361 in croplands, from 3999 to 4899 with a mean of 4396 in grasslands, from 5199 to 6216 with a mean of 5658 in shrublands, and from 3902 to 4508 with a mean of 4186 in forests (Supplementary Table S2). The lowest abundances for most of the C-degradation genes (40 out of 60 gene families) were found in the forests, while the highest abundances for most of the C-degradation genes (46 out of 60 gene families) were observed in the shrublands. For the lmo, vdh, heparinase, glucose_oxidase, and exochitinase genes, the highest abundances were detected in the croplands (Supplementary Table S1).
The number and abundances of genes involved in N-mineralization (i.e., gdh, ureC, amoA, and hao) varied across vegetation successional stage (Supplementary Tables S2, S3). The number of gdh genes was 101, 83, 106, and 78 in croplands, grasslands, shrublands, and forests, respectively (Supplementary Table S2). The abundances of gdh, ureC, and amoA in croplands and shrublands were significant higher than those in the grasslands and forests (Supplementary Table S3). The number of amoA gene was 17, 15, 16, and 14 in croplands, grasslands, shrublands, and forests, respectively, and the croplands had the highest abundances of amoA.
The results of ANOSIM showed that C-degradation gene (R = 0.67, P = 0.001) and N-mineralization gene composition (R = 0.71, P = 0.001) (based on signal intensity) differed significantly among the successional stages (Table 1). However, the dissimilarity between the cropland and shrubland was not significant (Table 1).
Soil microbial biomass and community composition
Vegetation successional stage significantly affected the amount of Gram- negative bacteria and Actinomycetes, as well as the Fungi/Bacteria ratio (Table 2). The highest mean values of Gram-negative bacteria was detected in the forests (15.09 nmol PLFAs g−1 soil), with the lowest mean values in the croplands (7.87 nmol PLFAs g−1 soil). The mean concentrations of total PLFAs ranged from 27.31 to 43.51 nmol PLFAs g−1 soil with lowest in croplands and highest in forests. However, the differences in the total PLFAs among successional stages were not significant statistically (P = 0.061). The Fungi/Bacteria ratio was significantly higher in croplands (0.1) than in shrublands (0.06) and forests (0.07) (Table 2).
The SON concentration was the main variables explaining the variations in the amount of total PLFAs, Gram-positive bacteria, AM Fungi, and Actinomycetes, while the total N concentration was the main variables explaining the variations in the amount of Gram-negative bacteria. The C/N ratio explained 66% of the variation in Fungi/Bacteria ratio (Supplementary Table S4). The total PLFAs positively correlated with SON, Total N, SOC, CEC, NH4-N, moisture, and pH, and negatively correlated with soil bulk density (Supplementary Table S5).
Soil physico-chemical properties
Vegetation successional stage significantly affected the soil moisture, CEC, SOC, SON, total N, NH4-N, Available P, C/N ratio, C/P ratio, N/P ratio, δ13C and δ15N (Table 2). The differences in the total P and total K concentrations among successional stages were not significant statistically (P > 0.05). The highest concentrations of SOC and SON were observed in forests, followed by shrublands, grasslands, and croplands (Table 2). The δ13C decreased significantly with the increasing of SOC concentration (Supplementary Fig. S2). Similarly, the δ15N decreased significantly with the increasing of SON concentration (Supplementary Fig. S2). Soil moisture in forests was significantly higher than that in grasslands and croplands, and soil moisture in shrublands was significantly higher than that in croplands (P < 0.05, Table 2).
Relationships among C and N mineralization rates, edaphic factors, and microbes
Soil C mineralization rate was negatively correlated with the abundance of amyA, apu, axe, beta_agarase, cdh, cellobiase, dextranase_fungi, endoglucanase, exochitinase, exoglucanase, lmo, mnp, phospholipase_C, pme, pme_Cdeg, rgl, tannase_Cdeg, vana, vdh, xylanase, and was not correlated with the other studied genes involved in C-degradation (Supplementary Table S6). Soil C mineralization rate was positively correlated with the total PLFAs, the amount of Gram-positive bacteria, Gram-negative bacteria, Actinomycetes, AM Fungi, soil moisture, SOC, Total N, NH4-N, SON, CEC, C/N ratio, C/P ratio, and N/P ratio, and was negatively correlated with the Gram-positive/ negative bacteria ratio, Fungi/Bacteria ratio, and soil bulk density, and total K (Table 3; Fig. 3).
Soil N mineralization rate was negatively correlated with the abundance of ureC and amoA genes, and was not correlated with gdh and hao genes (Table 4). Soil N mineralization rate correlated positively with the total PLFAs, the amount of Gram-negative bacteria, Actinomycetes, AM Fungi, SOC, Total N, SON, soil moisture, NH4-N, CEC, C/N ratio, C/P ratio, and N/P ratio, and was negatively correlated with the Fungi/Bacteria ratio, and soil bulk density, and total K (Table 3; Fig. 3).
For most of the studied C-degradation genes (38 out of 60 gene families), the gene abundances correlated positively with the soil NO3-N concentration (Supplementary Table S7). In addition, 23 out of 60 C-degradation genes correlated negatively with the soil NH4-N concentration (Supplementary Table S7). The abundance of alginase, apu, cda, cdh, endopolygalacturonase_fungi, exochitinase, glucose_oxidase_fungi, hyaluronidase, lmo, mannanase, vana, and vdh genes correlated positively with the soil available P concentration (Supplementary Fig. S7). The results of RDA showed that the NO3-N, SOC, and NH4-N were the most important environmental variables associated with the C-degradation gene composition, and together explained 34.1% of variations (Fig. 4). The NO3-N explained 18.6% of the total variation of C-degradation gene composition, which provided the greatest explanatory power. For the N-mineralization gene composition, the NO3-N, NH4-N, and pH were the most important environmental variables, and together explained 44.2% of variations (Fig. 5). The NO3-N concentration, the most important variable explained 31% of the total variation of N-mineralization gene composition.
RDA (Redundancy Analysis) ordination maps based on the signal intensity of C-degradation genes. Gene name: 1 beta_agarase, 2 alginase, 3 camdcab,4 axe, 5 cellobiase, 6 endoglucanase, 7 exoglucanase, 8 acetylglucosaminidase, 9 chitin_deacetylase, 10 chitinase, 11 endochitinase, 12 exochitinase, 13 cutinase, 14 glucose_oxidase, 15 ara, 16 mannanase, 17 xyla,18 xylanase, 19 xylose_isomerase, 20 heparinase, 21 hyaluronidase, 22 inulinase, 23 lactase, 24 glx, 25 ligninase, 26 mnp, 27 phenol_oxidase, 28 lipase, 29 vana,30 vdh, 31, endopolygalacturonase, 32 exopolygalacturonase, 33 pec_Cdeg, 34 pectin_lyase, 35 pectinase, 36 pel_Cdeg, 37 pme, 38 pme_Cdeg, 39 RgaE, 40 rgh, 41 rgl, 42 phospholipase_A2, 43 phospholipase_C, 44 phospholipase_D, 45 protease_aspartate, 46 protease_cysteine, 47 protease_serine, 48 amyA, 49 amyx, 50 apu, 51 cda, 52 glucoamylase, 53 pula, 54 dextranase_fungi, 55 invertase, 56 tannase_Cdeg, 57 cdh, 58 limeh, 59 lmo, 60 alpha_galactosidase
RDA (Redundancy Analysis) ordination maps based on the signal intensity of N-mineralization genes. N-mineralization genes (solid arrows), environmental variables (hollow arrows), and sampling sites (circles: Ca,Cb, Cc and Cd, Cropland; Ga, Gb, Gc, and Gd, Grassland; Sa, Sb, Sc and Sd, Shrubland; Fa, Fb, Fc and Fd, Forest) were showed in the plots of RDA
Discussion
To our knowledge, this is the first study to investigate the long-term (>100 years) impacts of vegetation succession following agricultural abandonment on microbial functional genes involved in C and N mineralization processes. As expected, the number and abundances of functional genes involved in C and N mineralization were significantly affected by vegetation succession (Supplementary Tables S1,S2,S3), which supported the argument that land use/cover change altered soil microbial functional genes (Paula et al. 2014; Zhang et al. 2017). We found that forests had the lowest number of functional genes, and for most genes, the lowest abundances were observed in forests and the highest abundances were observed in shrublands, indicating that different vegetation could harbor different functional gene contents (Paula et al. 2014). Different successional stages were characterized by different vegetation and edaphic variables in our study (Table 2), thus, the abundances of functional genes responded markedly to cropland-to-forest conversion.
We found that the concentrations of soil NO3-N and NH4-N were important features associated with the composition of the genes (Figs. 4, 5), and soil available P concentration correlated positively with the abundance of 12 genes (Supplementary Table S7), suggesting that soil nutrient availability shaped the abundances of soil microbial functional genes. The N and P are essential for soil microbes. The N occurs in amino acids (and thus proteins), nucleic acids (DNA and RNA) and adenosine triphosphate (ATP), and the P is a component of DNA, RNA, ATP, and phospholipids. Thus, the soil available P and NO3-N positively correlated with the abundances of most functional genes (Supplementary Tables S7, S8).
We found that soil C and N mineralization rates were significantly affected by vegetation succession following agricultural abandonment, and the soil C and N mineralization rates increased by sixfold and threefold respectively as croplands-to-forest conversion. The increasing patterns of soil C and N mineralization rates during secondary succession uncovered in this study indicated the recuperation of C and N cycling in abandoned agricultural lands following cessation of human disturbance and management. These results support the previous findings that land-use change altered soil C and N mineralization (Zeller et al. 2000; Jha et al. 2012; Strickland et al. 2010; Li et al. 2014). Similarly, Jha et al. (2012) also found that forest land-use system showed the highest SOC mineralization rates in comparison to agriculture lands (soybean-wheat system). However, our results were inconsistent with the findings of Nadal-Romero et al. (2016) who found that the differences in soil C mineralization rates between various land covers (i.e., bare soil, permanent pastureland, secondary succession lands, and planted forest) in the Central Spanish Pyrenees were not significant statistically. Li et al. (2014) observed that soil N mineralization rate was significantly lower in the forests than in the croplands, and Zeller et al. (2000) found that agricultural abandonment significantly decreased the soil net N mineralization rate in grasslands, which were also inconsistent with our results in this study. These inconsistencies indicated that the directions of changes in C and N mineralization rates caused by land-use change showed large variations and uncertainties (Zeller et al. 2000; Li et al. 2014).
The increasing patterns of soil C and N mineralization rates as nature restoration progresses in this study could be attributed to the changes in edaphic environments and soil microbial composition, due to mineralization rates were strongly correlated with soil physico-chemical properties (i.e., moisture, SOC, SON, CEC, C/P ratio, etc.) and microbial community features (Table 3). The positive correlations between SOC concentration and soil C mineralization rates, as well as the positive correlations between SON concentration and soil N mineralization rates (Fig. 3), further supported that the substrates accumulation during secondary succession controlled the soil C and N mineralization rates. As expected, we observed that the δ13C decreased along the vegetation succession gradient and also declined with SOC concentration, which could be attributed to the facts that the ecosystems in our study involved C4 vegetation (with higher δ13C) -to - C3 vegetation (with lower δ13C) conversion. After the croplands dominated by C4 plants (i.e., maize) were abandoned, the C3 plants grew spontaneously and produced litters and other residues with lower δ13C, and then consequently decreased the δ13C of soils (Zhang et al. 2015). Compared with the ‘heavy’ isotope 15N, the ‘light’ isotope 14N can be preferentially removed from the terrestrial ecosystem through some processes, e.g., nitrate leaching and ammonia volatilization, which could lead to an enrichment of 15N (Austin and Vitousek 1998; Swap et al. 2004; Wang et al. 2007). The declining δ15N along vegetation succession gradient (i.e., cropland→grassland→shrubland→forest) in our study indicated that the N cycling in abandoned ecosystems became less open than in the croplands (Table 2), and the vegetation utilized the recycled N as nature restoration progresses. Therefore, the changes in stable isotopic signals of soil C and N clearly evidenced that the SOC and SON were accumulated markedly due to the vegetation conversion in the studied abandoned croplands. The vegetation-derived SOC and SON provided substrates for mineralization progresses and microbial growth (Clement and Williams 1967; Zhu et al. 2014), thus, C mineralization rate increased significantly with SOC concentration, and N mineralization rate increased significantly with SON concentration (Fig. 3).
Meanwhile, shifts in soil microbial biomass and composition after agricultural abandonment may affect C and N mineralization rates (Table 3). The shifts in the total soil microbial biomass, the amount of Actinomycetes, and the Fungi/Bacteria ratio could be associated with the changes in edaphic environments because the SON concentration was the best predictor for the soil microbial biomass and Actinomycetes, and the C/N ratio was the best predictor for Fungi/Bacteria ratio as indicated by stepwise regression models (Supplementary Table S4). Our analysis showed that the amounts of Gram-negative bacteria, Actinomycetes, and AM fungi were positively linked with C and N mineralization rates, and the Fungi/Bacteria ratio were negatively linked with C and N mineralization rates, indicating that microbial composition may affect soil C and N mineralization processes in these studied ecosystems.
Interestingly, we found that soil C mineralization rate was negatively correlated with the abundances of 20 functional genes and was not correlated with the abundances of other 40 genes (Table S6), which was contrary to our hypothesis. Meanwhile, we did not observe positive correlations between soil N mineralization rate and the abundances of functional genes. Our results suggested that the linkage between microbial functional genes and ecosystem processes regulated by soil microbes could be unexpectedly complex. Thus, it is difficult to predict the biogeochemical processes by the metabolic potential of functional genes. In addition, the results of GeoChip just represent the functional potentials of microbial community rather than their real activities. To reveal the detailed relationships between functional genes and mineralization processes, the transcriptomic and proteomic characterization of genes should be studied in further research.
Conclusions
A novel contribution of our study comes from simultaneous measurements of soil C and N mineralization rates, edaphic characteristics, soil microbial community, and functional genes in ecosystems along a century of vegetation succession gradient following agricultural abandonment. Compared to the edaphic environments and microbial composition, the abundance of microbial functional genes played less important roles in regulating soil C and N mineralization rates. Soil nutrient availability shaped the abundances of soil microbial functional genes. Shifts in the edaphic environments (i.e., soil moisture, cation exchange capacity, and C/P ratio), vegetation-derived substrates (i.e., SOC and SON), and soil microbial composition (i.e., the Fungi/Bacteria ratio, the amounts of Gram-negative bacteria, Actinomycetes, and AM fungi) along with vegetation succession were determinants on the soil C and N mineralization rates. These findings highlight the importance of vegetation succession following agricultural abandonment in shaping the soil microbial composition, functional genes, and C and N processes, and also indicate that the soil nutrient availability and ecological processes could recovery following cessation of human disturbances.
References
Austin AT, Vitousek PM (1998) Nutrient dynamics on a precipitation gradient in Hawaii. Oecologia 113:519–529
Awiti AO, Walsh MG, Kinyamario J (2008) Dynamics of topsoil carbon and nitrogen along a tropical forest-cropland chronosequence: evidence from stable isotope analysis and spectroscopy. Agric Ecosyst Environ 127:265–272
Bedard-Haughn A, Matson AL, Pennock DJ (2006) Land use effects on gross nitrogen mineralization, nitrification, and N2O emissions in ephemeral wetlands. Soil Biol Biochem 38:3398–3406
Bracho R, Natali S, Pegoraro E, Crummer KG, Schädel C, Celis G, Hale L, Wu L, Yin H, Tiedje JM, Konstantinidis KT, Luo Y, Zhou J, Schuur EAG (2016) Temperature sensitivity of organic matter decomposition of permafrost-region soils during laboratory incubations. Soil Biol Biochem 97:1–14
Campbell JE, Lobell DB, Genova RC, Field CB (2008) The global potential of bioenergy on abandoned agriculture lands. Environ Sci Technol 42:5791–5794
Chatterjee A, Vance GF, Pendall E, Stahl PD (2008) Timber harvesting alters soil carbon mineralization and microbial community structure in coniferous forests. Soil Biol Biochem 40:1901–1907
Chen H, Li D, Xiao K, Wang K (2018) Soil microbial processes and resource limitation in karst and non-karst forests. Funct Ecol 32:1400–1409
Cheng X, Yang Y, Li M, Dou X, Zhang Q (2013) The impact of agricultural land use changes on soil organic carbon dynamics in the Danjiangkou reservoir area of China. Plant Soil 366:415–424
Clement CR, Williams TE (1967) Leys and soil organic matter: II. The accumulation of nitrogen in soils under different leys. J Agr Sci 69:133–138
Clivot H, Mary B, Valé M, Cohan JP, Champolivier L, Piraux F, Laurent F, Justes E (2017) Quantifying in situ and modeling net nitrogen mineralization from soil organic matter in arable cropping systems. Soil Biol Biochem 111:44–59
Cong J, Liu X, Lu H, Xu H, Li Y, Deng Y, Li D, Zhang Y (2015) Analyses of the influencing factors of soil microbial functional gene diversity in tropical rain- forest based on GeoChip 5.0. Genomics Data 5:397–398
Cramera VA, Hobbs RJ, Standisha RJ (2008) What's new about old fields? Land abandonment and ecosystem assembly. Trends in Ecol Evol 23:104–112
Davidson EA, Janssens IA (2006) Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173
Don A, Böhme IH, Dohrmann AB, Poeplau C, Tebbe CC (2017) Microbial community composition affects soil organic carbon turnover in mineral soils. Biol Fertil Soils 53:445–456
Frostegård Å, Bååth E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils 22:59–65
Graham EB, Knelman JE, Schindlbacher A, Siciliano S, Breulmann M, Yannarell A et al (2016) Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front Microbiol 7:214
Hallin S, Jones CM, Schloter M, Philippot L (2009) Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J 3:597–605
Jha P, De A, Lakaria BL, Biswas AK, Singh M, Reddy KS, Rao AS (2012) Soil carbon pools, mineralization and fluxes associated with land use change in Vertisols of Central India. Nat Acad Sci Lett 35:475–483
Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, Solaiman ZM, Murphy DV (2015) Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol 205:1537–1551
Kelley KR, Stevenson FJ (1995) Forms and nature of organic N in soil. Fertil Res 42:1–11
Li M, Zhou X, Zhang Q, Cheng X (2014) Consequences of afforestation for soil nitrogen dynamics in Central China. Agric Ecosyst Environ 183:40–46
Lu RK (2000) Soil agro-chemical analyses. Agricultural Technical Press of China, Beijing (in Chinese)
Marin-Spiotta ERIKA, Silver WL, Swanston CW, Ostertag R (2009) Soil organic matter dynamics during 80 years of reforestation of tropical pastures. Glob Change Biol 15:1584–1597
Meyer N, Welp G, Rodionov A, Borchard N, Martius C, Amelung W (2018) Nitrogen and phosphorus supply controls soil organic carbon mineralization in tropical topsoil and subsoil. Soil Biol Biochem 119:152–161
Miller EK, Johnson AH, Vann DR, Joshi AB (2003) Nitrogen availability and forest productivity along a climosequency on Whiteface Mountain, New York. Can J For Res 33:1880–1891
Morriën E, Hannula SE, Snoek LB, Helmsing NR, Zweers H, De Hollander M et al (2017) Soil networks become more connected and take up more carbon as nature restoration progresses. Nat Commun 8:14349
Nadal-Romero E, Cammeraat E, Pérez-Cardiel E, Lasanta T (2016) How do soil organic carbon stocks change after cropland abandonment in Mediterranean humid mountain areas? Sci Total Environ 566:741–752
Pathak H, Rao DLN (1998) Carbon and nitrogen mineralization from added organic matter in saline and alkali soils. Soil Biol Biochem 30:695–702
Paula FS, Rodrigues JL, Zhou J, Wu L, Mueller RC, Mirza BS, Bohannan BJM, Klaus NK, Deng Y, Tiedje JM, Pellizari VH (2014) Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol Ecol 23:2988–2999
Prasanna R, Kanchan A, Ramakrishnan B, Ranjan K, Venkatachalam S, Hossain F, Shivay YS, Krishnan P, Nain L (2016) Cyanobacteria-based bioinoculants influence growth and yields by modulating the microbial communities favourably in the rhizospheres of maize hybrids. Eur J Soil Biol 75:15–23
Development Core Team R (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. www.r-project. Org, Vienna
Reich PB, Grigal DF, Aber JD, Gower ST (1997) Nitrogen mineralization and productivity in 50 hardwood and conifer stands on diverse soils. Ecology 78:335–347
Rey Benayas JM, Martins A, Nicolau JM, Schulz JJ (2007) Abandonment of agricultural land: an overview of drivers and consequences. CAB reviews: perspectives in agriculture, veterinary science, nutrition and natural resources 2:1–14
Song XP, Hansen MC, Stehman SV, Potapov PV, Tyukavina A, Vermote EF, Townshend JR (2018) Global land change from 1982 to 2016. Nature 560:639–643
Stanturf JA, Palik BJ, Dumroese RK (2014) Contemporary forest restoration: a review emphasizing function. Forest Ecol Manag 331:292–323
Strickland MS, Callaham JMA, Davies CA, Lauber CL, Ramirez K, Richter JDD, Fierer N, Bradford MA (2010) Rates of in situ carbon mineralization in relation to land-use, microbial community and edaphic characteristics. Soil Biol Biochem 42:260–269
Sun S, Liu J, Chang SX (2013) Temperature sensitivity of soil carbon and nitrogen mineralization: impacts of nitrogen species and land use type. Plant Soil 372:597–608
Swap RJ, Aranibar JN, Dowty PR, Gilhooly WP III, Macko SA (2004) Natural abundance of 13C and 15N in C3 and C4 vegetation of southern Africa: patterns and implications. Glob Change Biol 10:350–358
Tardy V, Spor A, Mathieu O, Lévèque J, Terrat S, Plassart P, Regnier T, Bardgett RD, van der Putten WH, Roggero PP, Seddaiu G, Bagella S, Lemanceau P, Ranjard L, Maron PA (2015) Shifts in microbial diversity through land use intensity as drivers of carbon mineralization in soil. Soil Biol Biochem 90:204–213
Ter Braak CJF, Smilauer P (2012) CANOCO reference manual and User's guide: software for ordination. Version 5. Microcomputer power Ithaca, New York
Tian Q, Wang X, Wang D, Wang M, Liao C, Yang X, Liu F (2017) Decoupled linkage between soil carbon and nitrogen mineralization among soil depths in a subtropical mixed forest. Soil Biol Biochem 109:135–144
Trivedi P, Delgado-Baquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, Zhou J, Singh BK (2016) Microbial regulation of the soil carbon cycle: evidence from gene-enzyme relationships. ISME J 10:2593–2604
Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, Zhou A, Voordeckers J, Lee YJ, Qin Y, Hemme CL, Shi Z, Xue K, Yuan Y, Wang A, Zhou J (2014) GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol Ecol Resour 14:914–928
Van Nostrand JD, Yin H, Wu L, Yuan T, Zhou J (2016) hybridization of environmental microbial community nucleic acids by GeoChip. In microbial environmental genomics (MEG) (pp. 183-196). Humana press, New York. NY
Wang L, Shaner PJL, Macko S (2007) Foliar δ15N patterns along successional gradients at plant community and species levels. Geophys Res Lett 34:16403
Wang Q, Wang S, He T, Liu L, Wu J (2014) Response of organic carbon mineralization and microbial community to leaf litter and nutrient additions in subtropical forest soils. Soil Biol Biochem 71:13–20
Whitman T, Pepe-Ranney C, Enders A, Koechli C, Campbell A, Buckley DH, Lehmann J (2016) Dynamics of microbial community composition and soil organic carbon mineralization in soil following addition of pyrogenic and fresh organic matter. ISME J 10:2918–2930
Xiao W, Feng S, Liu Z, Su Y, Zhang Y, He X (2017) Interactions of soil particulate organic matter chemistry and microbial community composition mediating carbon mineralization in karst soils. Soil Biol Biochem 107:85–93
Xue K, Yuan MM, Shi ZJ, Qin Y, Deng Y, Cheng L, Wu L, He Z, Van Nostrand JD, Bracho R, Natali S, Schuur EAG, Luo C, Konstantinidis KT, Wang Q, Cole JR, Tiedje JM, Luo Y, Zhou J (2016) Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat Clim Chang 6:595–600
Zeller V, Bahn M, Aichner M, Tappeiner U (2000) Impact of land-use change on nitrogen mineralization in subalpine grasslands in the southern Alps. Biol Fert Soils 31:441–448
Zhang K, Cheng X, Dang H, Ye C, Zhang Y, Zhang Q (2013) Linking litter production, quality and decomposition to vegetation succession following agricultural abandonment. Soil Biol Biochem 57:803–813
Zhang K, Dang H, Zhang Q, Cheng X (2015) Soil carbon dynamics following land-use change varied with temperature and precipitation gradients: evidence from stable isotopes. Glob Change Biol 21:2762–2772
Zhang K, Cheng X, Shu X, Liu Y, Zhang Q (2018) Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant Soil 431:19–36
Zhang Y, Liu X, Cong J, Lu H, Sheng Y, Wang X, Li D, Liu X, Yin H, Zhou J, Deng Y (2017) The microbially-mediated soil organic carbon loss under degenerative succession in an alpine meadow. Mol Ecol 26:3676–3686
Zhu B, Gutknecht JL, Herman DJ, Keck DC, Firestone MK, Cheng W (2014) Rhizosphere priming effects on soil carbon and nitrogen mineralization. Soil Biol Biochem 76:183–192
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31770562, 31470499, 3192200779), Youth Innovation Promotion Association CAS (2018374), and the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDA05060500 and XDB15010200). We are grateful for the comments and constructive suggestions from the editors and anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Michael Luke McCormack.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, K., Li, X., Cheng, X. et al. Changes in soil properties rather than functional gene abundance control carbon and nitrogen mineralization rates during long-term natural revegetation. Plant Soil 443, 293–306 (2019). https://doi.org/10.1007/s11104-019-04212-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04212-9