Abstract
Biochar affects soil carbon (C) dynamics via shifting microbial community, but the active bacteria that regulate the rhizosphere-based C cycling remain to be identified. Here, a continuous 13CO2 labeling pot (Zea mays L.) experiment over 14 days, combined with RNA-based stable isotope probing (RNA-SIP), were used to characterize the active bacterial communities involved in the mineralization of rhizodeposits and soil organic C (SOC) in biochar-amended soil. Compared with the non-amended soil, biochar shifted the rhizosphere communities towards having lower richness and evenness, and particularly stimulated the growth of Actinobacteria (e.g., genus affiliated to Micrococcaceae) and other oligotrophs, most likely due to neutralizing soil acidity (from 4.53 to 6.17) and increasing content of recalcitrant organic C (from 10.69 to 25.77 g·kg−1). These enriched genera were associated with mineralization of both rhizodeposits and SOC, giving 35.09% and 87.28% increased mineralization of rhizodeposits and SOC. This led to much less (by 58.50% decrease) incorporation of 13C into biochar-amended soil. This study deciphered the active microorganisms in the biochar-soil–plant system that likely increased SOC and rhizodeposit mineralization (fewer rhizodeposits remaining), and thus diminished C sequestration by biochar per se.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil organic matter (SOC) plays an important role in maintaining soil quality and it stores the majority of terrestrial carbon (C) (Lal 2004). Atmospheric input of C into soils via rhizodeposition is an essential pathway for building SOC (Pausch and Kuzyakov 2017). Plant roots release about 20–30% of total photosynthetic C into the soil (Calvo et al. 2017), which can be subsequently utilized by the rhizosphere microbiome (Sasse et al. 2018). The structural dynamics of the rhizosphere bacterial community that assimilates rhizodeposited C can control (i) the quantity of C incorporated into soil pools in the form of metabolites or necromass, which are subsequently stabilized by minerals and aggregates (Liang et al. 2017; Jeewani et al. 2020; Luo et al. 2021), and (ii) the mineralization of both the rhizodeposits and SOC (Pausch and Kuzyakov 2017). Both stabilization and mineralization via rhizosphere microbiome are the keys to plant-derived C sequestration in soil, which are determined by climate, edaphic properties, and agriculture practices (Cotrufo et al. 2013; Keiluweit et al. 2015).
There has been a surge of interest over the last 15 years in the utilization of biochar for increasing soil C content due to its recalcitrance (Lehmann et al. 2008). While biochar can supply a stable C component to the soil, biochar and non-biochar interactions are essential while determining biochar half-life (Kuzyakov et al. 2009) and the decomposition of plant-derived C and native SOC (Liang et al. 2010). Biochar can either promote stabilization of plant-derived C, i.e., rhizodeposits (Weng et al. 2017), or result in faster rhizodeposit mineralization and larger SOC mineralization (Luo et al. 2017), which are mainly driven by biochar-induced changes to both soil abiotic and biotic properties (Yu et al. 2020).
The physicochemical properties of biochar can influence soil pH, nutrient availability, and C chemical composition (Dai et al. 2014; Blanco-Canqui 2017; Razzaghi et al. 2020), which in turn shape soil microbial communities and their functions (Chen et al. 2021; Wang et al. 2021). For example, biochar reduced the effect of protists on the bacterial communities by changing physicochemical properties (e.g., size of the micro-pores and nutrient content), thus altering the microbially mediated transformation of N in soil (Asiloglu et al. 2021). Zhang et al. (2017) found biochar increased pH and thus shifted AOA/AOB ratio and following N2O potential. Several studies highlight that the response of certain soil microbial communities to biochar is linked to altered soil C dynamics (Luo et al. 2013; Whitman et al. 2016, 2021; Campos et al. 2020). Previous studies reported that biochar amendment can decrease soil microbial activities, thus mitigating C loss and promoting soil C storage and fertility (Duan et al. 2020; Wu et al. 2021).
Quite recently, those biochar studies, including plants, have mainly emphasized either (i) biochar and rhizodeposit interactions and their impacts on soil effluxes and C sequestration (Whitman et al. 2014; Weng et al. 2015, 2017; Pei et al. 2020), but ignoring the role of microbial community underpinning C processes, or (ii) biochar-induced changes on rhizosphere microbiome and consequential effects on plant nutrient uptake and growth (Efthymiou et al. 2018; Fu et al. 2021), but omitting soil C processes. For example, greater microbial diversity and potential metabolism observed in the rhizosphere by biochar amendment enhanced plant performance (Kolton et al. 2017), but the microbial mechanisms (taxonomic guild and active microorganisms) underlying rhizosphere-C dynamics remain elusive (Liao et al. 2019; Joseph et al. 2021).
To predict the ecosystem function of the soil microbiome, it is vital to specifically target active members of the soil microbial community (Couradeau et al. 2019). Stable isotope probing (SIP) is a powerful tool for investigating specific active microorganisms by incorporating isotopically labeled substrates (e.g., 13C or 15 N) in situ, and thus link community and function. By using SIP of phospholipid fatty acids (PLFA-SIP), actinomycetes were shown to utilize rhizodeposits in a biochar-amended soil (Chen et al. 2021). To have detailed resolution, DNA-SIP is used to identify microorganisms that assimilate labeled organic substances and their functions in C cycling (Dumont and Murrell 2005). To target RNA as the biomarker, RNA-SIP has the advantage of greater sensitivity compared to DNA-SIP, and can reflect the active bacteria involved in C cycling (Lu and Conrad 2005). Some studies have applied the RNA-SIP technique to identify the active microorganisms in various ecosystems, including phenanthrene degraders in sandy soils (Schwarz et al. 2018), methylotrophs, and sulfate-reducing bacteria in paddy soils (Lueders et al. 2004; Liu et al. 2018). Combination of CO2 labeling and RNA-SIP can provide an approach to identify the microorganisms that are stimulated by plant rhizodeposits (Drigo et al. 2010; Hernández et al. 2015). This therefore may assist in unraveling the microbial mechanisms whereby biochar amendment controls or alters C dynamics in a plant-soil system.
The aims of this study were (i) to investigate the influence of biochar on active bacteria utilizing plant-derived C in the maize rhizosphere and (ii) to assess roles of active bacteria that may potentially regulate the mineralization of rhizodeposits and SOC. We conducted continuous labeling of maize (13CO2, 99% atom 13C) experiment coupled with RNA-SIP. We designed a two-compartment chamber to separate aboveground plant shoot respired CO2 from belowground root and SOC-derived CO2. CO2 from the belowground pools can be then separated by an isotopic signature using a mixed model (Fig. S1). This technique allowed us to understand changes to the mineralization of both maize rhizodeposits and SOC and active microbial community by rhizodeposits. We hypothesized that (i) biochar amendment will change rhizosphere bacterial communities that assimilate rhizodeposits due to altered soil properties such as pH and C recalcitrance, and (ii) the activated microorganisms, i.e., upregulated genera, will in turn enhance rhizodeposit loss and contribute to subsequent SOC mineralization via co-metabolism.
Materials and methods
Soil and biochar materials
Soil was collected from the 0–20-cm layer by using the five diagonal point sampling method from Wenling (28°170′ N, 121°126′ E), Zhejiang, China. The plant residues and stones were removed by hand. The soil was sieved (< 5 mm) and stored at 4 °C before use. The soil texture was loamy clay. Soil properties were total C (3.08 ± 0.21%), total N (0.32 ± 0.03%), pH of soil–water slurry (1:5, w/v) (4.86 ± 0.06), and δ13C (− 28.42 ± 0.03‰). Biochar was produced using swine manure and straw (manure:straw = 1:3) feedstock with the method described by Luo et al. (2011). In brief, materials were pyrolyzed with a heating rate of 26 °C per min, and the highest treatment temperature of 700 °C. This biochar contained 56.7 ± 0.67 total C (%), 0.67 ± 0.03 total N (%), pH of 9.60 ± 0.05, and the δ13C was − 29.8 ± 0.14 (‰). The biochar was sieved (2 mm) and stored at 25 °C before use.
Experimental setup
The experiment investigated one treatment (with biochar) and the control (non-biochar). Sixteen pots (four replicates each treatment) were set up, including (1) unlabeled plant in soil without biochar addition; (2) 13CO2 labeled plant in soil without biochar addition; (3) unlabeled plant in soil with biochar addition; and (4) 13CO2 labeled plant in soil with biochar addition.
The glasshouse experiment used a total of 16 polyvinyl chloride (PVC) pots (height 10.5 cm, diameter 11.3 cm) each filled with 400 g soil (dry weight basis). Biochar was applied at 5% (w/w all dry weight basis) (Wang et al. 2019) to 8 pots homogeneously. The soil in this study was adjusted to about 50% of water holding capacity (WHC) and pre-incubated in the greenhouse at 20 °C (night time) and 28 °C (day time) temperatures, and 70% relative humidity for 7 days before sowing.
Maize (Zea mays L.) seeds were sterilized with 30% H2O2 for 30 min, washed thoroughly with distilled water, and then sown in agar media. At 5 days after sowing the seeds, 1 maize seedling was selected and transplanted into each experimental pot. The pots were then placed in a climate-controlled greenhouse; 10-h dark (night) and 14-h light (day), at 20 °C night and 28 °C daytime temperatures, and 70% relative humidity (Chen et al. 2016). Four replicates of each treatment had 14 days of continuous labeling starting at day 18 (ripening stage) (Lu and Conrad 2005). Before the labeling, 100 mL of 1 M NaOH solution was put inside the chamber to exclude the 12CO2. For 13C labeling, maize was exposed to a 13CO2 enriched atmosphere at 400 ppm for 8 h by using the following procedure. A closed Perspex chamber (width 0.8 m, height 1.0 m, and length 1.5 m) housing 8 pots was used, and a glass beaker containing 100 mL H2SO4 (3 M) was put inside the chamber. At each labeling event, 25 mL of 13C labeled Na2CO3 (≥ 99% atom 13C, Cambridge Isotope Laboratories Inc, USA) solution (1 M) was then injected into a glass beaker containing H2SO4 solution through a tube to release 13CO2. The opening for injection of the 13C labeled Na2CO3 was sealed using Vaseline glue after each injection. The injection was repeated four times every day (every 1.5 h). An electric fan was placed in the chamber to deliver the homogenous distribution of CO2. The CO2 concentration was monitored by using a portable infrared sensor (PGD3-C-CO2, Shenzhen, China).
Chemical analyses
Soil pH was measured in suspension (1:5, soil: Millipore water) by using an ISFET electrode. TC content and TN content (air-dried, milled < 200 µm) were determined by using the PerkinElmer EA2400 (Shelton, CT, USA). The stable isotope was determined using an Elementar vario MICRO cube elemental analyzer coupled to a GV Isoprime 100 isotope ratio mass spectrometer (IRMS; GV Instruments, UK).
Chemical fractionations of SOC were measured according to Rovira and Vallejo (2002). Briefly, 500 mg of ground soil sample was hydrolyzed with 20 mL 2.5 mol L−1 H2SO4 for 30 min at the temperature of 105 °C. Then, the solution was centrifuged at 2795 × g min−1 for 10 min, and the supernatant hydrolysate was transferred to a 50-mL tube. The carbon content of the hydrolysate in the 50-mL tube was measured by using a Multi 3100 N/C TOC analyzer. This carbon content was considered labile organic C (LOC). The remaining soil residues were hydrolyzed and shacked overnight with 2 mL 13 mol L−1 H2SO4 at 25 °C, and then the H2SO4 concentration was diluted to 1 mol L−1 by adding deionized water. The diluted hydrolysate was kept at a temperature of 105 °C for 3 h, and centrifuged for 10 min. The C content of this supernatant hydrolysate was regarded as intermediate organic C (IOC). Finally, the C content of remaining soil residues was measured by using a Vario EL III Elemental Analyzer, which is treated as recalcitrant organic C (ROC).
Soil respiration and 13C-CO2
Total soil respiration was measured using a CO2 trap consisting of 10 mL 1.5 M NaOH solution. The amount of CO2 trapped was determined using a TIM840 auto titrator (Radiometer Analytical, Villeurbanne Cedex, France) with standard HCl (0.0501 M L−1). To determine the δ13C (‰) of the trapped CO2, 5 mL aliquots of each sample were added to 10 mL 1 M BaCl2 in a centrifuge tube. The precipitated BaCO3 was carefully rinsed 3 times with distilled water and dried overnight at 60 ℃ in the centrifuge tube. Finally, 1 mg BaCO3 was accurately weighed into tin caps and the δ13C was analyzed using an elemental analyzer-coupled-isotope ratio mass spectrometer (EA-IRMS) (Sercon Ltd, Crewe, UK). SOC-derived CO2 was separated from root-derived CO2 in planted treatments according to Lu et al. (2019):
where, CSoil, CRoot, and CTotal are the CO2-C derived from soil, CO2-C derived from roots, and total CO2-C derived from belowground in planted treatments, respectively. δ13CSoil, δ13CRoot, and δ13CTotal are defined as the δ13C values of CSoil, CRoot, and CTotal in planted treatments. The mean δ13C value of soil respiration in unplanted soil was used as δ13CSoil.
RNA extraction and isolation of 13C-labeled RNA
Soil RNA extraction of rhizosphere soil of each sample was completed using the RNeasy Power-Soil Total RNA kit (Qiagen) (Ding et al. 2015). The extracted RNA was added with DNase I (Ambion) and purified by using the RNA Clean and Concentrator kit (ZymoResearch). The integrity of the purified RNA was measured by Bio-Rad Experion™ and RNA HighSens Chips (Bio-Rad). Density gradient centrifugation was carried out to separate the 13C-labeled RNA from total RNA (Dumont et al. 2011; Li et al. 2019). Briefly, approximately 500 ng of extracted RNA was mixed with cesium trifluoroacetate (CsTFA) gradients to achieve an original buoyant density (1.790 g mL−1). The above mixtures were centrifuged at 130 000 g for 65 h at 20 °C. RNA fractionation was performed by displacing the gradient medium with sterile water from the top of the ultracentrifuge tube using a NE-1000 single syringe pump (New Era Pump Systems Inc. Farmingdale, NY, USA), with the controlled flow rate of 0.34 mL min−1. A total of 14 RNA gradient fractions with equal volumes of about 340 mL were generated, and the refractive index of these fractions was measured using an AR200 digital hand-held refractometer (Reichert, Inc., Buffalo, NY, USA). Each RNA fraction was converted to the complementary DNA (cDNA) according to the protocol provided by PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The copy numbers of bacterial 16S rRNA in each cDNA fraction were determined on an iCycleriQ 5 thermocycler (Roche Diagnostics, Meylan, France) using primer pairs 515F (5′-GTGYCAGCMGCCGCGGTAA) and 907R (5′-CCGYCAATTYMTTTRAGTTT) (Biddle et al. 2008). Each reaction was performed in a 20-μL volume containing 10 μL SYBR Premix Ex Taq (TaKaRa Biotechnology, Otsu, Shiga, Japan), 0.4 μM of each primer, and 1 μL cDNA template (1–10 ng). The thermocycling conditions were denaturation at 95 °C for 30 s, then 40 cycles of denaturation at 94 °C for 5 s, annealing at 58 °C for 15 s, and extension at 72 °C for 10 s. The bacterial 16S rRNA copy numbers of each cDNA fraction are shown in Fig. S3.
16S rRNA Illumina sequencing
cDNA from the “heavy” gradient fractions of each sample was subjected to 16S rRNA amplicon sequencing. Briefly, 16S rRNA fragments were amplified with barcoded and indexed universal prokaryotic V4-V5 primers 515f/907r, and the products were pooled and sequenced on the Illumina Miseq platform (Illumina, San Diego, USA), run by Majorbio, Inc. (Shanghai, China). PCR amplicons pooled from the triplicate reactions were purified using a QIAquick PCR purification kit (Qiagen, Shenzhen, China) and quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The amplicons from all samples were combined in equal mass. According to the Illumina MiSeq reagent kit preparation guide (Illumina, San Diego, CA, USA), the purified mixture was diluted and denatured to obtain the 8 pmol l−1 amplicon library and mixed with an equal volume of 8 pmol l−1 PhiX (Illumina). Finally, 600 μL of the mixture amplicons was loaded with read-1 and read-2, and the index sequencing primers and paired-end sequencing (each 250 bp) were completed on the Illumina MiSeq platform.
Statistical analysis
The quality control and OTU assignment from Quantitative Insights Into Microb Ecol (QIIME) might be outdated, but still reliable and valid, while some studies found that different bioinformatics pipelines (for example, QIIME vs QIIME2) compared in the microbiome were capable of discriminating samples by treatment, leading to similar biological conclusions (Allal et al. 2017; Moossavi et al. 2020). In our study, QIIME 1.9.0-dev pipeline was used to process the gene sequencing data (Caporaso et al. 2010a, b). In brief, reads which were less than length 200 bp and ambiguous bases were discarded. The sequences were then binned into operational taxonomic units (OTUs) based on 97% similarity by using UCLUST (Edgar 2010). Then chimeric sequences were identified and removed by using UCHIME (Edgar et al. 2011). The most highly connected sequence (i.e., the sequence with the highest similarity to all other sequences in the cluster) was chosen to represent each OTU. The representative OTU sequences were aligned by the PyNAST tool (Caporaso et al. 2010a, b). Taxonomy was assigned to bacterial phylotypes against the SILVA database (https://www.arb-silva.de/). Volcano plots were generated using two-sided t-tests to follow genera changes in response to the biochar amendment. Principal coordinate analysis (PCoA) was conducted using the “capscale” function. Shannon diversity was calculated using the “diversity” function from the Vegan package (Dixon 2003). Partial Mantel tests were also conducted to explore the effects of soil properties on the active bacterial community composition in R (vegan package). Phylogenetic trees were displayed using the “plot_tree” function from the PhyloSeq package (McMurdie and Holmes 2013). A heatmap was drawn by using the function “heatmap.2” in the R package “gplots.” All analyses were performed by using SPSS19.0 (SPSS Inc. Chicago, IL, USA).
Results
Soil physicochemical properties and CO2 efflux
At the completion of the experiment (37 days after sowing), we found that biochar amendment significantly increased soil pH soil–water slurry (1:5, w/v) from 4.53 to 6.17, ROC from 10.69 to 25.77 g·kg−1, and TOC from 14.32 to 29.94 mg·kg−1; in contrast, other properties (e.g., DOC, DON, LOC, IOC, and root biomass) had no significant differences between the soils with or without biochar amendment (Table S1).
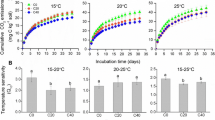
Biochar amendment significantly decreased the incorporation of 13C into the soil (by 58.50%), compared to the non-amended treatment (Fig. 1a) (p < 0.05). However, there were no statistical differences in the root/rhizodeposit-derived CO2 efflux or SOC-derived CO2 efflux (Fig. 1c).
Biochar amendment shifts the rhizodeposit-utilizing bacterial community
The diversity indices (richness and Shannon) of bacteria that utilize 13C-rhizodeposits remained unchanged between biochar-amended soil and the control (Fig. 2a). We observed a wide range of bacteria actively utilizing 13C-rhizodeposits (Fig. 2b). Biochar amendment had effect on the active bacterial communities utilizing rhizodeposits (p < 0.05) (Table S3). Actinobacteria abundance was enhanced by biochar (p < 0.05) (Fig. S4b). Proteobacteria dominated the microbial communities that utilize maize rhizodeposits with 39.64% of the communities accounted for in biochar-amended soils and 46.51% in the control (Fig. 2b; Fig. S4a). Firmicutes, Acidobacteria, Planctomycetes, and Bacteroidetes also participated in rhizodeposit utilization, but with a lesser abundance.
The most common genera utilizing 13C-rhizodeposits were the genus affiliated to Burkholderiaceae, Massilia, and Bacillus (Fig. 2c). Biochar amendment resulted in the upregulation of 45 genera that utilized 13C-rhizodeposits compared to the control (Fig. 3a). The genera with the greatest alteration (log2 FC ≥ 1, p < 0.05) were classified into Actinobacteria and Proteobacteria. Among these upregulated genera, members of Actinobacteria had high relative abundance in the biochar-amended soil, and included genera affiliated to Micrococcaceae, Oryzihumus, Nocardioides, and Methylophilus (Fig. 3b).
The volcano plot represents the filtering threshold for the t-test for differential expression analysis of genera utilizing rhizodeposits between non-amended treatment and biochar-amended treatment (a). The relative abundance of upregulated genera by biochar (b) and the Pearson correlation between these genera and environmental factors (c) and SOC-derived CO2 (d)
Correlation between soil properties, the bacterial community, and SOC mineralization
Bacterial communities in soil with biochar amendment were distinguished from the non-amended soil (Table S2). The largest source of variation (PCoA1) explained 61.96% of the variation in the soil’s active bacterial community, and the Mantel test showed a positive correlation between soil pH (r = 0.89, p < 0.05), TOC (r = 0.54, p < 0.05), ROC (r = 0.58, p < 0.05), and the active bacterial communities (Fig. S5a). Most of the upregulated genera were strongly and positively correlated with soil pH, TOC, and ROC (Fig. 3c).
In addition, we found that most upregulated genera showed a positive correlation with the mineralization of SOC, indicating the potential role of these genera in SOC mineralization (Fig. 3d).
Discussion
Changes in the rhizosphere bacterial community following biochar amendment
We investigated the bacterial community changes in biochar-amended soil using RNA-SIP, which revealed the microorganisms that were activated by rhizodeposits (Fig. 2). Although numerous studies have investigated the changes in the relative abundance of individual phylum using high-throughput sequencing in biochar-treated soils, the generalizations and consistent conclusions are far from being reached as the responses are a result of a wide range of factors, including the alteration of soil properties following biochar amendment, application rates, experimental conditions, and study duration (Lehmann et al. 2011; Luo et al. 2013; Whitman et al. 2016, 2021; Blanco-Canqui 2017; Campos et al. 2020). Considering (i) the large variation of biochar properties and (ii) overwhelmed effect by pre-biochar soil properties over biochar itself, the responders to biochar addition at either the phylum level or the genus level are not consistent across studies (Woolet and Whitman 2020). However, most studies found a positive response of the phylum Actinobacteria to biochar addition (Dai et al. 2017; Yu et al. 2018). Here, we found phylum Actinobacteria were the dominant taxa that utilized maize rhizodeposits in biochar-amended soil (Fig. 2; Fig. S4b). Previous studies that used PLFA found that biochar promotes the abundance of Actinobacteria relative to fungi and other bacteria (Chen et al. 2021), while sequencing analysis also revealed an increase in the abundance of phylum Actinobacteria in biochar-treated soils (Khodadad et al. 2011).
By adopting RNA-SIP and differential expression analysis, this study provided a much higher resolution to reveal the detailed taxonomic information of responders (genera utilizing rhizodeposits) to biochar (Fig. 3a). However, the 14-day continuous labeling and RNA-SIP have the potential to show cross-feeding of label from primary consumers to other soil microbes, resulting in the overestimation of label incorporation in RNA. Therefore, we discuss the more salient results below and compare the results with the previous literatures. As shown by the upregulated genera in biochar-amended soil, Actinobacteria (e.g., genus affiliated to family Micrococcaceae, Nocardioides, Janibacter, Corynebacteriaceae, Terrabacter) and Alphaproteobacteria (e.g., genus Caulobacter, Qipengyuania) were the main active microorganisms that utilized the rhizodeposits (Fig. 3a). The most heavily labeled genus was affiliated with Micrococcaceae, which was reported to be distributed in the rhizosphere of biochar-amended soil (Kolton et al. 2017). These two phyla (i.e., Actinobacteria and Proteobacteria) have also been previously identified in the post-fire soils (Cobo-Díaz et al. 2015; Mikita-Barbato et al. 2015). The changes in soil physical structure and increased abundance of recalcitrant components in pyrolyzed organic matter controlled the bacterial community assemblage of post-fire soils (Luo et al. 2016). Previous studies have widely reported changes in soil physicochemical properties (e.g., labile C and mineral nutrients, soil pH) following biochar amendment that influenced the growth, activity, distribution, and composition of the microbial community (Lehmann et al. 2011; Dai et al. 2021). By correlation analysis, the shift in the microbiome can be attributed to the difference in microbial adaptation ability to biochar itself (e.g., pore size and surface area) and changes in soil properties such as soil pH and chemical structure, e.g., aromatic components (Fig. 3c).
Drivers that shape the rhizosphere bacterial community
Biochar can have a vast range of properties that can alter a range of chemical and physical conditions (e.g., soil bulk density, porosity, pH, C and N availability) in soil (Bolan et al. 2021). These changed physicochemical properties may influence a range of biological interactions. We provide data on changes to available N and C substrates, as well as providing physical niches for colonization (Fig. S2; Table S1). The physical structure (e.g., surface area and porosity) of biochar was reported to be a determinant in shaping soil microbial community (Jaafar et al. 2015). Actinobacteria are particularly adapted to biochar structures, as they have a mycelium-like network. A previous study provides direct evidence that biochar was surrounded by a hyphal network of Actinobacteria (Luo et al. 2013), which are further identified (using PLFA-SIP) as the main microorganism in biochar-amended soil (Luo et al. 2018b).
The biochar-mediated increase of pH in soil from 4.53 to 6.17 (Table S1) may improve the viability of Actinobacteria and enhance their ability to compete with fungi. The optimal pH for Actinobacteria growth has been shown to range between 6.0 and 9.0 (Gohain et al. 2020), and increased pH enhances the bacteria to fungi ratio (Bååth and Anderson 2003). We found a significant positive correlation between upregulated genera of Actinobacteria and soil pH (Fig. 3c). For instance, new species of Nocardia pseudosphaeroides (phylum Actinobacteria) from highly saline and alkaline habitats account for more than 30% of the total number of published new species of Nocardia (He et al. 2007; Ding et al. 2010; Tian et al. 2013). This makes the natural environs a hotspot for the discovery of Nocardia pseudonose. As a response to increasing pH, we found a concomitant decrease in Acidobacteria which is consistent with other studies (Jenkins et al. 2017). Acidobacteria are usually acidophilic (Mao et al. 2012) and therefore less likely to compete in the more neutral soil pH environment following biochar addition. Biochar was found to adjust the competition and interactions between microbial groups via shifting soil pH (Luo et al. 2018a; Chen et al. 2019).
The upregulated genera following biochar amendment might be due to their affinity towards recalcitrant organic C (e.g., aromatic components) (Fabbri et al. 2010; Kolton et al. 2017). We found that the significantly upregulated genera show a positive correlation with ROC (Fig. 3c). Biochar favored the growth of some oligotrophs (e.g., genera belonging to Actinobacteria and Alphaproteobacteria) in the rhizosphere (Fig. 2). The genera affiliated with Micrococcaceae have been previously identified as fire-responders (Woolet and Whitman 2020; You et al. 2021). The post-fire soil contained increased quantities of phenolic compounds, which shaped the oligotrophs (Ling et al. 2021). These upregulated members of Actinobacteria (e.g., genus affiliated to family Micrococcaceae, Nocardioides, Janibacter, Corynebacteriaceae, Terrabacter) and Alphaproteobacteria (e.g., Caulobacter, Qipengyuania) were known to be oligotrophs (Mallory et al. 1977; Entcheva-Dimitrov and Spormann 2004; Khessairi et al. 2014; Huang and Shen 2016; Gao et al. 2019), which are better adapted to conditions where C and nutrient resources are limited, with a role therefore in the degradation of recalcitrant compounds. Biochar is considered to increase C and nutrients due to a small fraction of easily mineralizable C and nutrients. However, the labile C component of biochar is likely to be small when compared to inputs of rhizodeposits (Weng et al. 2020). Instead, the availability of DOC and nutrients might be lowered due to the absorption of these resources onto biochar (Bolan et al. 2021). Li et al. (2019) suggested that recalcitrant components are the main driver of the microbial community in biochar-amended soil, which was consistent with our study (Table S1, Fig. S5, and Fig. 3c). Taken together, a possible explanation for the significantly upregulated genera in the biochar-amended soil could be the adaption of these oligotrophs to changes in soil structure, pH, and particularly the affinity to aromatic components, which possibly further degrade more recalcitrant components from SOC.
Potential SOC decomposers in the biochar-plant-soil system
Short-term biochar-induced increase of SOC mineralization was both abiotically and biotically mediated. Physicochemical properties such as pH have some direct effects on SOC mineralization. For example, Wei et al. (2021) observed that a colloidal mobility was depressed in acidic environments, where SOM was stabilized by Al3+ in the absence of Ca2+. However, the transitory and large increase in SOC mineralization caused by biochar is mainly attributed to microorganisms activated by abiotic properties in our study. The enhanced taxa activated by rhizodeposits might utilize other C sources, e.g., SOC. Indeed, we found larger short-term SOC mineralization in biochar-amended than non-amended soil (Fig. 1c) which may be caused by the upregulation of some genera (Fig. 3a). Most of these dominant upregulated genera are correlated with SOC mineralization (Fig. 3d), including genera affiliated to Micrococcaceae and other oligotrophs such as the genera Oryzihumus, Caulobacter, Qipengyuania, and Dongia (Fig. 3b, d). Most of these genera that correlated with SOC decomposition belonged to the phylum Actinobacteria and Proteobacteria, which have been found in soil after wildfire (Woolet and Whitman 2020). Importantly, the abundance of Actinobacteria was associated with the degradation of SOC mostly from recalcitrant compounds (Blagodatskaya and Kuzyakov 2008). Positive interactions between dominant oligotrophs, e.g., Actinobacteria, and recalcitrant components, such as phenol-like substances, have been shown (Ling et al. 2021). The increased abundance of these upregulated genera might be due to their affinity and ability to degrade the aromatic components brought by biochar to soils, which in turn cause decomposition of SOC via the production of extracellular enzymes (Goodfellow and Williams 1983; Bao et al. 2021).
Microbial communities are generally considered to affect soil C stabilization in two ways: (1) they incorporate external C into their cellular biomass production, which may subsequently be stabilized by mineral associations, and (2) they supply enzymes that catalyze the decomposition of plant-derived and native soil C (Kögel-Knabner 2002; Malik et al. 2020). Different microbial groups with metabolic strategies likely influence these two pathways of C substrates. In our study, the co-metabolism stimulated in the biochar-amended soil caused both increased efflux of CO2 originating from rhizodeposits (Fig. 1b) and native SOC (Fig. 1c). This indicates these upregulated microbial genera mainly invested in enzyme production to increase the decomposition of C substrates, thus causing greater CO2 emissions and lower 13C incorporation in soil.
Specifically, the genus Nocardioides enriched in biochar-amended soil was identified as a putative degrader of recalcitrant components, including triazine herbicides (Topp et al. 2000) and crude oil (Schippers et al. 2005). Nocardiopsis can secrete cellulolytic enzymes to degrade carboxymethyl and microcrystalline cellulose (Saratale and Oh 2011). A variety of enzymes that can degrade starch, protein, cellulose, and xylan have been isolated from Nocardia pseudobacteria, showing their potential in the mineralization of organic matter in the soil. These extracellular enzymes including endoglucanases, exoglucanases, xylanase, and glucoamylase are highly involved in metabolisms of alkanes and phenanthrene, the benzene ring within straw and polycyclic aromatic hydrocarbon, and thus possibly drive decomposition of recalcitrant components of SOC via co-metabolism. It was reported that activated microorganisms following the addition of organic substances can accelerate SOC mineralization via co-metabolism (Fontaine et al. 2003). For instance, Johnson-Rollings et al. (2014) detected increases in the abundance of Nocardiopsis and associated chitinase of glycoside hydrolase family 18 (GH18) from soil extracts, and confirmed this enzyme is responsible for the degradation of chitin in the soil. Additionally, members of Micrococcaceae, being well known for high cellulolytic activity, are the primary decomposer of organic material (España et al. 2011). Other genera within the phylum Actinobacteria, such as Oryzihumus, were the dominant microorganism in the biochar-amended soil, but little information exists about their ecological role in SOC decomposition (Wang et al. 2020). The α-Proteobacteria may also produce SOC-degrading enzymes, enabling their growth by utilizing nutrients from the soil. For instance, the genus Caulobacter (Alphaproteobacteria), considered as facultative oligotrophs, is presumed to be responsible for considerable mineralization of organic matter through the production of cellulases, including endoglucanases and β-glucosidases (Song et al. 2013).
Taken together, amendment of swine manure biochar in soil stimulated the growth of oligotrophs (e.g., Actinobacteria and Alphaproteobacteria) by increasing pH (Fig. 3c), which accelerated SOC mineralization and offset C sequestration potential by biochar incorporation. To our knowledge, this is the first experimental evidence from the use of RNA-SIP to confirm the role of Actinobacteria in SOC mineralization in a biochar-plant-soil system. Here, we highlight the significance of oligotrophs, particularly Actinobacteria, in the biochar-amended soil and their potential role in SOC mineralization. This study demonstrates the value of RNA-SIP in providing targeted insights for coupling metabolic activity (i.e., decomposition of rhizodeposits) to phylogeny. However, to further investigate the physiological characteristic of active microorganisms, a combination of RNA-SIP with metatranscriptomic sequencing should be provided to get information on the metabolic potential produced by active taxa.
Conclusion
Combination of continuous 13CO2 labelling and RNA-SIP was used to detect the core rhizosphere microorganisms involved in soil C processes following swine manure biochar amendment to maize-planted soil. Here, we showed that (i) rhizosphere bacterial communities were dominated by Actinobacteria and Alphaproteobacteria in the biochar-amended soil, most likely due to the removal of soil acidity and the increased content of recalcitrant organic C and (ii) majority of these upregulated genera (e.g., genus affiliated to Micrococcaceae, Oryzihumus, Caulobacter) by biochar were found to be positively correlated with SOC-derived CO2, indicating the significance of these genera in C mineralization in biochar-amended soil. This study highlights the core rhizosphere-associated bacterial communities that function in SOC mineralization in the biochar-amended soil–plant system. Considering the short period in this study, further research should focus on the long-term effects of biochar on the temporal dynamics of the microbial communities and the expression of C-related genes during plant growth.
Abbreviations
- SOC:
-
Soil organic carbon
- LOC:
-
Labile organic carbon
- IOC:
-
Intermediate organic carbon
- ROC:
-
Recalcitrant organic carbon
- RNA-SIP:
-
RNA-based stable isotope probing
References
Allal I, Arnold JW, Roach J, Cadenas MB, Butz N, Hassan HM, Koci M, Ballou A, Mendoza M, Ali R, Azcarate-Peril MA (2017) A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 17:194
Asiloglu R, Sevilir B, Samuel SO, Aycan M, Akca MO, Suzuki K, Murase J, Turgay OC, Harada N (2021) Effect of protists on rhizobacterial community composition and rice plant growth in a biochar amended soil. Biol Fertil Soils 57:293–304
Bååth E, Anderson TH (2003) Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem 35:955–963
Bao Y, Dolfing J, Guo Z, Chen R, Wu M, Li Z, Lin X, Feng Y (2021) Important ecophysiological roles of non-dominant Actinobacteria in plant residue decomposition especially in less fertile soils. Microbiome 9:84
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH (2008) Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci 105:10583
Blagodatskaya E, Kuzyakov Y (2008) Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol Fertil Soils 45:115–131
Blanco-Canqui H (2017) Biochar and soil physical properties. Soil Sci Soc Am J 81:687–711
Bolan N, Hoang S, Beiyuan J, Gupta S, Hou D, Karakoti A, Joseph S, Jung S, Kim KH, Kirkham M, Kua H, Kumar M, Kwon E, Ok Y, Perera V, Rinklebe J, Shaheen S, Sarkar B, Sarmah A, Van Zwieten L (2021) Multifunctional applications of biochar beyond carbon storage. Int Mater Rev 67:150–200
Calvo OC, Franzaring J, Schmid I, Müller M, Brohon N, Fangmeier A (2017) Atmospheric CO2 enrichment and drought stress modify root exudation of barley. Global Change Biol 23:1292–1304
Campos P, Miller AZ, Prats SA, Knicker H, Hagemann N, De la Rosa JM (2020) Biochar amendment increases bacterial diversity and vegetation cover in trace element-polluted soils: a long-term field experiment. Soil Biol Biochem 150:108014
Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello E, Fierer N, Peña A, Goodrich J, Gordon J, Huttley G, Kelley S, Knights D, Koenig J, Ley R, Lozupone C, McDonald D, Muegge B, Pirrung M, Reeder J, Sevinsky J, Turnbaugh P, Walters W, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso J, Bittinger K, Bushman F, DeSantis T, Andersen G, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics (oxford England) 26:266–267
Chen L, Brookes P, Xu J, Zhang J, Zhang C, Zhou X, Luo Y (2016) Structural and functional differentiation of the root-associated bacterial microbiomes of perennial ryegrass. Soil Biol Biochem 98:1–10
Chen L, Jiang Y, Liang C, Luo Y, Xu Q, Han C, Zhao Q, Sun B (2019) Competitive interaction with keystone taxa induced negative priming under biochar amendments. Microbiome 7:77
Chen Z, Kumar A, Fu Y, Singh B, Ge T, Tu H, Luo Y, Xu J (2021) Biochar decreased rhizodeposits stabilization via opposite effects on bacteria and fungi: diminished fungi-promoted aggregation and enhanced bacterial mineralization. Biol Fertil Soils 57:533–546
Cobo-Díaz J, Fernández-González A, Villadas P, Robles A, Toro N, Fernández-López M (2015) Metagenomic assessment of the potential microbial nitrogen pathways in the rhizosphere of a Mediterranean forest after a wildfire. Microb Ecol 69:895–904
Cotrufo M, Wallenstein M, Boot C, Denef K, Paul E (2013) The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: do labile plant inputs form stable soil organic matter? Global Change Biol 19:988–995
Couradeau E, Sasse J, Goudeau D, Nath N, Hazen T, Bowen B, Chakraborty R, Malmstrom R, Northen T (2019) Probing the active fraction of soil microbiomes using BONCAT-FACS. Nat Commun 10:2770
Dai Z, Wang Y, Muhammad N, Yu X, Xiao K, Meng J, Liu X, Xu J, Brookes PC (2014) The effects and mechanisms of soil acidity changes, following incorporation of biochars in three soils differing in initial pH. Soil Sci Soc Am J 78:1606–1614
Dai Z, Barberan A, Li Y, Brookes PC, Xu J (2017) Bacterial community composition associated with pyrogenic organic matter (biochar) varies with pyrolysis temperature and colonization environment. MSphere 2:e00085-17
Dai Z, Xiong X, Zhu H, Xu H, Leng P, Li J, Tang C, Xu J (2021) Association of biochar properties with changes in soil bacterial, fungal and fauna communities and nutrient cycling processes. Biochar 3:239–254
Ding Z, Li M, Zhao J, Ren J, Huang R, Xie M, Cui X, Zhu H, Wen M (2010) Naphthospironone A: an unprecedented and highly functionalized polycyclic metabolite from an alkaline mine waste extremophile. Chemistry 16:3902–3905
Ding LJ, Su JQ, Xu HJ, Jia ZJ, Zhu YG (2015) Long-term nitrogen fertilization of paddy soil shifts iron-reducing microbial community revealed by RNA-13C-acetate probing coupled with pyrosequencing. The ISME J 9:721–734
Dixon P (2003) VEGAN a package of R functions for community ecology. J Veg Sci 14:927–930
Drigo B, Pijl AS, Duyts HK, Kielak AM, Kowalchuk GA (2010) Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. PNAS 107:10938–10942
Duan M, Wu F, Jia Z, Wang S, Cai Y, Chang SX (2020) Wheat straw and its biochar differently affect soil properties and field-based greenhouse gas emission in a Chernozemic soil. Biol Fertil Soils 56:1023–1036
Dumont MG, Murrell JC (2005) Stable isotope probing — linking microbial identity to function. Nat Rev Microbiol 3:499–504
Dumont MG, Pommerenke B, Casper P, Conrad R (2011) DNA- rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol 13:1153–1167
Edgar RC (2010) Search and Clustering Orders of Magnitude Faster than BLAST Bioinformatics (oxford England) 26:2460–2461
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (oxford England) 27:2194–2200
Efthymiou A, Grønlund M, Müller-Stöver DS, Jakobsen I (2018) Augmentation of the phosphorus fertilizer value of biochar by inoculation of wheat with selected Penicillium strains. Soil Biol Biochem 116:139–147
Entcheva-Dimitrov P, Spormann AM (2004) Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J Bacteriol 186:8254–8266
España M, Rasche F, Kandeler E, Brune T, Rodriguez B, Bending GD, Cadisch G (2011) Identification of active bacteria involved in decomposition of complex maize and soybean residues in a tropical Vertisol using 15N-DNA stable isotope probing. Pedobiologia 54:187–193
Fabbri D, Adamiano A, Torri C (2010) GC-MS determination of polycyclic aromatic hydrocarbons evolved from pyrolysis of biomass. Anal Bioanal Chem 397:309–317
Fontaine S, Mariotti A, Abbadie L (2003) The priming effect of organic matter: a question of microbial competition? Soil Biol Biochem 35:837–843
Fu Y, Kumar A, Chen L, Jiang Y, Ling N, Wang R, Pan Q, Singh BP, Redmile-Gordon M, Luan L, Li Q, Shi Q, Reid BJ, Fang Y, Kuzyakov Y, Luo Y, Xu J (2021) Rhizosphere microbiome modulated effects of biochar on ryegrass 15N uptake and rhizodeposited 13C allocation in soil. Plant Soil 463:359–377
Gao Y, Wu YH, Xu L, Cheng H, Wang CS, Xu XW (2019) Complete genome sequence of Qipengyuania sediminis CGMCC 112928T shed light on its role in matter-cycle and cold adaption mechanism of the genus Qipengyuania. Curr Microbiol 76:988–994
Gohain A, Manpoong C, Saikia R, De Mandal S (2020) Actinobacteria: diversity and biotechnological applications. In: De Mandal S, Bhatt P (eds) Recent advancements in microbial diversity. Press, London, pp 217–231
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Annu Rev Microbiol 37:189–216
He J, Roemer E, Lange C, Huang X, Maier A, Kelter G, Jiang J, Xu LH, Menzel KD, Grabley S (2007) Structure derivatization and antitumor activity of new griseusins from Nocardiopsis sp. J Med Chem 50:5168–5175
Hernández M, Dumont MG, Yuan Q, Conrad R (2015) Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl Environ Microbiol 81:2244–2253
Huang YS, Shen FT (2016) Bioprospecting of facultatively oligotrophic bacteria from non-rhizospheric soils. Appl Soil Ecol 108:315–324
Jaafar NM, Clode PL, Abbott LK (2015) Soil microbial responses to biochars varying in particle size, surface and pore properties. Pedosphere 25:770–780. https://doi.org/10.1016/S1002-0160(15)30058-8
Jeewani P, Gunina A, Tao L, Zhu Z, Kuzyakov Y, Van Zwieten L, Guggenberger G, Shen C, Yu G, Singh BP, Pan S, Luo Y, Xu JM (2020) Rusty sink of rhizodeposits and associated keystone microbiomes. Soil Biol Biochem 147:107840
Jenkins JR, Viger M, Arnold C, Harris ZM, Ventura M, Miglietta F, Girardin C, Edwards RJ, Rumpel C, Fornasier F, Zavalloni C, Tonon G, Alberti G, Taylor G (2017) Biochar alters the soil microbiome and soil function: results of next-generation amplicon sequencing across Europe. Glob Change Biol Bioen 9:591–612
Johnson-Rollings AS, Wright H, Masciandaro G, Wright H, Masciandaro G, Macci C, Doni S, Calvo-BaDo LA, Susan ES, Carlos VP, Elizabeth MHW (2014) Exploring the functional soil-microbe interface and exoenzymes through soil metaexoproteomics. The ISME J 8:2148–2150
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. The ISME J 3:442–453
Joseph S, Cowie AL, Van Zwieten L, Bolan N, Budai A, Buss W, Cayuela ML, Graber ER, Ippolito JA, Kuzyakov Y, Luo Y, Ok YS, Palansooriya KN, Shepherd J, Stephens S, Weng Z, Lehmann J (2021) How biochar works, and when it doesn’t: a review of mechanisms controlling soil and plant responses to biochar. GCB Bioenergy 13:1731–1764
Keiluweit M, Bougoure JJ, Nico PS, Pett-Ridge J, Weber PK, Kleber M (2015) Mineral protection of soil carbon counteracted by root exudates. Nat Clim Chang 5:588–595
Khessairi A, Fhoula I, Jaouani A, Turki Y, Cherif A, Boudabous A, Hassen A, Ouzari H (2014) Pentachlorophenol degradation by Janibacter sp, a new actinobacterium isolated from saline sediment of arid land. Biomed Res Int 4:296472
Khodadad CLM, Zimmerman AR, Green SJ, Uthandi S, Foster JS (2011) Taxa-specific changes in soil microbial community composition induced by pyrogenic carbon amendments. Soil Biol Biochem 43:385–392
Kögel-Knabner I (2002) The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol Biochem 34:139–162
Kolton M, Graber ER, Tsehansky L, Elad Y, Cytryn E (2017) Biochar-stimulated plant performance is strongly linked to microbial diversity and metabolic potential in the rhizosphere. New Phytol 213:1393–1404
Kuzyakov Y, Subbotina I, Chen H, Bogomolova I, Xu X (2009) Black carbon decomposition and incorporation into soil microbial biomass estimated by 14C labeling. Soil Biol Biochem 41:210–219
Lal R (2004) Soil carbon sequestration impacts on global climate change and food security. Science 304:1623–1627
Lehmann J, Skjemstad J, Sohi S, Carter J, Barson M, Falloon P, Coleman K, Woodbury P, Krull E (2008) Australian climate–carbon cycle feedback reduced by soil black carbon. Nat Geosci 1:832–835
Lehmann J, Rillig M, Thies J, Masiello C, Hockaday W, Crowley D (2011) Biochar effects on soil biota - a review. Soil Biol Biochem 43:1812–1836
Li Y, Li Y, Chang SX, Yang Y, Fu S, Jiang P, Luo Y, Yang M, Chen Z, Hu S, Zhao M, Liang X, Xu Q, Zhou G, Zhou J (2018) Biochar reduces soil heterotrophic respiration in a subtropical plantation through increasing soil organic carbon recalcitrancy and decreasing carbon-degrading microbial activity. Soil Biol Biochem 122:173–185
Li H, Su JQ, Yang XR, Zhou GW, Lassen SB, Zhu YG (2019) RNA stable isotope probing of potential Feammox population in paddy soil. Environ Sci Technol 53:4841–4849
Liang B, Lehmann J, Sohi SP, Thies JE, O’Neill B, Trujillo L, Gaunt J, Solomon D, Grossman J, Neves EG, Luizão FJ (2010) Black carbon affects the cycling of non-black carbon in soil. Org Geochem 41:206–213
Liang C, Schimel JP, Jastrow D (2017) The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol 2:17105
Liao H, Yaying L, Yao H (2019) Biochar amendment stimulates utilization of plant-derived carbon by soil bacteria in an intercropping system. Front Microbiol 10:1361
Liao H, Zheng C, Long J, Guzmán I (2021) Effects of biochar amendment on tomato rhizosphere bacterial communities and their utilization of plant-derived carbon in a calcareous soil. Geoderma 396:115082
Ling L, Fu Y, Jeewani P, Tang C, Pan S, Reid B, Gunina A, Li Y, Li Y, Cai Y, Kuzyakov Y, Li Y, Su WQ, Singh BP, Luo Y, Xu JM (2021) Organic matter chemistry and bacterial community structure regulate decomposition processes in post-fire forest soils. Soil Biol Biochem 160:108311
Liu P, Pommerenke B, Conrad R (2018) Identification of Syntrophobacteraceae as major acetate-degrading sulfate reducing bacteria in Italian paddy soil. Environ Microbiol 20:337–354
Liu YR, Delgado-Baquerizo M, Yang Z, Feng J, Zhu J, Huang Q (2020) Microbial taxonomic and functional attributes consistently predict soil CO2 emissions across contrasting croplands. Sci Total Environ 702:134885
Lu Y, Conrad R (2005) In situ stable isotope probing of methanogenic Archaea in the rice rhizosphere. Science 309:1088–1090
Lu J, Dijkstra FA, Wang P, Cheng W (2019) Roots of non-woody perennials accelerated long-term soil organic matter decomposition through biological and physical mechanisms. Soil Biol Biochem 134:42–53
Lueders T, Manefield M, Friedrich MW (2004) Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isoBiocharnic centrifugation gradients. Environ Microbiol 6:73–78
Luo Y, Durenkamp M, Nobili M, Lin Q, Brookes PC (2011) Short term soil priming effects and the mineralisation of biochar following its incorporation to soils of different pH. Soil Biol Biochem 43:2304–2314
Luo Y, Durenkamp M, De Nobili M, Lin Q, Devonshire BJ, Brookes PC (2013) Microbial biomass growth following incorporation of biochars produced at 350 °C or 700 °C in a silty-clay loam soil of high and low pH. Soil Biol Biochem 57:513–523
Luo Y, Yu Z, Zhang K, Xu J, Brookes P (2016) The properties and functions of biochars in forest ecosystems. J Soils Sediments 16:8
Luo Y, Zang H, Yu Z, Chen Z, Gunina A, Kuzyakov Y, Xu J, Zhang K, Brookes PC (2017) Priming effects in biochar enriched soils using a three-source-partitioning approach: 14C labelling and 13C natural abundance. Soil Biol Biochem 106:28–35
Luo Y, Dungait JAJ, Zhao X, Brookes PC, Durenkamp M, Li G, Lin Q (2018) Pyrolysis temperature during biochar production alters its subsequent utilization by microorganisms in an acid arable soil. Land Degrad Dev 29:2183–2188
Luo Y, Lin Q, Durenkamp M, Kuzyakov Y (2018) Does repeated biochar incorporation induce further soil priming effect? J Soils Sed 18:128–135
Luo Y, Xiao M, Yuan H, Liang C, Zhu Z, Xu J, Kuzyakov Y, Wu J, Ge T, Tang C (2021) Rice rhizodeposition promotes the build-up of organic carbon in soil via fungal necromass. Soil Biol Biochem 160:108345
Malik AA, Martiny JBH, Brodie EL, Martiny AC, Treseder KK, Allison SD (2020) Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. The ISME J 14:1–9
Mallory LM, Austin B, Colwell RR (1977) Numerical taxonomy and ecology of oligotrophic bacteria isolated from the estuarine environment. Can J Microbiol 23:733–750
Mao JD, Johnson RL, Lehmann J, Olk DC, Neves EG, Thompson ML, Schmidt-Rohr K (2012) Abundant and stable char residues in soils: implications for soil fertility and carbon sequestration. Environ Sci Technol 46:9571–9576
McMurdie P, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:4
Mikita-Barbato RA, Kelly JJ, Tate RL (2015) Wildfire effects on the properties and microbial community structure of organic horizon soils in the New Jersey Pinelands. Soil Biol Biochem 86:67–77
Moossavi S, Atakora F, Fehr K, Khafipour E (2020) Biological observations in microbiota analysis are robust to the choice of 16S rRNA gene sequencing processing algorithm: case study on human milk microbiota. BMC Microbiol 20:290
Pausch J, Kuzyakov Y (2017) Carbon input by roots into the soil: quantification of rhizodeposition from root to ecosystem scale. Global Change Biol 24:1
Pei Junmin Dijkstra FA, Li J, Fang C, Wu J (2020) Biochar-induced reductions in the rhizosphere priming effect are weaker under elevated CO2. Soil Biol Biochem 142:107700
Razzaghi F, Obour PB, Arthur E (2020) Does biochar improve soil water retention? A systematic review and meta-analysis. Geoderma 361:114055
Rovira P, Vallejo VR (2002) Labile and recalcitrant pools of carbon and nitrogen in organic matter decomposing at different depths in soil: an acid hydrolysis approach. Geoderma 107:109–141
Saratale GD, Oh SE (2011) Production of thermotolerant and alkalotolerant cellulolytic enzymes by isolated Nocardiopsis sp. Biodegradation 22:905–919
Sasse J, Martinoia E, Northen T (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23:25–41
Schippers A, Schumann P, Spröer C (2005) Nocardioides oleivorans sp, nov, a novel crude-oil-degrading bacterium. Int J Syst Evol Microbiol 55:1501–1504
Schwarz A, Adetutu EM, Juhasz AL, Aburto-Medina A, Ball AS, Shahsavari E (2018) Microbial degradation of phenanthrene in pristine and contaminated sandy soils. Microb Ecol 75:888–902
Song N, Cai HY, Yan ZS, Jiang HL (2013) Cellulose degradation by one mesophilic strain Caulobacter sp, FMC1 under both aerobic and anaerobic conditions. Bioresour Technol 131:281–287
Tian SZ, Pu X, Luo GY, Zhao LH, Xu LH, Li WJ, Luo Y (2013) Isolation and characterization of new p-terphenyls with antifungal antibacterial and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J Agric Food Chem 61:3006–3012
Topp E, Mulbry WM, Zhu H, Nour SM, Cuppels D (2000) Characterization of s-triazine herbicide metabolism by a nocardioides sp, isolated from agricultural soils. Appl Environ Microbiol 66:3134–3141
Wang Y, Zhong B, Shafi M, Ma J, Guo J, Wu J, Ye Z, Liu D, Jin H (2019) Effects of biochar on growth and heavy metals accumulation of moso bamboo (Phyllostachy pubescens) soil physical properties and heavy metals solubility in soil. Chemosphere 219:510–516
Wang M, Lan X, Xu X, Fang Y, Singh BP, Sardans J, Romero E, Peñuelas J, Wang W (2020) Steel slag and biochar amendments decreased CO2 emissions by altering soil chemical properties and bacterial community structure over two-year in a subtropical paddy field. Sci Total Environ 740:140403
Wang C, Chen D, Shen J, Yuan Q, Fan F, Wei W, Li Y, Wu J (2021) Biochar alters soil microbial communities and potential functions 3–4 years after amendment in a double rice cropping system. Agric Ecosyst Environ 311:107291
Wei L, Ge T, Zhu Z, Luo Y, Yang Y, Xiao M, Yan Z, Li Y, Wu J, Kuzyakov Y (2021) Comparing carbon and nitrogen stocks in paddy and upland soils: accumulation, stabilization mechanisms, and environmental drivers. Geoderma 398:115121
Weng Z, Van Zwieten L, Singh BP, Kimber S, Morris S, Cowie A, Macdonald LM (2015) Plant-biochar interactions drive the negative priming of soil organic carbon in an annual ryegrass field system. Soil Biol Biochem 90:111–121
Weng Z, Van Zwieten L, Singh BP, Tavakkoli E, Joseph S, Macdonald LM, Rose TJ, Rose MT, Kimber SWL, Morris S, Cozzolino D, Araujo JR, Archanjo BS, Cowie A (2017) Biochar built soil carbon over a decade by stabilizing rhizodeposits. Nat Clim Chang 7:371–376
Weng Z, Liu X, Eldridge S, Wang H, Rose T, Rose M, Rust J, Singh BP, Tavakkoli E, Tang C, Ou H, Van Zwieten L (2020) Priming of soil organic carbon induced by sugarcane residues and its biochar control the source of nitrogen for plant uptake: a dual 13C and 15N isotope three-source-partitioning study. Soil Biol Biochem 146:107792
Whitman T, Enders A, Lehmann J (2014) Biochar additions to soil counteract positive priming of soil carbon mineralization by plants. Soil Biol Biochem 73:33–41
Whitman T, Pepe-Ranney C, Enders A, Koechli C, Campbell A, Buckley DH, Lehmann J (2016) Dynamics of microbial community composition and soil organic carbon mineralization in soil following addition of pyrogenic and fresh organic matter. The ISME J 10:2918–2930
Whitman T, DeCiucies S, Hanley K, Enders A, Woolet J, Lehmann J (2021) Microbial community shifts reflect losses of native soil carbon with pyrogenic and fresh organic matter additions and are greatest in low-carbon soils. Applied and Environ Microbiol 87:8
Woolet J, Whitman T (2020) Pyrogenic organic matter effects on soil bacterial community composition. Soil Biol Biochem 141:107678
Wu Q, Lian R, Bai M, Bao J, Liu Y, Li S, Liang C, Qin H, Chen J, Xu Q (2021) Biochar co-application mitigated the stimulation of organic amendments on soil respiration by decreasing microbial activities in an infertile soil. Biol Fertil Soils 57:793–807
You X, Suo F, Yin S, Wang X, Zheng H, Fang S, Zhang C, Li F, Li Y (2021) Biochar decreased enantioselective uptake of chiral pesticide metalaxyl by lettuce and shifted bacterial community in agricultural soil. J Hazard Mater 417:126047
Yu Z, Chen L, Pan S, Li Y, Kuzyakov Y, Xu J, Brookes PC, Luo Y (2018) Feedstock determines biochar-induced soil priming effects by stimulating the activity of specific microorganisms. Eur J Soil Sci 69:521–534
Yu J, Pavia MJ, Deem LM, Crow SE, Deenik JL, Penton CR (2020) DNA-stable isotope probing shotgun metagenomics reveals the resilience of active microbial communities to biochar amendment in Oxisol soil. Front Microbiol 11:2718
Zhang K, Chen L, Li Y, Brookes PC, Xu J, Luo Y (2017) The effects of combinations of biochar, lime, and organic fertilizer on nitrification and nitrifiers. Biol Fertil Soils 53:77–87
Funding
This study was supported by Zhejiang Provincial Natural Science Foundation of China under Grant Number of R19D010005, and National Science Foundation of China (U1901601).
Author information
Authors and Affiliations
Contributions
Y.L. designed the research. Y.Y.F. contributed to the acquisition, analysis, or interpretation of data and drafted the manuscript, and all authors were involved in revising the manuscript and approving the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Open research statement
Sequencing data are available from the NCBI under the study accession number: SUB10465147.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fu, Y., Luo, Y., Auwal, M. et al. Biochar accelerates soil organic carbon mineralization via rhizodeposit-activated Actinobacteria. Biol Fertil Soils 58, 565–577 (2022). https://doi.org/10.1007/s00374-022-01643-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-022-01643-y