Abstract
Background and aims
Incorporation of biochar into the soil sequesters C for millennia, but the concomitant effects on plant rhizodepositions and nutrient (e.g., nitrogen; N) trade-offs via interactions of heterotrophic microbiota, might offset this sequestration.
Methods
Ryegrass (Lolium perenne L.) with and without biochar amendment were pulse labelled in a 13CO2 atmosphere and 15N fertilizer added. Ryegrass and soils were destructively sampled at 16 and 30 days after seedling emergence. Isotope analysis was coupled with MiSeq sequencing of bacterial (16s rRNA) and fungal (ITS) genes to identify the effect of biochar on the associated microbiota involved in 13C allocation into soil aggregates and promotion of 15N uptake by L. perenne.
Results
Biochar increased root biomass and 15N uptake but decreased rhizodeposited-13C recovery from large and small macroaggregates (by 12–57% and 57–72%, respectively). These changes in 13C flow and 15N uptake were accompanied by an increase in microbial biomass, and enhanced negative correlations between bacteria and fungi. O2PLS indicated members of seventeen genera that were correlated with soil stabilization of rhizodeposits in soil and plant N-uptake. For instance, Xanthomonadales (Proteobacteria) and RB41 (Acidobacteria), previously reported to be plant growth promoting rhizobacteria, were found to be positively correlated with 15N uptake by L. perenne.
Conclusions
Our research explored the genera associated with biochar-modified 15N uptake by Lolium perenne and photosynthate 13C allocation into soil aggregates. Future research with SIP is required to fully assess microbial turnover, the ubiquity of similar rhizosphere microbiota and their fundamental importance for sequestration in the plant-soil-microbe-biochar systems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Rhizodeposition, the processes of releasing organic compounds into the soil by living roots, represents an important flow of C from plant to soil and is inextricably linked to plant and soil microbial community development (Drigo et al. 2013). Indeed, root mediated alteration of microbial activity and community composition are dependent on plant phenology and growth stage (Cavaglieri et al. 2007; Houlden et al. 2008). Since the soil nutrient cycles are almost entirely regulated by microbes, changes in plant rhizodeposition can significantly impact rates of soil nutrient cycling (Rütting et al. 2009; Veresoglou et al. 2012). Plant allocation of C into rhizosphere soil is a vital factor influencing nitrogen (N) availability. Plants release rhizodeposits containing C for soil microorganisms in exchange for nutrients directly or indirectly (Laurent et al. 2013; Weng et al. 2017). Elevated CO2 increases rhizodeposition and enhance microbial N immobilization, thus increasing N limitation for plants (Graaff et al. 2007; Kuzyakov et al. 2019). Conversely, rhizodeposited C can stimulate microbial decomposition of SOM (soil organic matter) and thus release nutrients for plant growth (Jacoby et al. 2017; Sarker et al. 2017; Verburg et al. 2014). This interplay (between acquisition, storage, release) of C and nutrients - driven by specific microbiota - remains largely unexplored.
Toju et al. (Toju et al. 2018) introduced the concept of core microbiomes as key sets of microorganisms that moderate ecosystem functions directly, or through regulating interactions with other microorganisms. Another study adopted metagenomic data to link the core microbiome, e.g. Acidobacteria and Sphingobacteriales, and functions in CO2 and CH4 emissions and N cycling (Hester et al. 2018). AMF (arbuscular mycorrhizal fungi) stimulate releases C from plants, followed by assisting transfer of N back to the plant (Chen et al. 2019b). Increasing knowledge has been obtained to link plant-associated microbiomes with plant growth and health. However, attempts to identify key rhizosphere microbiomes using bioinformatic pipelines, especially to predict their role in modulating soil C flows and nutrient-uptake, are still rare (Toju et al. 2018).
The application of biochar to soils is a cost-effective technique to offset C emissions and improve soil fertility (Lehmann 2007). Biochar application to soil increases soil C stocks via the intrinsic recalcitrance of the biochar itself (Lehmann et al. 2006) and also via inhibit mineralization of SOM (Karhu et al. 2011; Luo et al. 2017b; Singh et al. 2010a). The holistic offset value linked to biochar is affected indirectly by i) stabilization of rhizodeposits from surface vegetation, and ii) subsequent effects on pre-existing pools of soil organic C (SOC) (Luo et al. 2011; Weng et al. 2017). In addition to SOC dynamics, investigating biochar’s impacts on a range of other soil processes, for example those related to N, will be vital to ultimately understanding biochars’ benefits (Gul et al. 2015). Since the soil nutrient cycle is almost completely regulated by microbes, there remains a need to better understand how plants and biochar interact via the soil, and specifically via the soil microbial community.
The mechanisms of biochar-effects on C and N processes vary through time, but are primarily attributed to direct modifications of the soil environment (e.g., pH and nutrient status) that in turn drive interactions between soil and microbial communities thus further affecting C/N dynamics in soil (Lehmann et al. 2011; Li et al. 2021; Major et al. 2010; Wang et al. 2019; Zhu et al. 2018). For example, changes in soil pH, soil cation exchange capacity and C/N ratio increased the relative abundances of Sphingomonadaceae and Hyphomicrobiaceae, consequently affecting C and N turnover and reducing N2O emissions (Xu et al. 2014). However, plants determine the amount and quality of rhizodeposited C available for soil microbial activity: affecting C metabolism and soil C content (Tian et al. 2016; Wang et al. 2016) and thus N uptake by plants (Lehmann et al. 2003). The mutual effects of living plant and biochar on the soil microbial community composition are determined by complex interactions and feedbacks.
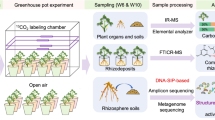
To study the effects of biochar application on the microbial community structure and the consequent changes in plant C allocation to soil and N uptake by plant, we used the model forage plant: ryegrass (Lolium perenne L.) at two growth stages (16 and 30 days after seedling emergence; DAE). 15N fertilizer and 13CO2 pulse labeling enabled photoassimilated 13C allocation into plant shoots, roots and soil micro- and macro-aggregates to be traced and 15N transport from soils into plant roots and shoots to be monitored. To identify the associated microbiomes involved in C/N processes, bidirectional orthogonal partial least squares (O2PLS) analysis was coupled with MiSeq sequencing of bacterial (16 s rRNA) and fungal (ITS) genes. We correlated microbial community composition and 13C (allocation in plant and soil aggregates) as well as 15N uptake by plants the specific microorganisms involved in these processes. We hypothesised that photoassimilated 13C incorporation into soil aggregates and 15N uptake by ryegrass would be modulated through plant growth and biochar application and accompanied by changes in the composition of soil microbial communities.
Materials and methods
Experimental setup
A sand loamy soil classified as Alfisol was collected from the Zhejiang university, China. Soil properties were as follow: organic C (1.3 ± 0.04%), total N (0.09 ± 0.01%), pH (6.8 ± 0.2), and δ13C (−18.98 ± 0.9‰). The sampled soil was sieved (< 5 mm) and plant residues and stones were removed. The sieved soil was stored at 4 °C for one week prior to use.
Biochar was produced by slow pyrolysis of rice straw (5–10 °C min−1, held at the highest temperature of 350 °C for one hour). Biochar had the following properties: organic C (59.6 ± 1.9%), total N (1.6 ± 0.1%), and δ13C (−26.17‰).
Biochar treated soil (1%) (Van Zwieten et al. 2014) was prepared by mixing biochar (3.235 g) with soil (323.5 g). Biochar passing through a 2 mm sieve was mixed evenly with the soil. Control soil was mixed to ensure parity in terms of mechanical disturbance. Totally 12 polyvinyl chloride pots (height 10 cm, diameter 11 cm) were filled with either control soil (323.5 g) or the biochar treated soil (323.5 g soil plus 3.235 g biochar). All the pots were adjusted to 80% water holding capacity (WHC) based on the control soil and incubated in the greenhouse for 2 days prior to sowing with ryegrass seeds.
Ryegrass (Lolium perenne L.) seeds were surface sterilized with 30% H2O2 for 30 min, washed thoroughly with distilled water and sown in the pots (100 seeds pot−1). Pots were incubated in an environmental control greenhouse with a 10 h dark (night; 20 °C) and 14 h light (day; 28 °C) at 70% relative humidity. During plant growth, the soils were watered every other day using sterilized deionized water. Weeds were removed manually from the pots if necessary. Seedlings emerged within 7 days of germination and at this time each pot was thinned to 20 seedlings. The soil water content of each pot was controlled gravimetrically, and maintained between 60%–70% of water holding capacity.
Pulse labeling
Plants were pulse-labeled at two growth stages (seedling stage and ripening stage) in independent pots. At seedling stage, the first 15N labeling took place at 9 days after seedling emergence and 13C labeling was performed on days 11. At ripening stage, the second 15N labeling took place at 23 days after seedling emergence and 13C labeling was performed on days 25. For 15N soil labeling, each pot received a 20 mL aliquot of (14NH4)2SO4 at a concentration of 57.5 mol L−1 (10.30 atom % 15N enrichment), injected at 10 locations, near the roots, 2 cm below the soil surface. For 13C labeling, plants were exposed to a 13CO2 enriched atmosphere for a period of 6 h using the following procedure. At each pulse-labeling event, 8 pots, with ryegrass (four with and four without biochar) were placed to a perspex chamber (height 1.0 m, width 0.8 m, and length 1.5 m), which provided a closed system, while 8 unlabeled pots (with or without biochar amendment) were exposed to the natural environment. A glass beaker containing 100 mL H2SO4 (3 M) was placed into the chamber. 25 mL of 13C labeled Na2CO3 (99% atom 13C, Cambridge Isotope Laboratories Inc., UCA) solution (1 M) was then periodically injected repeatedly (every 1.5 h) for 4 times into a glass beaker containing H2SO4 solution through a tube. The acid and carbonate reacted to liberate 13CO2. After each injection, the opening was sealed with glue. To ensure the 13CO2 was evenly distributed in the labeling chamber, an electric fan was fitted to mix the airmass. The CO2 concentration was monitored using a portable infrared sensor (PGD3-C-CO2, Shenzhen, China), when the CO2 concentration in chamber decreased below the original concentration, further 13Na2CO3 solution was injected. The ultimate atmospheric 13CO2 concentration at the end of each cycle (determined stoichiometrically) in the chamber was 400 ppm.
Harvesting
Plants were destructively harvested 5 days after each 13C labeling event constituting DAE16 (first harvest) and DAE30 (second harvest) days after seedling emergence (DAE). Plants were cut at the soil surface at each harvesting time. Visible roots were hand-picked and remaining roots were collected on a 0.5 mm sieve by washing with distilled water. The harvested plant material was washed with deionized water (0.1–1 μs cm−1) and dried at 60 °C for 12 h and then ground to a powder. After separating roots from soil, subsamples of soil were collected for isotopic analyses (13C and 15N), DNA extraction, microbial biomass C and N, and aggregate size fractionation.
Chemical analyses
Soil pH was determined in suspension (1:2.5, soil: Millipore water) using an ISFET electrode. Soil exchangeable H+ and Al3+ were extracted with 1 M KCl after adjusting the pH to neutral with 0.25 M NaOH (Pansu and Gautheyrou 2006). Total C and N in the soil after plant harvesting were measured by an elemental analyzer (Vario EL Cube, Elementary). The stable isotope was determined using an Elementar vario MICRO cube elemental analyzer coupled to the GV Isoprime 100 isotope ratio mass spectrometer (IRMS; GV Instruments, U.K.). For precision, the IRMS instrument was maintained at a controlled humidity (40%) and temperature (20°) at Zhejiang University. Isotope ratio calibration applied an international measurement standard, certified isotopic reference materials. In this experiment, IAEA-600 (Caffeine) and USGS41 (L-glutamic acid) were used as reference materials for carbon isotopic measurements (Crawford et al. 2008; Lollar et al. 2007). Microbial biomass C (Cmic) was determined by chloroform fumigation extraction (Hart 2015). Briefly, fresh soil (15 g; unfumigated) was shaken with 0.5 M K2SO4 (30 ml) for 1 h. After shaking, the suspension was filtered and organic C concentration in the filtrate was measured using a TOC analyzer (multi N/C 3100, Analytik Jena AG, Jena, Germany). The same extraction method was applied to the fumigated soil. For fumigation, fresh soil (15 g) was placed in a desiccator with chloroform (30 ml) for 24 h at a laboratory temperature. The Cmic was calculated as the difference in C between the fumigated and unfumigated soil samples (multiplied by 2.22 (KEC).
Aggregate size fractionation
Soil aggregates of three size classes were isolated (Kristiansen et al. 2006). Soil was spread on a thin layer of aluminum foil and dried to an optimal moisture content (20–25% WHC). Thereafter, 100 g of air-dried soil was transferred to a nest of sieves (2 and 0.25 mm) and shaken for 2 min. Aggregates remaining on 2 and 0.25 mm sieves were categorized as large macroaggregates (2–0.25 mm) and small macroaggregates (0.25–0.053 mm), whereas the aggregates passing through a 0.053 mm sieve were categorized as microaggregates (< 0.053 mm).
Determining potentially soluble organic matter in biochar
Dissolved organic matter (DOM) was extracted from biochar (N = 3) in 70% HPLC grade methanol and 30% nanopure water. The solutions were filtered and acidified, and dissolved organic matter (DOM) was determined on a 9.4 T custom-built FT-ICR-MS (Li et al. 2019). Samples (N = 3) were injected at 0.7 μL min−1, and 100 broadband scans were accumulated for each mass spectrum. Molecular formulae were assigned using EnviroOrg®TM software; these were categorized by compound class based on the elemental composition of molecular formulae. The groups of biochar referred are delineated by AImod (the modified aromaticity index), H/C, O/C, and elemental ratios (Dittmar and Koch 2006; Kellerman et al. 2014; Kim et al. 2003). Further groups were defined as lignin/CRAM-like, aromatic structures, unsaturated hydrocarbons carbohydrate, lipids, aliphatic/proteins and tannins (Kim et al. 2003).
DNA extraction
DNA of 12 samples (3 replicates per treatment, 2 treatments, 2 time points) was extracted from 0.50 g of moist soil using a FastDNA Spin Kit (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s protocol. The extracted DNA was dissolved in 50 μl of TE buffer, quantified using a spectrophotometer and stored at −20 °C until sequencing. Purity of the extracted DNA was determined by using Nanodrop-1000 absorbance (ratio 260/230 and 260/280) (SI Table. 5).
Sequencing of partial 16S rRNA genes and ITS genes
Bacterial and fungal genes present in the DNA extracts were identified with 16S and ITS-based probes, respectively. For bacterial identification, V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) (Vilo and Dong 2012) For fungal identification, ITS hypervariable regions of the fungal gene were amplified with primers ITS1F (5′- TCCGTAGGTGAACCTGCGG- 3′) and ITS2 (5′- GCT GCG TTC TTC ATC GAT GC- 3′) (Bazzicalupo et al. 2013). The PCR reactions were conducted using the following program: 3 min of denaturation at 95 °C; followed by 27 cycles of 30 s at 95 °C, 30 s at 55 °C, and 45 s at 72 °C; and a final extension at 72 °C for 10 min with a thermocycler PCR system (GeneAmp9700, ABI, USA). The PCR products were purified, combined, and sent to Majorbio, Inc. (Shanghai, China) for sequencing. Purified amplicons were collected in equimolar and paired-end sequenced (2*300) on an Illumina Miseq platform (Illumina, San Diego, USA).
Calculations of 13C and 15N in ryegrass shoots, roots and soil
The δ13C, δ15N, 13C atom%, and 15N atom% values in the plant and soil samples were used in 13C assimilation and 15N uptake calculation (Lu et al. 2002):
For calculations of 13C and 15N in labeled soil:
For calculations of 13C and 15N in labeled plant:
where, L and UL indicate labeled and unlabeled samples, respectively; 13Cx is the total 13C content in the plant and soil samples (mg 13C kg−1 dry soil), and 15Nx is the 15N content in different pools. Biochar itself also contains amount of carbon and nitrogen. To avoid the presence of error caused by biochar itself in the calculation of for 13C and 15N in soil, we used total C and N content of control pot as ‘Cx-control’ (mg C kg−1 dry soil) and ‘Nx-control’ (mg N kg−1 dry soil) for all treatment (soil with and without biochar amendment. For calculations of 13C and 15N in plant, total C and N content of treatment pots were designated as ‘Cx-treatment’ and ‘Nx-treatment’, respectively. 13Catom and 15Natom are the atom% of C and N samples, respectively. In order to make the process of calculation clearer, we provided some original data S1 Table 1. To unify the recovery of 13C in plants and soils with papers of the same type, we choose C/kg soil as units by calculating the 13C incorporation of different pools in each pot and multiplying the values by 3.07 (i.e., 1000/323.5).
Statistical analysis
Taxonomy The data of ITS and 16 s gene sequencing were processed using the Quantitative Insights Into Microbial Ecology (QIIME) 1.9.0-dev pipeline (Caporaso et al. 2010b). In brief, reads with less than 200 base pairs and ambiguous bases were discarded. The sequences were then combined into operational taxonomic units (OTUs) by UCLUST (Edgar 2010) based on 97% pairwise identity. Chimeric OTUs identified by USEARCH (Edgar et al. 2011) in QIIME were removed and the most abundant sequence in each OTU selected to represent that OTU. Identity was assigned to each OTUs against a subset of Silva 16S rRNA database (Cole et al. 2013) and the Unite database (Nilsson et al. 2015) by employing RDP Classifier. The representative OTU sequences were aligned using PyNAST (Caporaso et al. 2010a). Between 4682 and 51,150 valid sequences were obtained per sample for all experimental samples.
To avoid potential bias caused by sequencing depth, all sample datasets were rarefied to 60,271 and 53,702 sequences for the bacterial and fungal diversity analyses. Faith’s phylogenetic diversity (PD) was calculated to provide an integrated index of the phylogenetic breadth across taxonomic levels. A stochastic abundance model was also used to estimate the number of species (Anne and John 2015) .
Independent t-tests were performed to assess the effect of biochar amendment on various soil and plant parameters (plant biomass, 13C/15N allocations in each pool and percentage of aggregates) separately for each sampling time (DAE16 and DAE30) (Fig. 1a–d; SI Fig. 2; SI Fig. 4; SI Fig. 5). Abiotic and biotic variables correlating with plant 15N uptake and soil 13C allocation in LM and SM were calculated using the R package ‘random forest’. Distance-based linear model multivariate analysis (DISTLM) was applied to determine the relative effects of factors influencing the soil microbial communities (McArdle and Anderson 2001). The relative abundance of each protistan group in a given sample was calculated as a percentage value by dividing the raw number of sequences associated with the specific taxa by the total number of sequences in the sample. Circos, a visualization tool which was drawn by using “http://circos.ca/”, was used to generate circular link diagrams to illustrate the differences of bacterial and fungal community composition between treatments (Krzywinski et al. 2009). Linear discriminant analysis (LDA) effect size (LEfSe) was used to reveal the differentially abundant taxon in different treatments. The cladogram was drawn by using the Huttenhower Galaxy web application via the LEfSe (Segata et al. 2011). A permutational analysis of variance (PERMANOVA) was performed on Bray-Curtis distance of bacterial and fungal communities to explain the differences between samples by different grouping factors in R package ‘vegan’ (Gioria and Osborne 2009). The non-amended (Control DAE16 and Control DAE30) and biochar-amended (Biochar DAE16 and Biochar DAE30) samples were separated to examine the effects of biochar on soil microbial communities by soil microbial networks and LEfSe analysis.
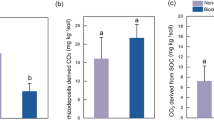
Biomass (g per pot), 13C incorporation (mg C per kg soil), 15N incorporation (mg N per kg soil) in soil, root and shoot and 13C incorporation (mg C per kg soil) within soil large- (0.25–2 mm; LM) and small- (0.053–0.25 mm; SM) macroaggregates, in biochar amended and biochar free treatments, at DAE16 and DAE30. Dissimilar letters indicate significant difference between biochar amended and biochar free treatments (independent t-test) at each harvesting time (DAE16 and DAE30)
Co-occurrence network (CoNet) While constructing networks for microbial assemblages, OTUs with relative abundances greater than 0.01% were kept. Networks were created based on a network meta-matrix combing both the standardized bacterial and fungal OTU tables. We then set the dissimilarity threshold to the maximum value of the KLD matrix and the Spearman’s correlation threshold to 0.8. For each edge and measure, permutation and bootstrap distributions were generated with 100 iterations. Measure-specific P values were computed as the area of the mean of the permutation distribution under a Gauss curve generated from the mean and standard deviation of the bootstrap distribution. The P values were adjusted using the Benjamini-Hochberg procedure (Benjamini and Hochberg 1995). Finally, only edges supported by two measures and with adjusted P-values below 0.05 were retained. The nodes in the constructed networks represent OTUs and edges represent strong and significant correlations between the OTUs. Network visualization was conducted using Gephi (Bastian 2009) and Cytoscape 3.5.1 (Shannon et al. 2003).
Bidirectional orthogonal partial least squares The O2PLS analysis (Daniel et al. 2010) is a new supervised classification analysis tool. By creating Y variables, the contribution of each species (X variables) to the class separation were quantitatively analyzed. This was performed using the SIMCAP 14 (Version 14.1.0.2047). The y-matrix was defined as the 13C allocation and 15N assimilation datasets and the x-matrix was defined as a meta-matrix combing both the standardized bacterial and fungal OTU tables at genus level. Here, variable importance in projection (VIP) coefficients represented the importance of each X variable in fitting both the X and Y variates because the Y-variates were predicted from the X-variates. The top significant features of genera were identified based on the VIP projection.
Results
Biochar characteristics
The relative abundances of chemical groups (weighed by signal magnitude normalized to the sum of all signals in each spectrum) (SI Fig. 1) indicated biochar contained a large proportion of lignin/CRAM-like, aromatic structures and unsaturated hydrocarbons. In contrast, the proportions of carbohydrate, lipids, aliphatic/proteins and tannins in biochar were minor.
Plant biomass
Root and shoot biomass increased more than two-fold between DAE16 and DAE30 in both the control and biochar treated soil. Biochar amendment had no effect on shoot biomass but significantly (p < 0.05) increased root biomass at both sampling times. Root biomass was increased through biochar amendment by a factor of 1.6 and 1.3 at DAE 16 and DAE 30, respectively (Fig. 1a).
Ryegrass uptake of 15N and rhizosphere allocation of photoassimilated 13C.
The 15N incorporation was higher in shoots than that in soil and roots at both sampling times (Fig. 1b). The 15N allocation in soil was independent of biochar amendment but increased significantly (p < 0.05) in roots with biochar amendment at DAE16 only. The 15N incorporation in shoots at DAE30 had increased by 70% compared to DAE16 (Fig. 1b). Random Forest analysis revealed that pH, DON and MBC had the greatest influence on 15N uptake by root (SI Fig. 6). The 13C allocation from plants to the soil was decreased by 76% in the presence of biochar compared to the control soil at DAE16 (Fig. 1c). With time, the suppressive effect of biochar on plant 13C allocation to soil lessened; being only 34% lower than the control at DAE30.
Incorporation of photoassimilated 13C into soil aggregates
Biochar amendment had no significant effect on proportion of large (LM) and small macroaggregates (SM) in soil except increasing the LM fraction at DAE30 (Fig.1d). Most of the total photosynthate-13C was allocated to SM and LM fractions while only a small part of photosynthate-13C was found in microaggregates (<0.053 mm) (Fig. 1d and SI Fig. 5). At DAE16 and DAE30, the mean 13C accumulation in small macroaggregates in biochar amended soil were 0.06 ± 0.019 mg C per kg soil (DAE16) and 0.08 ± 0.025 mg C per kg soil (DAE30); these being, respectively, 72% and 57% lower than in control treatment (Fig. 1d). 13C incorporation into large macroaggregates in biochar amended soil (1.54 ± 0.87 mg C per kg soil) was 57% lower at DAE 16 than in control soil (3.63 ± 1.08 mg C per kg soil) (Fig. 1d). Random Forest analysis revealed that pH, MBC and Root biomass had the greatest influence on photoassimilated 13C allocation into soil aggregates (SI Fig. 6).
Effects of biochar and plant growth stage on microbial community composition
At both growth stages, the observed richness (Sobs) and the Simpson diversity index of bacterial communities in biochar amended soil were lower than in control soil, whereas the changes observed in fungal diversity did not follow a discernable pattern across the experimental period (SI Fig. 7). The first two principal coordinates of principal co-ordinates analysis (PCoA) represented 37.9% (PC1) and 29.7% (PC2) of the variation in the bacterial communities, which showed distinct groupings according to biochar amendment and plant growth. There was no significant change in the fungal community between treatments (Fig. 2a and b). PERMANOVA indicated that bacterial composition was significantly affected by biochar addition (p < 0.01) whereas there was no effect on fungal composition between treatments (SI Table 3). The best multivariate model (DISTLM; Table 1) indicated that the bacterial composition was most influenced by soil and plant variables: root biomass (20%, p < 0.01), pH (10%, p < 0.01) and DOC (10%, p < 0.05).17% of the variation in the fungal community composition was accounted for by DON (p < 0.001) (Table 1).
Principal co-ordinates analysis (PCoA) of the bacterial and fungal community structures (a, b) and circus (c, d) of bacterial communities and fungal communities in biochar amended and biochar free treatments, at DAE16 and DAE30. The left semicircle indicates the microbial composition at phylum level of each treatment while the right semicircle represents the distribution of each phylum in the different treatments. The relative abundance (percentage of total) of sequences affiliated to each taxonomic group within a treatment is indicated by the thickness of the colored area at the perimeter of the circus
In all soil samples, the predominant bacteria phyla were Proteobacteria (24–31%), Actinobacteria (15–20%), Acidobacteria (16–23%), Chloroflexi (6–9%) and Firmicutes (6–15%) (Fig. 2c and SI Fig. 8). The fungal communities were mainly dominated by the phylum Ascomycota (95–96%) (Fig. 2d and SI Fig. 8). The independent-samples t-test showed that the relative abundance of Proteobacteria, Actinobacteria and Firmicutes decreased significantly, whereas Acidobacteria and Cyanobacteria increased significantly from DAE16 to DAE30. The relative abundance of and Cyanobacteria in the biochar amended soil increased compared to the control soil, at both DAE16 and DAE30 (SI Fig. 8).
Microbial communities in biochar amended and non-amended soil
Multiple network topological metrics consistently showed that the microbial co-occurrence pattern in the biochar amended soil differed from the un-amended soil (Fig. 3). Biochar increased the strength of negative connections, which created more intricate network patterns (higher clustering coefficient) in biochar amended soil (SI Table 7). Genera with the highest betweenness centrality scores were considered keystone species (González et al. 2010; Vickmajors et al. 2014). Mycobacterium, unclassified Hypocreales; unclassified Micrococcaceae; Bacillus and Arenimonas were identified as keystone species in control soil network. Whereas Cupriavidus, unclassified Gemmatimonadetes, Ciboria, Trichosporon; Fusarium and unclassified Acidimicrobiales were identified as keystone species in the biochar amended soils (Fig. 3).
Co-occurrence networks of microbial communities in biochar amended and biochar free treatments. A connection indicates a strong (Spearman’s ρ > 0.8) and significant (p value <0.01) correlation. The size of each node is proportional to the number of connections (that is, degree). The size of each edge is proportional to the weight of connections. Green colored circles are the fungi, orange colored circles are bacteria and purple colored circles are environment factors. (Step: Streptomyces; Myc: Mycobacterium; Hyp: unclassified Hypocreales; Mic: unclassified Micrococcaceae; Bac: Bacillus; Are: Arenimonas; Cup: Cupriavidus; Gem: unclassified Gemmatimonadetes; Cib: Ciboria; Tri: Trichosporon; Fus: Fusarium; Aci: unclassified Acidimicrobiales)
The primary taxa underpinning the differences in bacterial community composition between biochar amended and non-amended soil were assessed using linear discriminant analysis effect size (LEfSe) analysis. The cladogram revealed that 55 characteristics had significantly different abundances in soil (with and without biochar) at an LDA threshold of 2.0, including 5 classes, 9 orders, 18 families, and 28 genera (Fig. 4). At the genus level, biochar amended soil was enriched with the genera Cupriavidus, Sphingomonas; and, the LDA score of Cupriavidus was higher than that of the other genera (SI Fig. 9). In the fungal communities, the relative abundance of Trichosporon increased at DAE16 after biochar amendment.
Correlations between C allocation and N uptake by plants and rhizosphere microbial communities
Bidirectional orthogonal partial least squares (O2PLS) analysis was used to assess associations between microbial communities, C allocation and N uptake by plants. To identify the core functional microorganisms associated with C allocation and N uptake by plants, two conditions were considered: 1) VIP value ≥1; 2) correlation coefficient ≥ 0.7, p<0.05. Based on these criteria, members of 17 genera including Cupriavidus, Microvirga, unclassified Xanthomonadales, unclassified Latescibacteria, Bacillus, norank family Rhodobiaceae, Paenibacillus, unclassified SC-I-84, Variibacter, Gliocladiopsis, unclassified Anaerolineaceae, Gaiella, Kitasatospora, Pedomicrobium, Trichosporon, RB41 and 11–24 were identified as core functional bacteria. Most genera were related to 15N uptake within plants, especially in the ryegrass shoots (Table 2). Members of Xanthomonadales, Latescibacteria, unclassified Anaerolineaceae, Trichosporon, 11–24 and RB41 were positively correlated with 15N incorporation into shoots.
Compared to N uptake, fewer bacterial genera were correlated with photosynthetic 13C allocation to soil. Members of Bacillus were positively correlated to photosynthetic 13C allocation to soil, especially in the large macroaggregate while members of Cupriavidus were negatively correlated with 13C, especially in the small macroaggregates (Table 2).
Discussion
Aboveground ryegrass 15N uptake and belowground rhizodeposit-13C allocation
Consistent with our hypothesis, there were positive effects of biochar on 15N uptake by plants, which could be mainly attributed to biochar changes in soil abiotic and biotic properties. Specifically, our Random Forest analysis revealed that pH, DON and MBC had the great influence on 15N uptake by root (SI Fig. 6). Biochar benefits soil condition through neutralizing soil acidity in the rhizosphere (Rajkovich et al. 2012), thus increasing microbial biomass and affecting other N processes such as ammonification. Root biomass was strongly correlated with plant 15N uptake (SI Fig. 6). The observed enhanced root biomass might be due to biochar induced plant growth-promotion by enhanced nutrient availability (Atkinson et al. 2010) or regulatory compounds such as auxin (Lehmann et al. 2003) which in turn subsequently increase uptake of water and nutrients (Liu et al. 2013).
Biochar amendment reduced the recovery of root-derived 13C in the soil even though root biomass was increased (Fig. 1a and Fig. 1c). Increased root biomass typically releases more rhizodeposit-C (Pausch and Kuzyakov 2017), leading to increased C storage in soil (Johnson et al. 2006). The contradiction of high root biomass and less recovery of photoassimilated C (13C) in aggregates (Fig. 1a and d) might be, i) a consequence of the short experimental period, i.e. the 30 days of duration during plant growth may have been insufficient to allow the resulting microbial metabolites to be transferred into aggregates (Gunina and Kuzyakov 2014; Riederer et al. 2013), or ii) attributed to more rapid mineralization of root-derived C by the microbial biomass. Random Forest revealed that MBC, root biomass and pH were significantly affect the allocation of rhizo-C in soil, especially in large-macroaggregates (SI Fig. 6). Biochar accelerates microbial utilization of rhizodeposits by neutralization of soil acidity which can result in larger microbial biomass, faster C turnover and consequently, smaller 13C recovery in soil aggregates (0.25–2 mm and 0.053–0.25 mm), compared to the soil without biochar amendment (Fig. 1a, c and d). The larger microbial biomass and root C utilization by microorganisms after biochar amendment may accelerate the turnover of both new substrate C and native soil organic C (Luo et al. 2017a; Yu et al. 2018; Zhang et al. 2020). However, mineralization of rhizodeposit-C was not quantified in the present research and further investigations using isotope analysis to elucidate the efflux of 13CO2 (originating from rhizodeposits) and 12CO2 (originating from soil organic C) are required. Biochar addition decreased 13C content in soil aggregates (0.25–2 mm and 0.053–0.25 mm) but had very limited effects on soil aggregation. Current literature presents contrasting results concerning biochar’s effects on soil aggregation. The discrepancy depend on biochar surface properties and interactions with soil minerals. For instance, negative effects on aggregate formation were attributed to biochar’s low binding efficiency to soil particles relative to other soil constituents (Zhou et al. 2018). Though declined 13C allocation into aggregates was not because of any collapse of aggregation, further research to better understand biochar-aggregate-rhizodeposit interactions, is required.
Rhizosphere microbiome in biochar amended soil
Biochar amendment and plant growth stage both influenced the rhizosphere microbial community, especially biochar amendment (Fig. 5); this result supported our hypothesis. Plants interact with soils to shape rhizosphere microbiomes, with plant genotype, root architecture, rhizodeposition, soil structure and nutritional status influencing the outcome (Chaparro et al. 2014a; Qiao et al. 2017). DISTLM showed that soil pH, C and nutrients shaped the microbial community structure (Table 1). Plant roots and biochar had opposite effects on soil pH: biochar addition increase soil pH due to base cation content (Luo et al. 2011), while ryegrass reduced pH (by release of acidic rhizodeposits (Zhalnina et al. 2018) and unbalance NH4+ uptake (Badri and Vivanco 2009; Zamanian et al. 2018). Bacteria are typically abundant in neutral and alkaline soils (Feng et al. 2010; Sheng and Zhu 2018) and are more sensitive to low pH (Singh et al. 2010b). Bacterial composition was more negatively impacted than fungal composition by rhizosphere acidification during plant growth (Fig. 2a); this being buffered through biochar amendment. In addition to this environmental stress, resource availability is another key driving force in shaping the microbiome community (Malik et al. 2018). The interactions between plant and biochar may influence soil C/nutrient status and thus modulate the rhizosphere microbiome.
Schematic diagram summarizing biochar affected on root uptake of N (short-term) and allocation of ryegrass photoassimilated-C to soil aggregates. Observations revealed the inhibitory effect of biochar on downward photosynthetic 13C allocation within soil aggregates and upward increased nutrient (15N) assimilation by roots and shoots. In addition, co-occurrence network and O2PLS analysis indicated some bacterial genera were correlated to plant 13C allocation in soil and 15N uptake by ryegrass. For instance, members of genus Bacillus were positively correlated with 13C allocation in large macroaggregates, while genus Xanthomonadales (phyla Proteobacteria) and RB41 (phyla Acidobacteria) were positively correlated with Lolium 15N uptake
The content of C and nutrients in soil and their composition depend on i) quantity and quality of rhizodeposits, and ii) substances contained and adsorbed within biochar. A shift in microbial community composition in the ryegrass rhizosphere at both DAE16 and DAE30 was found: the relative abundance of Proteobacteria, Actinobacteria and Firmicutes decreased, whereas Acidobacteria and Cyanobacteria increased from DAE16 to DAE30 (Fig. 2c and SI Fig. 8). This observation highlights how plant growth stage is influential for soil microbiomes (Kuzyakov 2002) and specifically in relation to rhizodeposits. During the early growth stage (week 3), roots release easily metabolized substrates for a variety of microorganisms. For example, sucrose concentrations were higher in early growth of Avena barbata than at other stages of development and were allocated primarily to the area behind the root tips and decreasing as the root matured (Zhalnina et al. 2018). Also, for Arabidopsis, the release of sugar and alcohol decreased with growth stage (Chaparro et al. 2014b). Proteobacteria, Actinobacteria and Firmicutes, as r-strategists, prefer to utilize easily decomposable resources such as sucrose, this resulting in a relative reduction of these r-strategists as the plant matures (Ho et al. 2017). Moreover, plant demand for N increases with the development of advancing growth stages (Gastal and Lemaire 2002), which leads to the decreases of soil DON concentration, consequently decreasing these r-strategists.
Biochar indirectly affects rhizodeposited-C/nutrient availability for microorganisms via: i) biochar surface adsorption (Keith et al. 2015), and ii) interactions in the charosphere (Luo et al. 2013). Here, biochar might offer a hotspot for enhanced microbial mineralization of rhizodeposits. Most likely, the increased root-growth from biochar increased C availability to microorganisms (Johnson et al. 2006) and subsequently supported a larger microbial biomass, higher activity and more complex microbial interactions in biochar amended soil (Luo et al. 2013). This is indicated by the co-occurrence networks that highlighted more negative correlations in the biochar amended soil when compared with the control soil (Fig. 3). Also, as indicated by FT ICR-MS, biochar was characterized by both recalcitrant C (i.e., aromatics) and easily utilizable C (i.e., carbohydrates, aliphatics, proteins) (SI Fig. 1). Differences between bacterial communities in biochar amended soil and non-biochar amended soil might be the result of adaptation of microbiota to these substances. For instance, biochar increased the relative abundance of specific taxa, such as: Cupriavidus and Trichosporon, identified as keystone species in microbial network (Fig. 3). This observation may be linked to the possibility that these species could be more able to utilize or tolerate aromatics introduced into the soil with the biochar. It is reported that Cupriavidus pinatubonensis (ex c. necator) JMP134 (pJP4) was capable of degrading a variety of aromatic compounds (Ledger et al. 2011).
Core microbial groups regulate bi-directional transport of N and C
This study reveals the inhibitory effect of a largely stable C-based material (biochar) on downward photosynthetic 13C allocation to soil aggregates and upward increased nutrient (15N) assimilation by roots and shoots. To improve the understanding of C/N dynamics in the ryegrass soil amended with biochar, the correlations between rhizosphere microbiota and 13C allocation/15N uptake in the plant-soil pools were analyzed using a bidirectional orthogonal partial least square (O2PLS) (Table 2). This analysis provided insights into the biome-specific linkages to soil C-storage and plant nutrient uptake. To date, there has been only a limited number of studies linking microbial community composition and photosynthetic C allocation/nutrient-uptake by plants.
Seventeen microbial genera were correlated with 15N uptake by ryegrass. Members of Xanthomonadales (Proteobacteria), Latescibacteria, Anaerolineaceae (Chloroflexi), 11–24 and RB41 (Acidobacteria) and Gliocladiopsis (Ascomycota) and Trichosporon (Basidiomycota) which were found to be positively correlated to ryegrass 15N uptake, thus may have been more likely to enhance the nutrient status of plants and (or) synthesize signaling molecules (such as auxin, IAA, etc.) that affect plant growth and development. Xanthomonadales was identified as a key component of the core microbiome with members contributing to nitrogen fixation in the rhizosphere (Zhou et al. 2020). Stenotrophomonas rhizophila, of the order Xanthomonadales, are often found associated with plants and have capacity to promote plant growth and suppress colonization by plant pathogens (Berg and Martinez 2015). 11–24 have significant effects on plant morphology of G. longituba (Liu et al. 2020). RB41 is a member of the family Blastocatellaceae, the sibling genera from Blastocatellaceae were reported to have functions in soil metabolism related to N (Huber et al. 2017). Moreover, RB41 produces IAA, solubilizes nutrients and improves growth of plants by imparting resistance to pathogens (Suja et al. 2014). Members of Gliocladiopsis isolated from necrotic avocado roots in Australia increased the height of avocado seedlings (Dann et al. 2012) and have ability to produce bioactive metabolites which enhance plant’s medicinal properties (Refaei et al. 2011). Trichosporon asahii, members of Trichosporon, in the rhizosphere of sugar beet are reported for their ability to colonize roots and protect sugar beet from diseases (El-Tarabily 2004). These publications support that the microorganisms observed in this study (related to 15N uptake) are plant growth promoting, and this could be regulating increased 15N uptake.
Members of genus Bacillus and Cupriavidus were related to plant 13C allocation in soil aggregates (Table 2). Bacilli positively correlated with 13C allocation, especially to larger (more stable) soil macroaggregates. Bacilli are a group of well-characterized soil organisms known to produce a range of extracellular polymeric substances (EPS), forming cohesive biofilms that co-exist in intimate association with plant roots (Hera et al. 2013). EPS production utilizes labile C inputs that would otherwise be respired (Flemming et al. 2016; Malik et al. 2019) accordingly, EPS production in soil was found to be dependent on plentiful access to labile C inputs (Redmile-Gordon et al. 2015). EPS is an important mechanism for the stabilization of soil structure that can persist for several years (Redmile-Gordon et al. 2020). In contrast to Bacilli, members of Cupriavidus were negatively correlated with 13C especially in soil small-macroaggregates (Table 2) and were consistently identified as keystone species in the biochar amended soils (Fig. 3). They also had higher LDA scores than those of any other genera analyzed by LEfSe (SI Fig. 9). Members of Cupriavidus has been reported to utilize root exudates and organic compounds released by living plants’ roots in their surrounding environment (Koopman et al. 2010; Zhalnina et al. 2018). It is reported that phenolic resins were highly abundant in plant root exudates and tissues (Bertin et al. 2003) and these have some structural similarity to aromatic exogenous substances provided in the augmented biochar. As mentioned above, Cupriavidus pinatubonensis (ex c. necator) JMP134 (pJP4) was capable of breaking down a variety of aromatic compounds (Ledger et al. 2011). The members of Cupriavidus, that are capable of metabolizing a broad range of both rhizodeposit-C and biochar-C, were negatively correlated with 13C recovered in macroaggregates and might consequently limit sequestration in biochar amended soils.
Perspective
Considering that the effects of biochar and the rhizosphere microbiome on plant C allocation and N uptake are rarely investigated, this study has shed new light on the key role of these interactions in soil. Optimizing the core microbial communities will be key to managing agricultural ecosystems efficiently (Toju et al. 2018). This study reveals the inhibitory effect of a stable C-based product (biochar) on downward photosynthetic 13C allocation within to soil aggregates and upward increased nutrient (15N) assimilation by roots. In addition, by using a co-occurrence network and O2PLS analysis, seventeen bacterial genera were correlated to plant 13C allocation in soil and 15N uptake by ryegrass (Fig. 5). However, we have only established a very general framework to link plant-associated microbiomes and rhizosphere processes. To confirm these keystone microbiomes and their functions involved in C and N processes, more sophisticated techniques and approaches are required. For example, RNA/DNA-SIP could be used to confirm whether the rhizodeposited C is actively being utilized by specific microorganisms (Chen et al. 2019a). Also, a full assessment of 13C distribution into plant-soil-microbe-atmosphere pools to trace rhizodeposited-13C into not only microbial community (via assimilation) but also 13CO2 in atmosphere (via mineralization) would be beneficial. These approaches would help identify the keystone microbiota in soil based on the comprehensive understanding of their physiological characteristics and functions. Better understanding of soil-plant-microbe interactions, across contrasting pedospheres and rhizospheres, will open new windows of opportunity to manipulate plant C inputs for soil C storage and increased plant nutrient use efficiency.
Conclusions
This study revealed that biochar increased root biomass and N uptake by ryegrass (Lolium perenne L.) and decreased photosynthetic C incorporation into soil macroaggregates. By adopting O2PLS, seventeen bacterial genera were correlated with 13C allocation in soil aggregates and 15N uptake by Lolium (Fig. 5). For instance, members of genus Bacillus were positively correlated with 13C allocation in large macroaggregates. Genus Xanthomonadales (phyla Proteobacteria) and RB41 (phyla Acidobacteria) were positively correlated with Lolium 15N uptake. This indicated the significant role of these keystone microbiota in mediating soil C sequestration from Lolium rhizodeposits and root N assimilation. Future research adopting DNA/RNA-SIP is recommended to fully assess how these rhizosphere organisms assimilate and transfer rhizodeposits and/or nutrients directly, thus providing opportunity for improving soil-plant functions and interactions.
Abbreviations
- DAE16:
-
16 days after seedling emergence
- DAE30:
-
30 days after seedling emergence
- LM:
-
large macro-aggregate
- SM:
-
small macro-aggregate
- TC:
-
total carbon
- TN:
-
total nitrogen
- DOC:
-
dissolved organic carbon
- MBC:
-
microbial biomass carbon
- SIP:
-
Stable Isotope Probing
References
Anne C, John B (2015) Estimating the number of species in a stochastic abundance model. Biometrics 58:531–539
Atkinson CJ, Fitzgerald JD, Hipps NA (2010) Potential mechanisms for achieving agricultural benefits from biochar application to temperate soils: a review. Plant and Soil 337:1–18. https://doi.org/10.1007/s11104-010-0464-5
Badri D, Vivanco J (2009) Regulation and function of root exudates. Plant Cell Environ 32:666–681. https://doi.org/10.1111/j.1365-3040.2008.01926.x
Bastian MHSA (2009) Gephi: an open source software for exploring and manipulating networks. ICWSM:361–362
Bazzicalupo AL, Bálint M, Schmitt I (2013) Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol 6:102–109
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Berg G, Martinez J (2015) Friends or foes: can we make a distinction between beneficial and harmful strains of the Stenotrophomonas maltophilia complex? Front Microbiol 6:241. https://doi.org/10.3389/fmicb.2015.00241
Bertin C, Yang X, Weston LA (2003) The role of root exudates and allelochemicals in the rhizosphere. Plant and Soil 256:67–83. https://doi.org/10.1023/A:1026290508166
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. https://doi.org/10.1093/bioinformatics/btp636
Caporaso JG et al. (2010b) QIIME allows analysis of high-throughput community sequencing data Nature Methods 7:335-336 doi:https://doi.org/10.1038/nmeth.f.303
Cavaglieri L, Orlando J, Etcheverry M (2007) Rhizosphere microbial community structure at different maize plant growth stages and root locations. Microbiol Res 164:391–399. https://doi.org/10.1016/j.micres.2007.03.006
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. Isme J 8:790–803. https://doi.org/10.1038/ismej.2013.196
Chen L et al. (2019a) Competitive interaction with keystone taxa induced negative priming under biochar amendments 7
Chen S, Waghmode TR, Sun R, Kuramae EE, Hu C, Liu B (2019b) Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 7:136. https://doi.org/10.1186/s40168-019-0750-2
Cole JR et al (2013) Ribosomal DATABASE Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:42–D642. https://doi.org/10.1093/nar/gkt1244
Crawford K, McDonald R, Bearhop S (2008) Application of stable isotope techniques to the ecology of mammals. Mamm Rev 38:87–107. https://doi.org/10.1111/j.1365-2907.2008.00120.x
Daniel E, Miyako K, Thomas M, Johan T (2010) Data integration in plant biology: the O2PLS method for combined modeling of transcript and metabolite data. Plant J 52:1181–1191
Dann E, Cooke A, Forsberg L, Pegg K, Tan YP, Shivas R (2012) Pathogenicity studies in avocado with three nectriaceous fungi, Calonectria ilicicola, Gliocladiopsis sp. and Ilyonectria liriodendri. Plant Pathol 61:896–902. https://doi.org/10.1111/j.1365-3059.2011.02579.x
Dittmar T, Koch BP (2006) Thermogenic organic matter dissolved in the abyssal ocean. Mar Chem 102:208–217
Drigo B, Kowalchuk G, Knapp B, Pijl A, Boschker H, Veen J (2013) Impacts of 3 years of elevated atmospheric CO2 on rhizosphere carbon flow and microbial community dynamic. Glob Chang Biol 19:621–636. https://doi.org/10.1111/gcb.12045
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461%Jbioinformatics
Edgar RC, Haas BJ, Clemente JC, Christopher Q, Rob K (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194
El-Tarabily K (2004) Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J Appl Microbiol 96:69–75. https://doi.org/10.1046/j.1365-2672.2003.02043.x
Feng X, Simpson A, Schlesinger W, Simpson M (2010) Altered microbial community structure and organic matter composition under elevated CO2 and N fertilization in the Duke Forest. Glob Chang Biol 16:2104–2116. https://doi.org/10.1111/j.1365-2486.2009.02080.x
Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice S, Kjelleberg S (2016) Biofilms: An emergent form of bacterial life. Nature Rev Microbiol 14:563–575. https://doi.org/10.1038/nrmicro.2016.94
Gastal F, Lemaire G (2002) N uptake and distribution in crops: An agronomical and ecophysiological perspective. J Exp Bot 53:789–799. https://doi.org/10.1093/jexbot/53.370.789
Gioria M, Osborne B (2009) Assessing the impact of plant invasions on soil seed bank communities: use of univariate and multivariate statistical approaches. J Veg Sci 20:547–556. https://doi.org/10.1111/j.1654-1103.2009.01054.x
González M, Murature DA, Fidelio GD (2010) Thermal stability of human immunoglobulins with sorbitol: a critical Evaluation. Vox Sanguinis 68:1–4
Graaff M-A, Six J, Kessel C (2007) Elevated CO2 increases nitrogen rhizodeposition and microbial immobilization of root-derived nitrogen. New Phytol 173:778–786. https://doi.org/10.1111/j.1469-8137.2006.01974.x
Gul S, Whalen JK, Thomas BW, Sachdeva V, Deng H (2015) Physico-chemical properties and microbial responses in biochar-amended soils: Mechanisms and future directions. Agric Ecosyst Environ 206:46–59
Gunina A, Kuzyakov Y (2014) Pathways of litter C by formation of aggregates and SOM density fractions: Implications from 13C natural abundance. Soil Biol Biochem 71:95–104
Hart M (2015) Laboratory manual of the soil microbial biomass group. Research, Rothamsted
Hera V, Yunrong C, Pascale B, Richard L, Roberto K (2013) Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168
Hester E et al (2018) Linking nitrogen load to the structure and function of wetland soil and rhizosphere Microbial Communities. mSystems 3:3. https://doi.org/10.1128/mSystems.00214-17
Ho A, Di Lonardo DP, Bodelier PLE (2017) Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiology Ecology:93. https://doi.org/10.1093/femsec/fix006
Houlden A, Timms-Wilson TM, Day MJ, Bailey MJ (2008) Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops & Fems. Microbiol Ecol 65:193–201
Huber KJ et al (2017) Aridibacter nitratireducens sp. nov., a member of the family Blastocatellaceae, class Blastocatellia, isolated from an African soil. Int J Syst Evol Microbiol 67:4487–4493
Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S (2017) The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Front Plant Sci 8:1617. https://doi.org/10.3389/fpls.2017.01617
Johnson JMF, Allmaras RR, Reicosky DC (2006) Estimating source carbon from crop residues, roots and rhizodeposits using the national grain-yield database. Agron J 98:622–636
Karhu K, Mattila T, Bergström I, Regina K (2011) Biochar addition to agricultural soil increased CH4 uptake and water holding capacity – results from a short-term pilot field study. Agricul Ecosyst Environ 140:309–313
Keith A, Singh B, Dijkstra FA (2015) Biochar reduces the rhizosphere priming effect on soil organic carbon. Soil Biol Biochem 88:372–379
Kellerman A, Dittmar T, Kothawala D, Tranvik L (2014) Supplementary information for Chemodiversity of dissolved organic matter in lakes driven by climate and hydrology
Kim S, Kramer RW, Hatcher PG (2003) Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the Van Krevelen diagram. Anal Chem 75:5336–5344
Koopman F, Wierckx N, de Winde H, Ruijssenaars H (2010) Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proceedings of the National Academy of Sciences of the United States of America 107:4919–4924. https://doi.org/10.1073/pnas.0913039107
Kristiansen SM, Schjønning P, Thomsen IK, Olesen JE, Kristensen K, Christensen BT (2006) Similarity of differently sized macro-aggregates in arable soils of different texture. Geoderma 137:147–154. https://doi.org/10.1016/j.geoderma.2006.08.005
Krzywinski M et al (2009) CIRCOS: an information aesthetic for comparative genomics. Genome Research 19:1639–1645. https://doi.org/10.1101/gr.092759.109
Kuzyakov Y (2002) Model for rhizodeposition and CO2 efflux from planted soil and its validation by 14 C pulse labelling of ryegrass. Plant Soil 239:87–102
Kuzyakov Y, Horwath WR, Dorodnikov M, Blagodatskaya E (2019) Review and synthesis of the effects of elevated atmospheric CO2 on soil processes: no changes in pools, but increased fluxes and accelerated cycles. Soil Biol Biochem 128:66–78. https://doi.org/10.1016/j.soilbio.2018.10.005
Laurent P, Raaijmakers JM, Philippe L, Putten WHVD (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799
Ledger T, Zúñiga A, Kraiser T, Dasencich P, Donoso R, Pérez-Pantoja D, Gonzalez B (2011) Aromatic compounds degradation plays a role in colonization of Arabidopsis thaliana and Acacia caven by Cupriavidus pinatubonensis. JMP 134 vol 101
Lehmann J (2007) A handful of carbon. Nature 447:143–144. https://doi.org/10.1038/447143a
Lehmann J, Silva JPD, Steiner C, Nehls T, Zech W, Glaser B (2003) Nutrient availability and leaching in an archaeological Anthrosol and a Ferralsol of the Central Amazon basin: fertilizer, manure and charcoal amendments. Plant Soil 249:343–357
Lehmann J, Gaunt J, Rondon M (2006) Bio-char sequestration in terrestrial ecosystems – a review. Mitigation and Adaptation Strategies for Global Change 11:11–427. https://doi.org/10.1007/s11027-005-9006-5
Lehmann J, Rillig MC, Thies J, Masiello CA, Hockaday WC, Crowley D (2011) Biochar effects on soil biota – A review. Soil Biol Biochem 43:1812–1836
Li X-M et al (2019) Organic carbon amendments affect the chemodiversity of soil dissolved organic matter and its associations with soil microbial communities. Environ Sci Technol 53:50–59
Li Y, Shahbaz M, Zhu Z, Deng Y, Tong Y, Chen L, Wu J, Ge T (2021) Oxygen availability determines key regulators in soil organic carbon mineralisation in paddy soils. Soil Biol Biochem 153:108106. https://doi.org/10.1016/j.soilbio.2020.108106
Liu X et al (2013) Biochar’s effect on crop productivity and the dependence on experimental conditions—a meta-analysis of literature data. Plant Soil 373:583–594. https://doi.org/10.1007/s11104-013-1806-x
Liu L, Jin L, Guo Q (2020) Effects of soil microbiomes and enzymatic activities on Glechoma longituba. Hort Science: 1-7, https://doi.org/10.21273/HORTSCI14659-19
Lollar B, Hirschorn S, Chartrand M, Lacrampe-Couloume G (2007) An approach for assessing total instrumental uncertainty in compound-specific carbon isotope analysis: implications for environmental remediation studies. Anal Chem 79:3469–3475. https://doi.org/10.1021/ac062299v
Lu Y, Watanabe A, Kimura M (2002) Contribution of plant-derived carbon to soil microbial biomass dynamics in a paddy rice microcosm
Luo Y, Durenkamp M, Nobili M, Lin Q, Brookes PC (2011) Short term soil priming effects and the mineralisation of biochar following its incorporation to soils of different pH. Soil Biol Biochem 43:2304–2314. https://doi.org/10.1016/j.soilbio.2011.07.020
Luo Y, Durenkamp M, Nobili M, Lin Q, Devonshire BJ, Brookes PC (2013) Microbial biomass growth, following incorporation of biochars produced at 350 °C or 700 °C, in a silty-clay loam soil of high and low pH. Soil Biol Biochem 57:513–523. https://doi.org/10.1016/j.soilbio.2012.10.033
Luo Y, Dungait J, Zhao X, Brookes P, Durenkamp M, Li G, Lin Q (2017a) Pyrolysis temperature during biochar production alters its subsequent utilization by microorganisms in an acid arable soil. Land Degrad Dev. https://doi.org/10.1002/ldr.2846
Luo Y et al (2017b) Priming effects in biochar enriched soils using a three-source-partitioning approach: 14C labelling and 13C natural abundance. Soil Biol Biochem 106:28–35. https://doi.org/10.1016/j.soilbio.2016.12.006
Major J, Rondon M, Molina D, Riha SJ, Lehmann J (2010) Maize yield and nutrition during 4 years after biochar application to a Colombian savanna oxisol. Plant & Soil 333:117–128
Malik A, Martiny J, Brodie E, Martiny A, Treseder K, Allison S (2018) Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. https://doi.org/10.1101/445866
Malik A, Martiny J, Brodie E, Martiny A, Treseder K, Allison S (2019) Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J 14:1–9. https://doi.org/10.1038/s41396-019-0510-0
McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290–297. https://doi.org/10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
Nilsson RH, Tedersoo L, Ryberg M, Kristiansson E, Hartmann M, Unterseher M, Porter TM, Bengtsson-Palme J, Walker DM, de Sousa F, Gamper HA, Larsson E, Larsson KH, Kõljalg U, Edgar RC, Abarenkov K (2015) A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environ advpub 30:145–150. https://doi.org/10.1264/jsme2.ME14121
Pansu M, Gautheyrou J (2006) Analysis of extractable and Total elements
Pausch J, Kuzyakov Y (2017) Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob Chang Biol 24:24–12. https://doi.org/10.1111/gcb.13850
Qiao Y, Miao S, Han X, Yue S, Tang C (2017) Improving soil nutrient availability increases carbon rhizodeposition under maize and soybean in Mollisols Science of the Total Environment 603-604: 416
Rajkovich S, Enders A, Hanley K, Hyland C, Zimmerman AR, Lehmann J (2012) Corn growth and nitrogen nutrition after additions of biochars with varying properties to a temperate soil. Biol Fertil Soils 48:271–284. https://doi.org/10.1007/s00374-011-0624-7
Redmile-Gordon M, Evershed R, Hirsch P, White RP, Goulding K (2015) Soil organic matter and the extracellular microbial matrix show contrasting responses to C and N availability. Soil Biol Biochem 88:88–267. https://doi.org/10.1016/j.soilbio.2015.05.025
Redmile-Gordon M, Gregory AS, White RP, Watts CW (2020) Soil organic carbon, extracellular polymeric substances (EPS), and soil structural stability as affected by previous and current land-use. Geoderma 363:114143. https://doi.org/10.1016/j.geoderma.2019.114143
Refaei J, Jones E, Sakayaroj J, Santhanam J (2011) Endophytic fungi from Rafflesia cantleyi: species diversity and antimicrobial activity 2:429–447
Pausch J, Tian J, Riederer M, Kuzyakov Y (2013) Estimation of rhizodeposition at field scale: upscaling of a 14C labeling study. Plant Soil 364, 273–285
Rütting T, Clough T, Müller C, Lieffering M, Newton P (2009) Ten years of elevated atmospheric carbon dioxide alters soil nitrogen transformations in a sheep-grazed pasture. Glob Chang Biol 16:2530–2542. https://doi.org/10.1111/j.1365-2486.2009.02089.x
Sarker J, Singh BP, He X, Fang Y, Li G, Collins D, Cowie A (2017) Tillage and nitrogen fertilization enhanced belowground carbon allocation and plant nitrogen uptake in a semi-arid canola crop–soil system. Sci Rep 7:7. https://doi.org/10.1038/s41598-017-11190-4
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol
Shannon P et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks
Sheng Y, Zhu L (2018) Biochar alters microbial community and carbon sequestration potential across different soil pH. Sci Total Environ 622-623:1391-1399 . https://doi.org/10.1016/j.scitotenv.2017.11.337
Singh B, Bardgett R, Smith P, Reay D (2010a) Microorganisms and climate change: Terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8:779–790. https://doi.org/10.1038/nrmicro2439
Singh BK, Munro S, Reid E, Ord B, Potts JM, Paterson E, Millard P (2010b) Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur J Soil Sci 57:72–82
Suja S, Hegde V, T Makeshkumar, IP Anjanadevi (2014) Screening of rhizobacteria associated with cassava for plant growth promotion and biocontrol potential. Journal of Root Crops 40:66–73
Tian J, Wang J, Dippold M, Gao Y, Blagodatskaya E, Kuzyakov Y (2016) Biochar affects soil organic matter cycling and microbial functions but does not alter microbial community structure in a paddy soil. Sci Total Environ 556:89–97. https://doi.org/10.1016/j.scitotenv.2016.03.010
Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET (2018) Core microbiomes for sustainable agroecosystems. Nat Plants 4:4–257. https://doi.org/10.1038/s41477-018-0139-4
Van Zwieten L et al. (2014) An incubation study investigating the mechanisms that impact N2O flux from soil following biochar application 191: 53-62
Verburg P, Kapitzke S, Stevenson B, Bisiaux M (2014) Carbon allocation in Larrea tridentata plant-soil systems as affected by elevated soil moisture and N availability. Plant Soil 378:227–238. https://doi.org/10.1007/s11104-013-2017-1
Veresoglou S, Chen B, Rillig M (2012) Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol Biochem 46:53–62. https://doi.org/10.1016/j.soilbio.2011.11.018
Vickmajors TJ, Priscu JC, Amaralzettler LA (2014) Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. Isme J 8:778
Vilo C, Dong Q (2012) Evaluation of the RDP classifier accuracy using 16S rRNA gene variable regions. Metagenomics 1: a1-5
Wang J, Xiong Z, Yan X, Kuzyakov Y (2016) Carbon budget by priming in a biochar-amended soil. Eur J Soil Biol 76:26–34. https://doi.org/10.1016/j.ejsobi.2016.07.003
Wang R et al. (2019) Biochar significantly alters rhizobacterial communities and reduces Cd concentration in rice grains grown on Cd-contaminated soils. Science of The Total Environment 676
Weng Z et al (2017) Biochar built soil carbon over a decade by stabilizing rhizodeposits. Nature Climate Change 7:7–376. https://doi.org/10.1038/nclimate3276
Xu H-J, Wang X, Yao H, Su JQ, Li H (2014) Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ Sci Technol 48:48–9399. https://doi.org/10.1021/es5021058
Yu Z et al. (2018) Feedstock determines biochar-induced soil priming effects by stimulating the activity of specific microorganisms: feedstock of biochar determines priming effects. Eur J Soil Sci
Zamanian K, Zarebanadkouki M, Kuzyakov Y (2018) Nitrogen fertilization raises CO2 efflux from inorganic carbon: a global assessment. Glob Chang Biol 24:2810–2817. https://doi.org/10.1111/gcb.14148
Zhalnina K et al (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480
Zhang Q, Song Y, Wu Z, Yan X, Gunina A, Kuzyakov Y, Xiong Z (2020) Effects of six-year biochar amendment on soil aggregation, crop growth, and nitrogen and phosphorus use efficiencies in a rice-wheat rotation. J Clean Prod 242:118435. https://doi.org/10.1016/j.jclepro.2019.118435
Zhou H et al (2018) Biochar enhances soil hydraulic function but not soil aggregation in a sandy loam: biochar effects on soil hydraulic function and aggregation. Eur J Soil Sci. https://doi.org/10.1111/ejss.12732
Zhou J, Yu L, Zhang J, Zhang X, Xue Y, Liu J, Zou X (2020) Characterization of the core microbiome in tobacco leaves during aging. MicrobiologyOpen 9:e984. https://doi.org/10.1002/mbo3.984
Zhu Z et al (2018) Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol Biochem 121:67–76. https://doi.org/10.1016/j.soilbio.2018.03.003
Acknowledgments
This study was supported by the National Science Foundation of China (41520104001; 41671233), and Fundamental Research Funds for the Central Universities (2019QNA6012).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Lucas Silva
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCM 1020 kb)
Rights and permissions
About this article
Cite this article
Fu, Y., Kumar, A., Chen, L. et al. Rhizosphere microbiome modulated effects of biochar on ryegrass 15N uptake and rhizodeposited 13C allocation in soil. Plant Soil 463, 359–377 (2021). https://doi.org/10.1007/s11104-021-04845-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04845-9