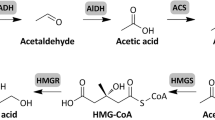
Abstract
Confronted with the gradual and inescapable exhaustion of the earth’s fossil energy resources, the bio-based process to produce platform chemicals from renewable carbohydrates is attracting growing interest. Escherichia coli has been chosen as a workhouse for the production of many valuable chemicals due to its clear genetic background, convenient to be genetically modified and good growth properties with low nutrient requirements. Rational strain development of E. coli achieved by metabolic engineering strategies has provided new processes for efficiently biotechnological production of various high-value chemical building blocks. Compared to previous reviews, this review focuses on recent advances in metabolic engineering of the industrial model bacteria E. coli that lead to efficient recombinant biocatalysts for the production of high-value organic acids like succinic acid, lactic acid, 3-hydroxypropanoic acid and glucaric acid as well as alcohols like 1,3-propanediol, xylitol, mannitol, and glycerol with the discussion of the future research in this area. Besides, this review also discusses several platform chemicals, including fumaric acid, aspartic acid, glutamic acid, sorbitol, itaconic acid, and 2,5-furan dicarboxylic acid, which have not been produced by E. coli until now.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Confronted with the gradual and inescapable exhaustion of the earth’s fossil energy resources, industrial biotechnologies for the production of platform chemicals, which can be either directly used or further processed for the production of large-volume and high value-added products in the chemical industry, have recently gained tremendous interests and attentions (Hatti-Kaul et al. 2007).
Biosynthesis of chemicals and biomaterials from recombinant microorganisms has arisen as a competitive alternative to the traditional chemistry-based routes. The development of recombinant DNA technology promotes the advancement of metabolic engineering in which we can get genetically modified strains to improve the yields of end products (Vemuri and Aristidou 2005; Bianco et al. 2006).
Among the candidate biocatalysts, Escherichia coli, a gram-negative bacterium, has been widely used in various biotechnological processes. Compared with other microbiocatalysts to produce organic acids or alcohols, E. coli has several advantages: (1) many feasible genetic tools have been developed in E. coli for over 30 years; (2) sensitive to many antibiotics which can be used as engineering tags to chase the genetically modified strains (Nelson and Cox 2000); (3) clear genomic, proteomic, and metabolic information which benefit the metabolic engineering efforts (Harrington et al. 2000); (4) a wealth of knowledge on its central carbon metabolism (Sauer and Eikmanns 2005) and physiology; (5) a specific advantage—it grows quickly in minimal media and maintains the ability to metabolize both 5 and 6 carbon sugars (Zaldivar et al. 2001); and (6) its high tolerance to organic acids. Due to these advantages, artificial metabolic pathways have been constructed in E. coli by recruiting and integrating recombinant enzymes from a variety of resources to produce compounds which do not have metabolic pathways in wild microorganisms.
In the previous reviews, only several high-value platform chemicals were summarized with different microcatalysts. These reviews focused on the development of metabolic engineering for one or several compounds production with many detailed description of products (Chotani et al. 2000; Wee et al. 2006; Wendisch et al. 2006; Park and Lee 2008; Shanmugam and Ingram 2008; Jiang et al. 2009; Okabe et al. 2009; Saxena et al. 2009; Song and Vieille 2009). However, scientists engineering E. coli strains were not particularly concerned with finding the relationship in all high-value platform chemical metabolic pathways including native and constructed.
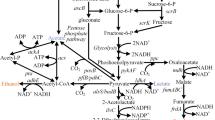
In this review, we summarize the application of E. coli strains for efficient production of a group of high-value organic acids or alcohols such as succinic acid, 3-hydroxypropanoic acid, glucaric acid, xylitol, and glycerol, which were identified as “top value-added chemicals from biomass” in a report of the Pacific Northwest National Laboratory and the National Renewable Energy Laboratory (Werpy and Petersen 2004). We also review the production of other important platform organic acid or alcohols like lactic acid, 1,3-propanediol, and mannitol. Meanwhile, the most recent commercial information of all these compounds is given. In addition, the review proposes the possibility that several platform chemicals, including fumaric acid, aspartic acid, glutamic acid, sorbitol, and itaconic acid, are produced by E. coli in the near future. The pathway interrelation of all high-value chemicals is given in Fig. 2 except three chemicals: 13, 14, and 15 [see Fig. 1, chemicals (13), (14), and (15)].
Succinic acid
Succinic acid [see Fig. 1, chemical (1)] is a platform chemical which can be converted to 1,4-butanediol and related products, tetrahydrofuran, γ-butyrolactone, n-methyl pyrrolidinone, and 2-pyrrolidinone, or other chemicals that are used to make a wide assortment of products. Succinic acid has the potential to replace important chemical intermediates (Kleff 2007). These chemicals are requested by American markets total almost 1 billion pounds or more than $l.3B every year from the information of Pacific Northwest National Laboratory. The industrial potential for succinic acid fermentations was recognized as early as 1980 (Zeikus 1980).
E. coli can utilize multiple pathways to form succinic acid (der Werf et al. 1997). Under anaerobic conditions, wild-type E. coli can produce a small amount of succinic acid (7.8%) by mixed-acid fermentation because of insufficient reducing power (Lin et al. 2005c; Wendisch et al. 2006). In order to improve succinic acid production, genetically modified metabolic pathways are first constructed to enhance the key enzyme activity. For example, succinic acid production from glucose by E. coli was significantly increased by overexpression of endogenous phosphoenolpyruvate carboxylase (ppc, PEPPc), while overexpression of endogenous phosphoenolpyruvate carboxykinase(pck, PEPCk) had no effect (Millard et al. 1996). However, overexpression of Actinobacillus succinogenes PEPCk increased the production of succinic acid as much as 6.5-fold (Kim et al. 2004). Using E. coli AFP111 as biocatalyst in which the pyc gene (pyruvate carboxylase) was overexpressed (AFP111/pTrc99A-pyc), a final succinic acid concentration and productivity reached 99.2 g/l and 1.3 g/l/h (Vemuri et al. 2002). Simultaneous overexpression of genes encoding PEPPc from Sorghum vulgare and pyruvate carboxylase (pyc) from Lactococcus lactis in E. coli increased the succinic acid yield, with a concomitant decrease of the lactate yield (Lin et al. 2005d). As fumarate reductase (frdABCD) is also a key enzyme in this metabolic pathway, two recombinant E. coli strains with amplified frdABCD activity have been constructed for the conversion of fumarate to succinic acid (Goldberg et al. 1983; Wang et al. 1998). In E. coli strains with recombinant frdABCD, when the concentration of glucose was 23.7 g/l, conversion rate of fumarate to succinic acid could reach 93%. However, malate could be accumulated when glucose was absent or cell density in the cultures was quite low. To improve the yield of succinic acid, further studies have been done, e.g., overexpressing of cyanobacterium Anabaena sp. 7120 ecaA (encoding carbonic anhydrase) in E. coli, to enhance the supply of HCO3ˉ for the improvement of succinic acid production (Wang et al. 2009).
The other metabolic engineering strategy to increase the production of succinic acid was to knock out or inhibit the enzyme in succinic acid competition pathways. Chatterjee et al. obtained AFP111 by spontaneous chromosomal mutation of the ptsG (glucose phosphotransferase) gene in strain NZN111, which is unable to ferment glucose due to the inactivation of the genes encoding pyruvate: formate lyase and lactate dehydrogenase. In a batch reactor, this strain AFP111 could produce 36 g/l of succinic acid (Chatterjee et al. 2001). Sánchez et al. constructed E. coli strain, SBS550MG, that inhibited the central metabolic pathway by deactivating adhE (encoding aldehyde dehydrogenase), ldhA (encoding lactate dehydrogenase), and ack-pta (encoding acetate kinase-phosphate acetyltransferase) from the central metabolic pathway and activating the glyoxylate pathway through the inactivation of iclR (encoding isocitrate lyase). In SBS550MG, the yield of succinic acid from glucose is about 1.6 mol/mol with an average anaerobic productivity rate of 10 mM/h (Sánchez et al. 2005). Because anaerobic fermentation of succinic acid has a number of disadvantages, Lin et al. constructed a mutant E. coli strain by deletion of sdh (encoding succinic acid dehydrogenase), poxB (encoding pyruvate oxidase), pta-ack, iclR, and ptsG genes, and overexpression of ppc gene. Fed-batch fermentation of Lin’s strain resulted in a concentration of up to 58.3 g/l of succinic acid with an overall yield of 0.85 mol/mol glucose (Lin et al. 2005a, b). Considering the cost of construction, materials, purification, and waste disposal, novel strains of E. coli C were constructed by deletion of five genes, ldhA, adhE, ackA, focA (formate transporter), and pflB (encoding pyruvate-formate lyase), to produce high titers of succinic acid (600–700 mM) in simple batch fermentations (10% sugar) using mineral salts medium without any complex nutrients (Jantama et al. 2007). In addition, the effects of different carbon sources were investigated on the production of succinic acid in E. coli strain AFP184, which lacks functional genes coding for pyruvate formate lyase, fermentative lactate dehydrogenase, and the glucose phosphotransferase system. The result showed that carbon sources could significantly affect the yield of succinic acid with only small amounts of byproducts formed (Andersson et al. 2007).
Engineering E. coli strains were tested as biocatalysts for the scale-up production of succinic acid even though their strain information was limited. In 2008, DSM and France's Roquette Frères developed biotechnical methods for the production of kilo-scale succinic acid, using E. coli as catalyst and glucose as feedstock. Meanwhile, Myriant from the USA tested the scale-up production of succinic acid in the 20,000-l bioreactor, also using E. coli as catalyst but unrefined sugar as feedstock (http://www.rsc.org/chemistryworld/News/2010/January/21011003.asp).
The biological production processes need to be economically feasible including a yield around 0.88 g/g, a rate between 1.8 and 2.5 g/l/h and a titer around 80 g/l (Beauprez 2010). To date, none of the developed microbial strains which have been reported has reached all of these standards. However, developments in the metabolic engineering methods mentioned above showed great promise for further improvements in the near future. Next efforts should be done to optimize the current metabolic engineering towards succinic acid rather than to set up new metabolic routes.
Hydroxy-propanoic acids
Lactic acid [see Fig. 1, chemical (2)] and 3-hydroxypropanoic acid [3-HP, see Fig. 1, chemical (3)] are industrially relevant microbial products. They receive significant attention due to their application as platform chemicals and building blocks for “bio-based” polymers. Lactic acid, as the monomer for biodegradable polymer, will replace various petrochemical industry-based polymers in applications ranging from packaging to fibers (Tullo 2000) and be used in some new fields such as cosmetics, chemical industry, food, and pharmaceutics (Wee et al. 2006). 3-HP has similar applications but without the side-chain methyl groups, which influence the strength of the materials (Zhu et al. 2010). Besides, it is a precursor for the synthesis of many high-volume commercial intermediates such as 1,3-propanediol (1,3-PDO) and acrylates (Brown 2003; Werpy and Petersen 2004).
Production of lactic acid
The microorganisms selected for the biotechnological production of lactic acid include two groups: bacteria and fungi (Wee et al. 2006). Lactobacilli are commonly used for industrial production of lactic acid, but the fermentations rarely yield an optically pure product, while in E. coli, lactic acid production is one of the most successful large-scale fermentations to produce organic acids. The productivity for lactic acid differs upon various E. coli strains. Wild-type E. coli produces a mixture of organic acids including lactate. Engineering constructed E. coli for the production of lactic acid has several advantages compared to many other microorganisms available for lactic acid production. For example, E. coli can be metabolically engineered to produce optically pure lactic acid with less other fermentation byproducts, and Belgian scientists have reported that lactic acid has little effect on the proliferation of E. coli (http://www.foodnavigator.com).
Optical lactic acid production was greatly developed in the recent years. Chang et al. demonstrated that an E. coli RR1 pta (encoding phosphotransacetylase) mutant could be used as the host for the production of optically pure d- or l-lactic acid. They introduced l-lactate dehydrogenase genes from Lactobacillus casei into a pta ldhA (encoding lactate dehydrogenase) mutant strain, which lacked phosphotransacetylase and d-lactate dehydrogenase. The results suggested that the central fermentation metabolism of E. coli could be reoriented to the production of d(−)- or l(+)-lactic acid (Chang et al. 1999). Similarly, Dien et al. constructed recombinant E. coli for l(+)-lactic acid production from hexose and pentose sugars. They metabolically engineered E. coli including a ptsG − but glucose+ mutant that carried mutations in the genes encoding PFL (pyruvate-formate lyase) and D-LDH (lactate dehydrogenase) and expressed a gene encoding L-LDH for the construction of carbon catabolite repression mutants (Dien et al. 2001, 2002). Zhou et al. constructed derivatives of E. coli W3110 as new biocatalysts for the production of d-lactic acid. d-Lactic acid production by these new strains (SZ40, SZ58, and SZ63) approached the theoretical maximum yield of two molecules per glucose molecule. Competing pathways were eliminated by chromosomal inactivation of genes encoding fumarate reductase (frdABCD), alcohol/aldehyde dehydrogenase (adhE), and pyruvate formate lyase (pflB). The cell yield and lactate productivity were increased by a further mutation in the acetate kinase gene (ackA) (Zhou et al. 2003). Although capable of efficient fermentation of 5% (w/v) glucose or sucrose, higher sugar concentrations were incompletely metabolized by this biocatalyst and continuous antibiotic selection was required for plasmid maintenance. Hence, Zhou et al. constructed KO11-based biocatalysts (strain SZ132, produced by metabolic evolution from SZ110) that fermented glucose and sucrose to produce over 1 mol d(−)-lactate per liter of fermentation broth (10% w/v glucose or sucrose) (Zhou et al. 2005). Although SZ132 rapidly fermented 10% (w/v) sugars to completion, rich medium was required and performance was poor in mineral salts medium. Therefore, further improvements were done on SZ132. A non-recombinant mutant of E. coli B, strain SZ194 was developed from SZ132 that produced over 1 M d-lactate from glucose (or sucrose) in 72 h using mineral salts medium supplemented with 1 mM betaine in simple anaerobic fermentations. Rates and yields were highest at pH 7.5 with only trace amounts of co-products (Zhou et al. 2006). What is more, altering NADH utilization pathways had effects on the distribution of metabolic products. d-Lactate was the primary product in the ndh nuo (NADH-dehydrogenase-encoding genes) adhE inactivation strain (Yun et al. 2005).
Expanding the substrate range was also concerned in engineering E. coli strain improvements. A cluster of sucrose genes (three adjacent chromosomal operons, cscR′, cscA, and cscKB, encoding a repressor protein, invertase, fructokinase, and anion symport) were cloned and characterized from E. coli KO11. The resulting plasmid was functionally expressed in strain SZ63, which produced over 500 mM d(−)-lactate from sucrose (Shukla et al. 2004). By combining two substrate-selective strains of E. coli, knockout in pflB (encoding pyruvate-formate lyase), the xylose-selective strain with deletions of the glk (encoding glucokinase), ptsG, and manZ (encoding mannose PTS permease) genes and the glucose-selective strain with a xylA (encoding xylose isomerase) deletion in a single process, xylose and glucose in a mixed sugar solution were simultaneously converted to lactate (Eiteman et al. 2008, 2009).
All engineered E. coli strains mentioned above have various problems for industrial applications, thus we need to optimize the lactic acid biocatalysts with increased fermentation rates, product titer, and yields to reduce costs. In a recent patent, E. coli was constructed by deleting the genes that encode competing pathways followed by a growth-based selection for mutants with improved performance for the production of lactate, which could ferment 10% glucose or sucrose to produce over 1 mol d(−)-lactate of fermentation broth, with yields based on metabolized sugar ranging from about 88% to about 95%. Over 100 g/l in 48 h of chirally pure l(+)- and d(−)-lactate (>99.9% chiral purity) can be readily produced by recombinant E. coli B in mineral salts medium supplemented with 1 mM betaine (Shengde et al. 2010), showing great potential for scale-up production of lactic acid.
Production of 3-hydroxypropanoic acid
3-Hydroxypropionic acid (3-HP) production has the potential to be the next chemical intermediate produced by fermentation following lactic acid and succinic acid. Recently, Jiang et al. reviewed all 12 known synthetic pathways to produce 3-HP from sugars, glycerol, or carbon dioxide (Jiang et al. 2009). In this review, seven possible biosynthetic routes were summarized for the formation of 3-HP from glucose. Considering the thermodynamical advantage and the balance of reducing power and ATP, the pathway [see Fig. 2, enzymes used—pgt (encoding pyruvate-glutamate transaminase), aam (encoding alanine 2,3-aminomutase), aoat (encoding β-alanine-2-oxoglutarate aminotransferase), and hid (encoding 3-hydroxyisobutyrate dehydrogenase)] from pyruvate to 3-HP is most attractive. The point of this pathway is to increase the activity of the enzyme aoat. E. coli cannot metabolize glycerol into 3-HP because it lacks glycerol dehydratase (dhaB) and the native expression of aldehyde dehydrogenase (aldH) is also very weak (Jo et al. 2008). In 2001, formation of low concentrations of 3-HP (0.2 g/l) from glycerol, via 3-hydroxypropanal, was demonstrated with genetically modified E. coli expressing a dhaB from Klebsiella pneumoniae and a non-specific aldH from Saccharomyces cerevisiae (Suthers and Cameron 2001). A recombinant E. coli strain was developed by cloning dhaB and aldH genes, and the production of 3-HP from glycerol was demonstrated (Raj et al. 2008). After optimizing the physicochemical parameters, the recombinant E. coli strain, expressing dhaB and aldH genes, can produce 3-HP at 31 g/l with a yield of 35% when glycerol is used as the sole carbon and energy source (Mohan Raj et al. 2009). However, the productivity is far behind its commercial application. Hence, further work will remain to be done. Excitedly, Cargill and Novozymes predicted that 3-HP would be produced and sold within 5 years (http://www.cargill.com/news-center/news-releases/2008/NA3007665.jsp).
The metabolic pathways for the production of the high-value chemical building blocks by using E. coli. The solid lines indicate E. coli native pathways while the dotted lines refer recombinant pathway by metabolic engineering strategies. Enzymes encoded by the genes shown are: ino1: myo-inositol-1-phosphate synthase; miox: myo-inositol oxygenase; udh: uronate dehydrogenase; xr: xylose reductases; mpd: mannitol-6-phosphate dehydrogenase; mk: mannitol kinase; spd: sorbitol-6-phosphate dehydrogenase; sk: sorbitol kinase; gdht: glycerol dehydrogenase; por: 1,3-propanediol oxidoreductase; aldh: aldehyde dehydrogenase; pyk: pyruvate kinase; ldh: lactate dehydrogenase; CoAt: Co-enzyme A transferase; lcd: lactyl-CoA dehydratase; pgt: pyruvateglutamate transaminase; aam: alanine 2,3-aminomutase; aoat: β-alanine-2-oxoglutarate aminotransferase; pdc: pyruvate dehydrogenase complex; acc: acetyl-CoA carboxylase; mcr: malonyl-CoA reductase; pyc: pyruvate carboxylase; cs: citrate synthesase; acn: aconitase; adc: aconitate decarboxylase; icd: isocitrate dehydrogenase; gdh: glutamate dehydrogenase; kgd: α-ketoglutarate dehydrogenase; scs: succinyl-CoA synthetase; ssd: succinic semialdehyde dehydrogenase; sud: succinic acid dehydrogenase; frd: fumarate reductase; fum: fumarase; mdh: malate dehydrogenase; ast: aspartate aminotransferase; asd: aspartate decarboxylase; abt: 4-aminobutyrate aminotransferase; hid: 3-hydroxyisobutyrate dehydrogenase
Glucaric acid
Glucaric acid [see Fig. 1, chemical (4)] has been studied for therapeutic purposes including cholesterol reduction (Walaszek et al. 1996) and cancer chemotherapy (Singh and Gupta 2007). It also has potential applications as a building block for a number of polymers such as new nylons and hyperbranched polyesters.
Moon et al. constructed a synthetic pathway for the production of d-glucaric acid in E. coli by co-expression of the genes encoding myo-inositol-1-phosphate synthase (Ino1) from S. cerevisiae, myo-inositol oxygenase (MIOX) from mice, and uronate dehydrogenase (Udh) from Pseudomonas syringae. This is a simple and economical pathway for the production of d-glucaric acid compared with the long pathway of more than ten conversion steps from glucose in mammals. Glucaric acid productivity of over 1 g/l was observed (Moon et al. 2009). To further improve the productivity, they used modular, synthetic scaffolds for producing high-value compounds economically, efficiently, and cleanly, thus improving the maximum titers to 2.5 g/l (Moon et al. 2010). There is no industrial supply of glucaric acid by microbial fermentation so far. However, Rivertop Renewables is currently converting renewable glucose into glucaric acid. The company hopes to build a commercial plant and put out small production volumes by September 2011 (http://cleantech.com/news/5613/startup-pursues-glucaric-acid-poten).
Glycerol
Glycerol [see Fig. 1, chemical (5)], 1,2,3-propanetriol, is a commodity chemical used in cosmetics, liquid soaps, food, pharmaceuticals, lubricants, antifreeze solutions, and tobacco. The further chemical modification of glycerol is also carried out in industry, especially the synthesis of some esters, polyethers, and alkyd resins. Behr et al. reviewed the improved utilization of glycerol (Behr et al. 2008).
The most important microbial biocatalyst is S. cerevisiae but other yeast species (Taherzadeh et al. 2002). E. coli does not have a natural efficient pathway to produce glycerol. In early-stage research, a Genencorv International and Dupont team had demonstrated obvious glycerol production in E. coli. Expression of either glycerol-3-phosphate dehydrogenase (GPD1) or glycerol-3-phosphatase (GPP2) in E. coli resulted in a low level of glycerol production (Bulthuis et al. 2002). When GPD1 and GPP2 were co-expressed in E. coli, the yield of glycerol increased 10–20-fold. Because E. coli can utilize glycerol as carbon source through glycerol kinase (glpk) and glycerol dehydrogenase (gldA) pathways, Nair et al. removed these dissimilation pathways, which resulted in minimal glycerol consumption and increased carbon yield to glycerol with near theoretical yield and over 200 g/l of glycerol (Nair et al. 2005). Recently, in order to demonstrate, under appropriate “metabolic pressure”, the evolutionary adaptation of a heterogeneous pathway in a strain and validate this concept, the S. cerevisiae glycerol pathway (GPD1 and GPP2) was evolved in vivo in E. coli. Three key modifications were introduced in the central metabolism of E. coli. Without any optimization of the fermentation conditions, high yield (1 mol/mol), titer (130 g/l), and productivity (16 g/l/day) were obtained in glucose-limited fed-batch cultures (Meynial Salles et al. 2007).
Recently, the study on its microbial production is not very popular because of its rising inevitable formation as a by-product of biodiesel production. However, with growing interest of glycerol as a bulk platform chemical, microbial production of glycerol still could be a concern based on the consideration of biomass from waste residual, and in that case, engineered E. coli due to its special features could be one alternative to S. cerevisiae.
1,3-Propanediol
1,3-Propanediol [1,3-PD; see Fig. 1, chemical (6)] is suitable for fiber and textile applications. It has the potential to replace the traditional polyethylene terephthalate and polybutylene terephthalate as well as being used in solvents, adhesives, laminates, resins, detergents, and cosmetics as a bifunctional organic compound (Zeng and Biebl 2002).
1,3-Propanediol production needs two enzymes, glycerol dehydratase (encoded by the gene dhaB1-3) and 1,3-propanediol oxidoreductase (encoded by the gene dhaT) in natural organisms (Nakamura and Whited 2003). Experiments to overexpress these genes in E. coli have been successful, but the 1,3-PD concentrations obtained with these constructs could originally not be raised above 9 g/l (Daniel and Gottschalk 1992). By expressing a non-specific alcohol dehydratase instead of dhaT for the conversion of 3-hydroxypropionaldehyde, a final concentration of 129 g/l was obtained with transformed E. coli in a patent (Zeng and Biebl 2002).
Recently, Wang et al. (2007) co-expressed in E. coli three genes including dhaB, which encodes glycerol dehydratase; dhaT, which encodes 1,3-PD oxidoreductase; and gdrAB, which encodes glycerol dehydratase. In a fed-batch fermentation of glycerol and glucose, the recombinant E. coli consumed 14.3 g/l of glycerol and produced 8.6 g/l of 1,3-propanediol. In the substitution case of yqhD (encoding alcohol dehydrogenase from E. coli) for dhaT, the final 1,3-propanediol concentration of the recombinant E. coli could reach 13.2 g/l. To improve 1,3-PD yield, several efforts were done by expressing three genes (dhaB1, dhaB2, and yqhD) in a recombinant E. coli and constructing a novel 1,3-PD operon of these three genes randomly arrayed under the control of a constitutive, temperature-sensitive promoter in the vector pBV220 for heterologous expression in E. coli. Under a two-stage fermentation process, the overall 1,3-PD yield and productivity reached 104.4 g/l and 2.61 g/l/h (Tang et al. 2009), which show a good potential for the economic and effective production of 1,3-PD. For scale-up production, Dupont and Genencor have commercialized 1,3-PD by engineered E. coli, and 1,3-PD-based polyester which is estimated to be 1–2 billion pounds per year over the next 10 years can also be produced by Dupont and Genencor using glucose as the feedstock (Nakamura and Whited 2003).
Xylitol
Xylitol [see Fig. 1, chemical (7)] is the first rare sugar that has global markets. Xylitol can be used as a raw material for a number of different bioconversions for other rare sugar production. It has beneficial health properties and is used as a nutritive sweetener and food additive (Granstrom et al. 2007). Its major use is for the prevention of dental caries as xylitol prevents the growth of microorganisms responsible for tooth decay. The xylitol market is increasing and is estimated to be $340 million year−1 and priced at $4–5 kg−1 according to Prakasham et al. (2009).
E. coli has the ability to assimilate both hexose and pentose sugars besides the advantages mentioned above. In order to produce xylitol in an economical and eco-friendly manner, metabolically engineered E. coli has been studied as an alternative for industrial production of xylitol. Xylose reductases catalyze the initial reaction in the xylose utilization pathway, the NAD(P)H-dependent reduction of xylose to xylitol. Häcker et al. cloned and successfully expressed the xylose reductase gene from Candida tenuis in E. coli (Häcker et al. 1999). In 1999, the d-xylose reductase (XR) gene (xyrA) of Candida tropicalis was also expressed in E. coli, which successfully converted d-xylose to xylitol. When d-xylose (50 g/l) and d-glucose (5 g/l) were added to IPTG-induced cells, 13.3 g/l of xylitol was produced during 20 h of cultivation (Suzuki et al. 1999).
Cirino et al. replace E. coli’s native cyclic AMP receptor protein (CRP) with a cyclic AMP-independent mutant (CRP*) which facilitated xylose uptake and xylitol production from mixtures of glucose and xylose, with glucose serving as the growth substrate and source of reducing equivalents. Overexpression of NADPH-dependent CbXR (xylose reductases from Candida boidinii) produced the highest concentrations of xylitol in shake-flask cultures (Cirino et al. 2006). Subsequent deletion of the xylB gene (encoding xylulokinase) and expression of xylose reductase from C. boidinii resulted in a strain which produced xylitol from glucose–xylose mixtures. Khankal et al. engineered E. coli W3110 to produce xylitol from a mixture of glucose plus xylose by expressing xylose reductase and deleting xylulokinase (DxylB), combined with either plasmid-based expression of a xylose transporter (XylE or XylFGH) (Khankal et al. 2008). More researches were done by heterologous expression of d-xylulokinase from Pichia stipitis with high levels of xylitol production by engineered E. coli co-expressing xylose reductase during growth on xylose (Akinterinwa and Cirino 2009). In current research, Cheng et al. cloned the NAD-dependent d-xylulose-forming d-arabitol dehydrogenase gene (aArDH) from an acetic acid bacterium and heterogeneously expressed it in E. coli for the bio-conversion of d-arabitol to xylitol (Cheng et al. 2009).
Although other microorganisms such as yeasts were tested as xylitol producers, E. coli offers the increasing possibilities of economic production due to its special advantages. Furthermore, E. coli can reduce required energy comparing to the relative chemical methods. For the production of xylitol via biotechnological process at an economic industrial scale, focus should be maintained on a common platform of understanding of the hydrolysate material, hydrolysis procedure, microbial performance, and bioconversion environment, and downstream processing is one of the most essential aspects for development of integrated technological solution (Prakasham et al. 2009).
Mannitol
Mannitol [see Fig. 1, chemical (8)] has a variety of applications in pharmaceutical products, food industry, and medicine (Song and Vieille 2009). E. coli was engineered to produce mannitol from fructose by constructing a recombinant oxidation/reduction cycle. The recombinant strain co-expressed mdh gene (encoding malate dehydrogenase) from Leuconostoc pseudomesenteroides, fdh gene (encoding formate dehydrogenase) from Mycobacterium vaccae N10, and glf gene (encoding the glucose facilitator protein) from Zymomonas mobilis. Under pH control mode(by the addition of formic acid), the concentration of mannitol was 362 mM within 8 h and the yield was 84 mol% (Kaup et al. 2004). Further research was done by additional expression of extracellular glucose isomerase in E. coli, leading to the formation of 800 mM mannitol from 1,000 mM glucose. Co-expression of the xylA gene of E. coli in this strain led to a mannitol concentration of 420 mM from 1,000 mM glucose (Kaup et al. 2005). Considering the restriction of long-term stability of recombinant E. coli which produced mannitol from fructose, fupL gene, encoding a putative mannitol permease, was cloned and expressed in E. coli. The productivity for the biotechnical production of mannitol was enhanced by 20% (Heuser et al. 2009). More researches still need to be done to optimize the E. coli strains for the commercial production of mannitol. At the same time, other biocatalysts like L. intermedius NRRL B-3693 was used to commercially produce mannitol by the American company zuChem. However, compared with E. coli, L. intermedius-based process has too many by-products in the final products (http://www.foodnavigator-usa.com/Financial-Industry/ZuChem-gears-up-for-first-mannitol-sweetener).
Other top value-added platform chemicals
-
1.
Fumaric acid [see Fig. 1, chemical (9)] has many potential industrial applications, e.g., precursors of unsaturated polyester resins and plasticizers, miscellaneous including lubricating oil, inks, and lacquers, carboxylating agent for styrenebutadiene rubber, personal care additives, and food and beverage additives (Roa Engel et al. 2008). Fungi have been used in fermentation processes for fumaric acid production.
-
2.
Aspartic acid [see Fig. 1, chemical (10)] and glutamic acid [see Fig. 1, chemical (11)] are building blocks for active ingredients required in the pharmaceutical and cosmetics industries. The biosynthesis of aspartic acid and glutamic acid in some bacteroids has been researched, such as Rhizobium lupine and Rhizobium japonicum (Lillich and Elkan 1971; Kretovich et al. 1981). The aspartate pathway has also been researched in plants. Single and often multiple genes have been cloned and expressed successfully in E. coli (Azevedo et al. 2006). The metabolic engineering of glutamic acid production by Corynebacterium glutamicum has been investigated in depth (Kimura 2002; Schultz et al. 2007).
-
3.
Sorbitol [see Fig. 1, chemical (16)] has broad applications not only in the food industry but also in pharmaceutical production (Silveira and Jonas 2002). Two recombinant strains of Lactobacillus plantarum (Ladero et al. 2007) and Lactobacillus casei (Yebra and Perez-Martinez 2002; Nissen et al. 2005) were constructed to product sorbitol.
-
4.
Itaconic acid [IA; see Fig. 1, chemical (12)] has been used in synthetic resins, synthetic fibers, plastics, rubbers, surfactants, oil additives, and biomedical fields (Okabe et al. 2009). Aspergillus terreus is now the most frequently used commercial producer of IA, and the IA production yield from sugar is higher than 80 g/l (Willke and Vorlop 2001). However, the production costs are relatively high.
-
5.
2,5-Furan dicarboxylic acid [see Fig. 1, chemical (13)] can be used to produce numerous furan derivatives, succinic acid, esters, levulinic acid, furanoic polyamines, and polyethylene terephthalate analogs. Its biosynthesis has not been reported so far as well as levulinic acid [LA; see Fig. 1, chemical (14)] and 3-hydroxybutyrolactone [see Fig. 1, chemical (15)]. The researches still need to be further enhanced.
Conclusions
Compared with conventional petrochemical production, bio-based platform chemicals, which can be either directly used or further processed for the production of large-volume and value-added products in the chemical industry, have gained more concern. E. coli strains could be engineered for overproduction of native and non-native small molecules with obvious advantages over other bacteria. Improved E. coli strains have been developed that are well adapted to fermentation processes and produce desired products with high yields. With recent successes of metabolic engineering of E. coli, several other platform chemicals [see Fig. 1, chemicals (3)–(8)] have the potential to be scaled up in the near future. However, the biotransformation route of some top value-added chemicals has not been researched such as chemicals (12) and (16) in Fig. 1. New tools such as modulating expression of chromosomal genes or creating multiple chromosomal deletions will certainly improve the efficiency and the fine-tuning of metabolic engineering for E. coli to overcome its native limitation. Furthermore, all of genomic, proteomic, and metabolic modeling tools could be used in E. coli to enhance the production of the present organic acids/alcohols products, and on this base to further develop innovative organic acids/alcohols products through the creation and optimization of novel metabolic pathways.
References
Akinterinwa O, Cirino P (2009) Heterologous expression of d-xylulokinase from Pichia stipitis enables high levels of xylitol production by engineered Escherichia coli growing on xylose. Metab Eng 11:48–55
Andersson C, Hodge D, Berglund K, Rova U (2007) Effect of different carbon sources on the production of succinic acid using metabolically engineered Escherichia coli. Biotechnol Prog 23:381–388
Azevedo R, Lancien M, Lea P (2006) The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 30:143–162
Beauprez J (2010) Metabolic engineering and modelling of Escherichia coli for the production of succinate. Dissertation, Ghent University
Behr A, Eilting J, Irawadi K, Leschinski J, Lindner F (2008) Improved utilisation of renewable resources: new important derivatives of glycerol. Green Chem 10:13–30
Bianco C, Imperlini E, Calogero R, Senatore B, Pucci P, Defez R (2006) Indole-3-acetic acid regulates the central metabolic pathways in Escherichia coli. Microbiology 152:2421–2431
Brown SF (2003) Bioplastic fantastic. Fortune 148:92–94
Bulthuis B, Gatenby A, Haynie S, Hsu A, Lareau R (2002) Method for the production of glycerol by recombinant organisms. Patent application no. US6358716
Chang DE, Jung HC, Rhee JS, Pan JG (1999) Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl Environ Microbiol 65:1384–1389
Chatterjee R, Millard CS, Champion K, Clark DP, Donnelly MI (2001) Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl Environ Microbiol 67:148–154
Cheng H, Li Z, Jiang N, Deng Z (2009) Cloning, purification and characterization of an NAD-dependent d-arabitol dehydrogenase from acetic acid bacterium, Acetobacter suboxydans. Protein J 28:263–272
Chotani G, Dodge T, Hsu A, Kumar M, LaDuca R, Trimbur D, Weyler W, Sanford K (2000) The commercial production of chemicals using pathway engineering. Biochim Biophys Acta 1543:434–455
Cirino PC, Chin JW, Ingram LO (2006) Engineering Escherichia coli for xylitol production from glucose–xylose mixtures. Biotechnol Bioeng 95:1167–1176
Daniel R, Gottschalk G (1992) Growth temperature-dependent activity of glycerol dehydratase in Escherichia coli expressing the Citrobacter freundii dha regulon. FEMS Microbiol Lett 79:281–285
der Werf MJV, Guettler MV, Jain MK, Zeikus JG (1997) Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch Microbiol 167:332–342
Dien BS, Nichols NN, Bothast RJ (2001) Recombinant Escherichia coli engineered for production of L-lactic acid from hexose and pentose sugars. J Ind Microbiol Biotechnol 27:259–264
Dien BS, Nichols NN, Bothast RJ (2002) Fermentation of sugar mixtures using Escherichia coli catabolite repression mutants engineered for production of L-lactic acid. J Ind Microbiol Biotechnol 29:221–227
Eiteman MA, Lee SA, Altman E (2008) A co-fermentation strategy to consume sugar mixtures effectively. J Biol Eng 2:3–11
Eiteman M, Lee S, Altman R, Altman E (2009) A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose. Biotechnol Bioeng 102:822–827
Goldberg I, Lonberg-Holm K, Bagley EA, Stieglitz B (1983) Improved conversion of fumarate to succinate by Escherichia coli strains amplified for fumarate reductase. Appl Environ Microbiol 45:1838–1847
Granstrom T, Izumori K, Leisola M (2007) A rare sugar xylitol. Part II: biotechnological production and future applications of xylitol. Appl Microbiol Biotechnol 74:273–276
Häcker B, Habenicht A, Kiess M, Mattes R (1999) Xylose utilisation: cloning and characterisation of the xylose reductase from Candida tenuis. Biol Chem 380:1395–1403
Harrington C, Rosenow C, Retief J (2000) Monitoring gene expression using DNA microarrays. Curr Opin Microbiol 3:285–291
Hatti-Kaul R, Tornvall U, Gustafsson L, Borjesson P (2007) Industrial biotechnology for the production of bio-based chemicals—a cradle-to-grave perspective. Trends Biotechnol 25:119–124
Heuser F, Marin K, Kaup B, Bringer S, Sahm H (2009) Improving d-mannitol productivity of Escherichia coli: impact of NAD, CO2 and expression of a putative sugar permease from Leuconostoc pseudomesenteroides. Metab Eng 11:178–183
Jantama K, Haupt M, Svoronos S, Zhang X, Moore J, Shanmugam K, Ingram L (2007) Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng 99:1140–1153
Jiang X, Meng X, Xian M (2009) Biosynthetic pathways for 3-hydroxypropionic acid production. Appl Microbiol Biotechnol 82:995–1003
Jo J, Mohan Raj S, Rathnasingh C, Selvakumar E, Jung W, Park S (2008) Cloning, expression, and characterization of an aldehyde dehydrogenase from Escherichia coli K-12 that utilizes 3-hydroxypropionaldehyde as a substrate. Appl Microbiol Biotechnol 81:51–60
Kaup B, Bringer-Meyer S, Sahm H (2004) Metabolic engineering of Escherichia coli: construction of an efficient biocatalyst for D-mannitol formation in a whole-cell biotransformation. Appl Microbiol Biotechnol 64:333–339
Kaup B, Bringer-Meyer S, Sahm H (2005) D-Mannitol formation from D-glucose in a whole-cell biotransformation with recombinant Escherichia coli. Appl Microbiol Biotechnol 69:397–403
Khankal R, Chin J, Cirino P (2008) Role of xylose transporters in xylitol production from engineered Escherichia coli. J Biotechnol 134:246–252
Kim P, Laivenieks M, Vieille C, Zeikus JG (2004) Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl Environ Microbiol 70:1238–1241
Kimura E (2002) Metabolic engineering of glutamate production. Adv Biochem Eng Biotechnol 79:37–58
Kleff S (2007) The integrated biorefinery: conversion of corn fiber to value-added chemicals. Conference report from 4th World Congress on Industrial Biotechnology and Bioprocessing Orlando, FL March 21–24, 2007
Kretovich W, Kariakina T, Weinova M, Sidelnikova L, Kazakova O (1981) The synthesis of aspartic acid in Rhizobium lupini bacteroids. Plant Soil 61:145–156
Ladero V, Ramos A, Wiersma A, Goffin P, Schanck A, Kleerebezem M, Hugenholtz J, Smid E, Hols P (2007) High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering. Appl Environ Microbiol 73:1864–1872
Lillich T, Elkan G (1971) The biosynthesis of aspartic acid, glutamic acid, and alanine in Rhizobium japonicum. Can J Microbiol 17:683–688
Lin H, Bennett GN, San KY (2005a) Effect of carbon sources differing in oxidation state and transport route on succinate production in metabolically engineered Escherichia coli. J Ind Microbiol Biotechnol 32:87–93
Lin H, Bennett GN, San KY (2005b) Fed-batch culture of a metabolically engineered Escherichia coli strain designed for high-level succinate production and yield under aerobic conditions. Biotechnol Bioeng 90:775–779
Lin H, Bennett GN, San KY (2005c) Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab Eng 7:116–127
Lin H, San KY, Bennett GN (2005d) Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli. Appl Microbiol Biotechnol 67:515–523
Meynial Salles I, Forchhammer N, Croux C, Girbal L, Soucaille P (2007) Evolution of a Saccharomyces cerevisiae metabolic pathway in Escherichia coli. Metab Eng 9:152–159
Millard C, Chao Y, Liao J, Donnelly M (1996) Enhanced production of succinic acid by overexpression of phosphoenolpyruvate carboxylase in Escherichia coli. Appl Environ Microbiol 62:1808
Mohan Raj S, Rathnasingh C, Jung W, Park S (2009) Effect of process parameters on 3-hydroxypropionic acid production from glycerol using a recombinant Escherichia coli. Appl Microbiol Biotechnol 84:649–657
Moon TS, Yoon SH, Lanza AM, Roy-Mayhew JD, Prather K (2009) Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl Environ Microbiol 75:589–595
Moon TS, Dueber JE, Shiue E, Prather KLJ (2010) Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab Eng 12:298–305
Nair R, Payne M, Trimbur D, Valle F (2005) Method for the production of glycerol by recombinant organisms. Patent application no. US 2006/0286653 A1
Nakamura CE, Whited GM (2003) Metabolic engineering for the microbial production of 1, 3-propanediol. Curr Opin Biotechnol 14:454–459
Nelson DL, Cox MM (2000) Lehninger principles of biochemistry. Worth, New York
Nissen L, Pérez-Martínez G, Yebra M (2005) Sorbitol synthesis by an engineered Lactobacillus casei strain expressing a sorbitol-6-phosphate dehydrogenase gene within the lactose operon. FEMS Microbiol Lett 249:177–184
Okabe M, Lies D, Kanamasa S, Park E (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol 84:597–606
Park JH, Lee S (2008) Towards systems metabolic engineering of microorganisms for amino acid production. Curr Opin Biotechnol 19:454–460
Prakasham R, Rao R, Hobbs P (2009) Current trends in biotechnological production of xylitol and future prospects. Curr Trends Biotechnol Pharm 3:8–36
Raj SM, Rathnasingh C, Jo J-E, Park S (2008) Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 strain. Process Biochem 43:1440–1446
Roa Engel C, Straathof A, Zijlmans T, van Gulik W, van der Wielen L (2008) Fumaric acid production by fermentation. Appl Microbiol Biotechnol 78:379–389
Sánchez A, Bennett G, San K (2005) Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng 7:229–239
Sauer U, Eikmanns B (2005) The PEP–pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29:765
Saxena PK, Anand P, Saran S, Isar J (2009) Microbial production of 1, 3-propanediol: recent developments and emerging opportunities. Biotechnol Adv 27:895–913
Schultz C, Niebisch A, Gebel L, Bott M (2007) Glutamate production by Corynebacterium glutamicum: dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl Microbiol Biotechnol 76:691–700
Shanmugam KT, Ingram LO (2008) Engineering biocatalysts for production of commodity chemicals. J Mol Microbiol Biotechnol 15:8–15
Shengde Z, Lonnie ON, Shanmugam KT, Yomano L, Tammy BG, Jonathan CM (2010) Materials and methods for efficient lactic acid production. Patent application no: 20100203602
Shukla V, Zhou S, Yomano L, Shanmugam K, Preston J, Ingram L (2004) Production of d (−)-lactate from sucrose and molasses. Biotechnol Lett 26:689–693
Silveira M, Jonas R (2002) The biotechnological production of sorbitol. Appl Microbiol Biotechnol 59:400–408
Singh J, Gupta KP (2007) Induction of apoptosis by calcium D-glucarate in 7, 12-dimethyl benz [a] anthracene-exposed mouse skin. J Environ Pathol Toxicol Oncol 26:63–73
Song SH, Vieille C (2009) Recent advances in the biological production of mannitol. Appl Microbiol Biotechnol 84:55–62
Suthers PF, Cameron DC (2001) Production of 3-hydroxypropionic acid in recombinant organisms. Patent application no. PCT WO 01-16346
Suzuki T, Yokoyama S, Kinoshita Y, Yamada H, Hatsu M, Takamizawa K, Kawai K (1999) Expression of xyrA gene encoding for D-xylose reductase of Candida tropicalis and production of xylitol in Escherichia coli. J Biosci Bioeng 87:280–284
Taherzadeh M, Adler L, Liden G (2002) Strategies for enhancing fermentative production of glycerol—a review. Enzyme Microb Technol 31:53–66
Tang X, Tan Y, Zhu H, Zhao K, Shen W (2009) Microbial conversion of glycerol to 1, 3-propanediol by an engineered strain of Escherichia coli. Appl Environ Microbiol 75:1628–1634
Tullo A (2000) Plastic found at the end maize. Chem Eng News 78:13
Vemuri G, Aristidou A (2005) Metabolic engineering in the o-mics era: elucidating and modulating regulatory networks. Microbiol Mol Biol Rev 69:197–216
Vemuri G, Eiteman M, Altman E (2002) Succinate production in dual-phase Escherichia coli fermentations depends on the time of transition from aerobic to anaerobic conditions. J Ind Microbiol Biotechnol 28:325–332
Walaszek Z, Szemraj J, Hanausek M, Adams A, Sherman U (1996) D-glucaric acid content of various fruits and vegetables and cholesterol-lowering effects of dietary D-glucarate in the rat. Nutr Res 16:673–681
Wang X, Gong C, Tsao G (1998) Bioconversion of fumaric acid to succinic acid by recombinant E. coli. Appl Biochem Biotechnol 70:919–928
Wang F, Qu H, Zhang D, Tian P, Tan T (2007) Production of 1, 3-propanediol from glycerol by recombinant E. coli using incompatible plasmids system. Mol Biotechnol 37:112–119
Wang D, Li Q, Li W, Xing J, Su Z (2009) Improvement of succinate production by overexpression of a cyanobacterial carbonic anhydrase in Escherichia coli. Enzyme Microb Technol 45:491–497
Wee Y, Kim J, Ryu H (2006) Biotechnological production of lactic acid and its recent applications. Food Technol Biotechnol 44:163
Wendisch VF, Bott M, Eikmanns BJ (2006) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol 9:268–274
Werpy T, Petersen G (2004) Top value added chemicals from biomass: volume I—results of screening for potential candidates from sugars and synthesis gas. US Department of Energy http://www.osti.gov/bridge
Willke T, Vorlop K (2001) Biotechnological production of itaconic acid. Appl Microbiol Biotechnol 56:289–295
Yebra M, Perez-Martinez G (2002) Cross-talk between the L-sorbose and D-sorbitol (D-glucitol) metabolic pathways in Lactobacillus casei. Microbiology 148:2351–2359
Yun N, San K, Bennett G (2005) Enhancement of lactate and succinate formation in adhE or pta-ackA mutants of NADH dehydrogenase-deficient Escherichia coli. J Appl Microbiol 99:1404–1412
Zaldivar J, Nielsen J, Olsson L (2001) Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 56:17–34
Zeikus J (1980) Chemical and fuel production by anaerobic bacteria. Annu Rev Microbiol 34:423–464
Zeng A, Biebl H (2002) Bulk-chemicals from biotechnology: the case of 1, 3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74:239–260
Zhou S, Causey T, Hasona A, Shanmugam K, Ingram L (2003) Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol 69:399
Zhou S, Yomano L, Shanmugam K, Ingram L (2005) Fermentation of 10%(w/v) sugar to D(−)-lactate by engineered Escherichia coli B. Biotechnol Lett 27:1891–1896
Zhou S, Shanmugam K, Yomano L, Grabar T, Ingram L (2006) Fermentation of 12%(w/v) glucose to 1.2 M lactate by Escherichia coli strain SZ194 using mineral salts medium. Biotechnol Lett 28:663–670
Zhu Y, Eiteman M, Lee S, Altman E (2010) Conversion of glycerol to pyruvate by Escherichia coli using acetate- and acetate/glucose-limited fed-batch processes. J Ind Microbiol Biotechnol 37:307–312
Acknowledgments
This work was financially supported by National Natural Science Foundation (No. 20872075) and CAS 100 Talents Program (KGCXZ-YW-801).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, C., Cao, Y., Zou, H. et al. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl Microbiol Biotechnol 89, 573–583 (2011). https://doi.org/10.1007/s00253-010-2970-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2970-z