Abstract
Sclerotinia stem rot caused by Sclerotinia sclerotiorum is one of the major fungal diseases of Brassica napus L. To develop resistance against this fungal disease, the defensin gene from Raphanus sativus and chimeric chit42 from Trichoderma atroviride with a C-terminal fused chitin-binding domain from Serratia marcescens were co-expressed in canola via Agrobacterium-mediated transformation. Twenty transformants were confirmed to carry the two transgenes as detected by polymerase chain reaction (PCR), with 4.8 % transformation efficiency. The chitinase activity of PCR-positive transgenic plants were measured in the presence of colloidal chitin, and five transgenic lines showing the highest chitinase activity were selected for checking the copy number of the transgenes through Southern blot hybridisation. Two plants carried a single copy of the transgenes, while the remainder carried either two or three copies of the transgenes. The antifungal activity of two transgenic lines that carried a single copy of the transgenes (T4 and T10) was studied by a radial diffusion assay. It was observed that the constitutive expression of these transgenes in the T4 and T10 transgenic lines suppressed the growth of S. sclerotiorum by 49 % and 47 %, respectively. The two transgenic lines were then let to self-pollinate to produce the T2 generation. Greenhouse bioassays were performed on the transgenic T2 young leaves by challenging with S. sclerotiorum and the results revealed that the expression of defensin and chimeric chitinase from a heterologous source in canola demonstrated enhanced resistance against sclerotinia stem rot disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sclerotinia stem rot caused by Sclerotinia sclerotiorum is one of the most important diseases of canola (Brassica napus L.). This ascomycete fungus is distributed worldwide and significantly reduces the yield and quality of some important crops, including canola (Kirkegaard et al. 2006). Genetic engineering as a promising method for the effective control of plant diseases has the advantage of incorporating genes which produce resistance proteins from any species to any crop (van der Biezen 2001). Among the antifungal proteins, pathogenesis-related (PR) proteins such as defensins and chitinases are well known to provide resistance to fungal infection in various plants (Thomma et al. 2002).
Defensins are a class of evolutionarily and structurally related small (45 to 54 amino acids), low-molecular-weight (5 kDa), highly basic and cysteine-rich peptides (Thomma et al. 2002). They have been detected in different organs of plants, such as leaves, flowers, pods, seeds and tubers (Wong and Ng 2005). They cause permeabilisation of fungal membranes, leading to the inhibition of fungal growth without having any concomitant of toxicity to either mammalian or plant (Thomma et al. 2002). Different plant defensins exhibit inhibitory activity against a broad range of fungal pathogens; for example, the inhibitory activity of Tfgd1 from Trigonella foenum-graecum against Rhizoctonia solani (Olli and Kirti 2006), PgD5 from Picea glauca against R. solani (Picart et al. 2012) and Rs-AFP2 from Raphanus sativus against Alternaria longipes, Magnaporthe oryzae and R. solani (Terras et al. 1995; Jha and Chattoo 2010).
Chitinases are low-molecular-weight PR proteins that are often extracellular, acid-soluble and protease-resistant (Graham and Sticklen 1994). They catalyse the hydrolysis of b-1,4 linkages of N-acetyl-D D-glucosamine chitin polymer of fungal mycelial walls into N-acetyl glucosamine oligomers and, consequently, result in the cell wall degradation and death of pathogenic fungi. They are found in bacteria, fungi, insects and higher plants (Kramer and Muthukrishnan 1997). In particular, chitinase-encoding genes from Trichoderma species have received considerable attention, as they have much higher antifungal capacities than the corresponding plant genes (Lorito et al. 1998). The antifungal activity of different chitinases from Trichoderma species has been demonstrated before; for example, the antifungal activity of chit33 from T. harzianum against R. solani (Limón et al. 1995; de las Mercedes Dana et al. 2006), chit42 from T. harzianum against Alternaria alternata, A. solani, Botrytis cinerea and R. solani (Lorito et al. 1998; de las Mercedes Dana et al. 2006) and a chitinase gene from T. virens against R. solani (Kumar et al. 2009).
Genes encoding defensins and chitinases were separately overexpressed in different crops, including tobacco (Saitoh et al. 2001), potato (Khan et al. 2008), rice (Jha et al. 2009; Jha and Chattoo 2010), wheat (Li et al. 2011) and cotton (Ganesan et al. 2009; Tohidfar et al. 2005), which resulted in enhanced tolerance of transgenic plants to fungal pathogens.
There have been certain evidences suggesting that the co-expression of more than one gene which exhibits different modes of action induced synergistic enhancement of disease resistance against fungal pathogens in different transgenic plants (Kim et al. 2003; Liu et al. 2004; Ntui et al. 2011; Chhikara et al. 2012; Khan et al. 2014).
Here, we present the co-expression of Rs-AFP1 from R. sativus that suppresses fungal growth (Terras et al. 1995) and chimeric chit42 from T. atroviride, which is expected to play a key role in biocontrol activities against phytopathogenic fungi (Limón et al. 2004), with a C-terminal fused chitin-binding domain from Serratia marcescens that facilitates chitinase binding (Hardt and Laine 2004) against S. sclerotiorum in B. napus. The novelty of this study is to introduce different fungal pathogen-resistant genes in canola to create new resistance sources and is likely to reduce dependence on chemical fungicides.
Materials and methods
Enzymes and chemicals
All chemicals, culture media, plant growth regulators and antibiotics were purchased from Merck (Darmstadt, Germany) and Sigma (USA) at the highest purity available, unless stated otherwise. Restriction enzymes and other DNA-modifying enzymes were obtained from Roche (Applied Science GmbH, Mannheim, Germany) and Fermentas (Burlington, Canada).
Plant material, microorganisms and growth conditions
Canola (B. napus) R line Hyola 308, was used as the receptor, which was kindly provided by the Oilseed Research and Development Co., Tehran, Iran. Sclerotinia sclerotiorum was kindly provided by H. Afshari-Azad, Iranian Research Institute of Plant Protection, Tehran, Iran. The fungal strain was grown on potato dextrose agar (PDA) medium (potatoes, infusion 200 g/L, dextrose 20 g/L and agar 15 g/L) at 22 °C for 24 h and potato dextrose broth (PDB) medium used for liquid culturing of the fungal mycelia (Ayers et al. 1976). Escherichia coli DH5α was used in all molecular experiments and A. tumefaciens strain LBA4404 was used for the plant transformation procedure. Bacteria were grown in Luria–Bertani (LB) medium (Bacto Tryptone 10 g/L, Bacto Yeast Extract 5 g/L, NaCl 10 g/L) at appropriate temperatures (37 °C for E. coli and 28 °C for A. tumefaciens) with shaking (150 rpm). Media were supplemented, when required, with ampicillin and kanamycin (Sigma, 100 and 50 μg/mL), respectively.
Plasmid construction
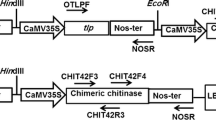
Plasmid pJETMZ1 (containing chimeric chit42 from T. atroviride with a C-terminal fused chitin-binding domain from S. marcescens) was digested with EcoRI restriction enzyme and subsequently ligated into the homologous site in plasmid pBINZP1 (containing Rs-AMP1 cDNA from R. sativus). The recombinant plasmid pBINZP2 contains, within the T-DNA region, the neomycin phosphotransferase II (nptII) gene as a selectable marker that is a kanamycin-resistant gene for plant selection; Rs-AMP1 and chimeric chit4 genes. The nptII gene is regulated by the nopaline synthase (NOS) promoter and terminator; the Rs-AMP1 and the chimeric chit42 are regulated by the cauliflower mosaic virus 35S promoters (CaMV 35S) and terminated by the NOS terminators (Fig. 1). The recombinant plasmid was then mobilised into Agrobacterium by the freeze–thaw method.
Schematic representation of the T-DNA region of plasmid pBINZP2. The position of recognition sites of restriction enzymes is indicated. RB right border; LB left border; CaMV35S-P cauliflower mosaic virus 35S promoter; NOS-P and NOS-T nopaline synthase promoter and terminator, respectively; nptII neomycin phosphotransferase II
Preparation of the explants and bacterial strain for transformation
Seeds were surface sterilised by treating with 70 % ethanol for 2 min and 0.1 % (v/v) sodium hypochlorite for 10 min, and rinsed three times with sterile distilled water. The seeds were then germinated on 1/2 × Murashige and Skoog (MS; Murashige and Skoog 1962) medium and incubated in the presence of light for 5 days. After germination, the cotyledonary petioles were cut and placed on the MS solid medium with 3.5 mg/L benzylaminopurine (BAP) for pre-culture. After 2 days, the explants were used for transformation. Single colonies of the A. tumefaciens-harbouring strain were grown in the LB medium supplemented with 50 mg/L kanamycin and allowed to grow overnight at 28 °C with constant shaking (180 rpm) to mid-log phase. The bacterial culture was then transferred to a fresh medium and cultivated till OD600 = 0.4 with liquid medium. The bacterial cells were collected by centrifugation (6000 rpm for 5 min) and re-suspended in 1/2 × MS medium for the subsequent inoculation step.
Canola transformation and selection procedure
The explants were immersed in the bacterial suspension for 5 min with constant shaking and placed on sterile filter paper to remove the excessive moisture and then placed on the MS solid medium with 3.5 mg/L BAP in Petri dishes for co-cultivation at 25 °C for 3 days in the dark. After co-cultivation, the explants were washed with the sterile water containing 200 mg/L cephatoxime to inhibit the growth of A. tumefaciens attached to the explants and then transferred to the MS solid medium with 3.5 mg/L BAP, 8 mg/L kanamycin and 200 mg/L cephatoxime. After shoot initiation, the explants were transferred to MS solid medium with 15 mg/L of kanamycin and 200 mg/L of cephatoxime. The regenerated shoots (about 3 cm in length) were excised from the explants and transferred to MS solid medium with 2 mg/L of 3-indolebutyric acid (IBA) and 200 mg/L of cephatoxime for rooting. All the above media contained 3 % (w/v) sucrose and 8 g/L agar. The pH of the media was adjusted to 5.6–5.8 prior to the addition of agar and autoclaving at 121 °C for 20 min. All the explants were cultured at 25 ± 2 °C and 16 h of day time with a light intensity of 2000 lux.
PCR analysis of the transgenic canola
The leaf material from the transgenic and non-transgenic canola was harvested, lyophilised and ground into a fine powder for the extraction of genomic DNA. Polymerase chain reaction (PCR) amplification was used for initial evidence of the presence of transgene in the putative transgenic plants. DNA fragments containing the defensin and chimeric chitinase genes were amplified by PCR using the genomic DNA and RDeff/NOSR primers for amplifying defensin and chit42F4/NOSR primers for amplifying chimeric chitinase (Table 1). PCR was carried out as follows: an initial denaturation at 94 °C for 15 min, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. The resulting PCR products were separated by electrophoresis on 1 % (w/v) agarose gel.
Chitinase activity assay
Young leaves from putative transgenic lines as well as untransformed canola plants were frozen in liquid nitrogen and ground to fine powder. The soluble proteins were then extracted in 50 mM of sodium acetate buffer (pH 7.0). In this assay, chitinase activity was generally measured in the reaction mixture (total 500 mL) containing colloidal chitin as a substrate (3.8 mg) and the crude of enzymes from transgenic plants (20 μg/mL). The reaction was performed at 37 °C for 1 h and then centrifuged at 6000 rpm for 5 min. The supernatant was boiled with 100 μL of potassium tetra borate buffer for 3 min. Then, 3 mL of DMAB reagent [10 g of dimethylaminobenzaldehyde in 100 mL of glycial acetic acid (87.5 % v/v) and 10 N chloridric acid (12.5 % v/v)] was added to the reaction, incubated at 37 °C for 20 min and the amount of N-acetylglucosamine (GLcNAc) produced in the supernatant was determined by the method described by Zeilinger et al. (1999), using GLcNAc as a standard. One unit of enzyme activity was defined as the amount of enzyme that catalyses the release of 1 μmol GLcNAc in 60 min at 37 °C. The assay for each sample was performed three times. The total soluble protein concentration was measured according to Bradford (1976), using bovine serum albumin (BSA) as a standard.
Southern blot analysis
Genomic DNA was extracted from fresh leaves of putative transgenic plants and untransformed control plants by the cetyltrimethylammonium bromide (CTAB) method. PCR-positive plants and untransformed control plants were analysed by Southern blot analysis to confirm the integration of the introduced genes. Genomic DNA (20 μg) was digested with HindIII (Fig. 1). The digested genomic DNAs were fractioned on 0.8 % (w/v) agarose gels, transferred onto a nylon membrane (Amersham Hybond NTM+; Amersham International plc, Amersham, UK) and hybridised to the Dig-dUTP labelled CaMV35S promoter probe. A partial internal fragment (631 bp in size) was obtained from PCR amplification by using CaMV35SF/CaMV35SR primers (Table 1) and plasmid pBINZP2 containing the CaMV35S promoter as a template, which was subjected to DIG DNA labelling (Roche Applied Science GmbH, Mannheim, Germany) and used as a probe in hybridisation experiments.
SYTOX Green uptake assay
The ability of defensin gene in transgenic canola to cause plasma membrane permeabilisation was measured by the uptake of SYTOX Green (Invitrogen Corp., Carlsbad, CA, USA). An agar disc (5 mm in diameter) of S. sclerotiorum, which was derived from the fungi in an actively growing state supplemented with 750 μL half-strength PDB, 5 mM MgCl2 and equal aliquots (45–50 μg) of the crude protein plants, extracted with 50 mM Tris-HCl (pH 8.8), from transgenic plants was incubated at 28 °C with shaking (100 rpm) for 24 h. Crude protein from non-transgenic canola plant was used as a control. After incubation, the samples were washed with 0.1 M Tris-HCl (pH 8) and stained with 0.2 μM SYTOX Green at 28 °C for 6 h, with periodic agitation. Images were captured with a fluorescence microscope (Zeiss, Germany) using an excitation wavelength of 460–490 nm.
Radial diffusion assay
An agar disc (5 mm in diameter) containing S. sclerotiorum, which was derived from the fungi in an actively growing state, previously cultured on PDA, was placed at the centre of Petri dishes (100 × 15 mm) containing 25 mL PDA. The plates were then incubated at 28 °C. After incubation for 48 h, wells were subsequently punched into the agar at a distance of 1 cm away from the rim of the mycelia colony. Equal aliquots (45–50 μg) of the crude protein, extracted with 50 mM Tris-HCl (pH 8.8), from transgenic plants containing defensin and chimeric chitinase were added to the wells. Crude protein from non-transgenic canola plant was used as a control. The plates were incubated for a further 24 h, after which the inhibition of hyphal extension around the crude protein was observed. The inhibition rate (%) was calculated as follows: [(the distance between the control hyphal edges to the centre − the distance between the treated hyphal edges to the centre)/the distance between the control hyphal edges to the centre in the control] × 100 %. The assay for each sample was performed three times.
Progeny segregation
Segregation of the transgenes in the canola progeny was evaluated from the expression of the kanamycin resistance gene. A number of 50–60 seeds of the T2 transgenic lines were cultured on MS solid medium with 30 mg/L kanamycin for 10 days. The green seedlings were considered resistant, whereas non-germinated/purple seedlings were considered sensitive. The events having only green seedlings were considered as homozygous plants, whereas the events having non-germinated or purple seedlings besides green seedlings were considered as hemizygous plants. Also, the events having only non-germinated or purple seedlings were considered as non-transgenic plants.
Greenhouse assay for resistance to S. sclerotiorum
The bioassay was based on the inoculation of leaves on 30-day-old intact plants. For this purpose, a fully expanded leaf of normal colour and shape was selected from each plant and marked for inoculation. The inoculation site for each selected leaf was the first one-third from the leaf tip. Leaves were inoculated with 4-mm-diameter agar plugs containing hyphal of S. sclerotiorum which had been transferred from the margins of 3-day-old cultures. The mycelium-containing surface of the plugs was laid on the adaxial side of leaves. Plugs of 1/4 × PDA without the fungal mycelium were used on the control plants. All pots were immediately covered with transparent plastic bags to keep the humidity high. The experiment was established in the evening to provide dark conditions in the greenhouse. A completely randomised experimental design with three replications was used. Infection progress was measured as the diameter of necrotic areas at the inoculation points after 72 h of incubation.
Statistical analysis
Statistical differences were assessed based on analysis of variance (ANOVA) using SPSS (version 15, USA). Differences were considered significant at a probability level of p < 0.05.
Results
Transformation, selection and regeneration of transgenic plants
In order to construct a double expression vector harbouring defensin and chimeric chitinase genes, the complete expression cassette of chimeric chit42 gene from the pJETMZ1 plasmid was subcloned into the EcoRI site, downstream of the complete expression cassette of the defensin gene in the pBINZP1 plasmid. The recombinant plasmid (pBINZP2) was then mobilised into the A. tumefaciens (LBA4404) and, subsequently, was transformed to the cotyledonary petioles of the B. napus, R line Hyola 308. A total of 230 plantlets from 410 co-cultured explants (56 %) showed regeneration on selection medium containing 8 mg/L kanamycin and as much as 39 % (89 out of 230) of plantlets remained green on selection medium containing 15 mg/L kanamycin. A number of 71 elongated shoots were successfully rooted and then transferred to the soil and let flower and set seeds under the greenhouse conditions (Fig. 2).
Production of transgenic canola plants harbouring Rs-AFP1 and chimeric chit42 genes. a Co-cultivated cotyledonary petioles with Agrobacterium tumefaciens harbouring pBINZP2. b Regenerated shoots in the medium containing 8 mg/L kanamycin. c Plantlet with elongated shoot and induced roots. d Acclimated plantlet in non-aseptic environment. e, f Putative transgenic plant with flowers and pods in the greenhouse
Molecular analysis of the transgenic plants
The kanamycin-resistant plants were initially screened by PCR for the presence of defining and chimeric chitinase genes. Twenty putative transgenic lines showed amplification of a 500-bp fragment for Rs-AMP1 and a 900-bp fragment for chimeric chit42 using appropriate primers (Fig. 3). The successful introduction of these genes was calculated as 4.8 % transformation efficiency. No amplification was detected in wild-type plants. A set of virG primers (Table 1) was used for the detection of Agrobacterium contamination if there were any that might have escaped the selection. PCR of transgenic plants DNA with virG primers showed no band (data not shown).
a Polymerase chain reaction (PCR) analysis of genomic DNA from putative pBINZP2-transformed plants in the T0 generation using Rdeff/NOSR primers; lanes 1–20 transformed plants; C + positive control (pBINZP2 plasmid); WT wild-type plant (negative control); M molecular marker. b PCR analysis of genomic DNA from putative pBINZP2-transformed plants in the T0 generation using chit42F4f/NOSR primers; lanes 1–20 transformed plants; C + positive control (pBINZP2 plasmid); WT wild-type plant (negative control); M molecular marker
Chitinase activity assay
The chitinase activity from leaf tissues of all PCR-positive transgenic plants was measured in the presence of colloidal chitin. The specific enzyme activity of transgenic lines varied from 6.228 ± 0.38 to 0.63 ± 0.23 U/μg (pne unit of enzyme activity was defined as the amount of enzyme that catalyses the release of 1 μmol GLcNAc in 60 min at 37 °C). Five transgenic lines showing the highest chitinase activity, T20 (6.228 ± 0.38 U/μg), T4 (5.527 ± 0.36 U/μg), T10 (4.798 ± 0.074 U/μg), T2 (4.404 ± 0.22 U/μg) and T15 (3.976 ± 0.19 U/μg), were selected for checking the copy number of transgenes through Southern blot hybridisation.
Southern blot analysis
Integration and the copy number of transgenes in five PCR-positive plants with the highest chitinase activity (T20, T4, T10, T2 and T15) were defined through Southern blot hybridisation. Genomic DNA prepared from the mentioned transgenic lines were digested with HindIII. The results of Southern blot hybridisation with the CaMV35S promoter probe indicated that two of the five tested plants had only one copy (T4 and T10), and the rest of the plants had more than one copy, including two copies (T2 and T15) and three copies (T20) (Fig. 4). Also, no hybridisation signal occurred in the non-transgenic control plants. The lines with only one copy of inserted T-DNA were selected for further analyses.
Southern blot analysis of HindIII-digested genomic DNA from representative pBINZP2-transformed plants (PCR-positive plants having the highest enzyme activity) in the T0 generation; lane 1 transgenic line T4; lane 2 transgenic line T20; lane 3 transgenic line T2; lane 4 transgenic line T10; lane 5 transgenic line T15; C + positive control (pBINZP2 plasmid); WT wild-type plant (negative control); M molecular marker
Radial diffusion assay
The antifungal activity of expressed defensin and chimeric chitinase in single copy transgenic lines in the T0 generation were examined through a radial diffusion assay. The growth inhibition ability of the extracted proteins from leaves of the T4 and T10 transgenic lines were tested on the actively growing S. sclerotiorum, a phytopathogenic fungus. The results indicated that the crude protein from T4 and T10 showed remarkable inhibition against hyphal growth of fungus compared to wild-type plant. The growth inhibition rate of transgenic lines T4 and T10 were obtained as 49 % and 47 %, respectively (Fig. 5).
SYTOX Green uptake assay
The ability of defensin gene in transgenic lines (T4 and T10) to cause plasma membrane permeabilisation of S. sclerotiorum was measured by the SYTOX Green uptake assay. This study showed that the total protein from T4 and T10 induced fungal membrane permeability and uptake of SYTOX Green, but the total protein from wild-type plant did not change the fugal plasma membrane permeability and, so, no uptake of SYTOX Green occurred (Fig. 6).
SYTOX Green uptake assay of S. sclerotiorum hyphae in the presence of total protein from transgenic plants harbouring pBINZP2.a White light images. b Fluorescent images; T4 fungal hyphae after treatment with total protein from transgenic line T4; T10 fungal hyphae after treatment with total protein from transgenic line T10; WT fungal hyphae after treatment with total protein from wild-type plant; scale bar = 20 μm
Greenhouse assay for resistance to S. sclerotiorum
Transgenic lines (T4 and T10) were permitted for self-pollination to produce the T1 and T2 generations. T2 seeds were then cultured on medium containing 30 mg/L kanamycin to select homozygous plants. The events having 100 % green seedlings were considered as homozygous plants, which were transferred to the soil to evaluate the reaction of intact plants to S. sclerotiorum. After 72 h of inoculation, all leaves showed symptoms of necrosis and grey-coloured lesions. The lesions had a mostly necrotic soft and ovoid-shaped expansion. The results revealed that there was a remarkable difference between transgenic lines and wild-type plant with respect to lesion size (Fig. 7). The lesion diameter for T4, T10 and wild-type plants were measured as 8.85 ± 1.17 mm, 9.53 ± 1.15 mm and 23.4 ± 1.12 mm, respectively.
Discussion
Genetic engineering as an important and alternative approach in the control of fungal pathogens has facilitated the integration of beneficial genes into plants. There are several reports on enhanced resistance against fungal pathogens by transgenic crop plants expressing defensins and chitinases either alone or in combination with other genes, e.g. expression of a defensin from R. sativus (Rs-AFP2) in wheat against F. graminearum and R. cerealis (Li et al. 2011), a defensin from Dahlia merckii (Dm-AMP1) and Rs-AFP2 separately in rice against Magnaporthe oryzae and R. solani (Jha et al. 2009; Jha and Chattoo 2010), bean chitinase in cotton against V. dahlia (Tohidfar et al. 2005), co-expression of a barley class II chitinase and type I ribosome inactivating protein in B. juncea against A. brassicae (Chhikara et al. 2012), pyramiding chitinase from Streptomyces griseus strain and defensin from Wasabia japonica in potato against F. oxysporum and A. solani (Khan et al. 2014).
In this study, a binary expression vector carrying defensin and chimeric chit42 (pBINZP2) was introduced to B. napus via the Agrobacterium-mediated method. The putative transgenic plants were initially screened by PCR for the presence of the two transgenes with 4.8 % transformation efficiency, which is similar to that also reported in B. napus (Liu et al. 2011; Wang et al. 2005) and other Brassica species, B. rapa and B. juncea (Cho et al. 2001; Das et al. 2006).
The chitinase activity of chimeric chit42 in all PCR-positive transgenic plants were measured in the presence of colloidal chitin and showed a range from 0.63 ± 0.23 U/μg to 6.228 ± 0.38 U/μg, which might be due to the site effects of integration, gene silencing, the extent of methylation in the transgene loci and the inherent mechanisms of plants against foreign DNA invasion and expression (Srivastava et al. 1996; Demeke et al. 1999; Alvarez et al. 2000; De Buck et al. 2004).
Five transgenic lines showing the highest enzyme activity were selected for checking the copy number of transgenes through Southern blot hybridisation. According to the results, the genes were integrated into all of the selected PCR-positive plants. In addition, two transgenic lines (T4 and T10) had one copy of the transgene. Southern analysis showed a dissimilar banding pattern, indicating that the transgenic plants originated from independent transformation events. As an increased copy number has often been associated with transgene silencing (Finnegan and McElroy 1994), the lines with only one copy of the inserted T-DNA were selected for further assays.
In order to check the antifungal ability of Rs-AFP1 and chimeric chit42, the extracted protein from single copy transgenic lines (T4 and T10) were evaluated by a radial diffusion assay. The results showed that the constitutive expression of these genes was sufficient to inhibit the growth of S. sclerotiorum as compared to non-transformed plants. The results of several in vitro inhibition assays also revealed that defensins and chitinases exhibited inhibitory activity against a broad range of fungal pathogens (Tohidfar et al. 2005; Jha et al. 2009; Jha and Chattoo 2010; Picart et al. 2012; Chhikara et al. 2012).
Further evidence for enhanced resistance in transgenic lines comes from the SYTOX Green uptake assay. SYTOX Green is a high affinity nucleic acid stain that fluoresces upon nucleic acid binding and penetrates cells with compromised plasma membranes (Broekaert et al. 1995; Matsuzaki et al. 1997). The results from the SYTOX Green uptake assay in the current study confirmed the ability of extracted proteins from transgenic lines (T4 and T10) to permeate the fugal pathogen plasma membrane. The use of SYTOX Green staining in the membrane permeabilising action of plant defensins has been reported previously, including the study of Nicotiana alata defensin on F. oxysporum (van der Weerden et al. 2008), Medicago sativa defensin on F. graminearum (Sagaram et al. 2011) and Picea glauca defensin on B. cinerea, F. oxysporum and V. dahliae (Picart et al. 2012).
Transgenic lines (T4 and T10) were then let to self-pollinate to produce the T2 generation. The greenhouse assay was performed on the homozygous plants by challenging with S. sclerotiorum. The results revealed that overexpression of these genes in transgenic canola improved resistance to this fungal pathogen, as compared to that of untransformed plants. Similar reports on enhanced resistance against fungal pathogen in greenhouse texts by transgenic plants expressing defensins and chitinases either alone or in combination with other proteins have been documented (Schaefer et al. 2005; Tohidfar et al. 2005; Jha et al. 2009; Portieles et al. 2010; Chhikara et al. 2012; Huang et al. 2013).
In conclusion, the results of the current study indicate that the simultaneous deployment of different fungal pathogen-resistant genes, Rs-AFP1 and chimeric chit42, enhances resistance against sclerotinia stem rot disease in canola, and is likely to reduce the costs, heath risks and environmental concerns of using chemical fungicides. These transgenic lines (T4 and T10) demonstrate a promising potential for the development of a variety of canola lines with enhanced resistance against S. sclerotiorum and they may also provide a potential germplasm source for enhanced resistance to some other important fungal pathogens.
References
Alvarez ML, Guelman S, Halford NG, Lustig S, Reggiardo MI, Ryabushkina N, Schewry P, Stein J, Vallejos RH (2000) Silencing of HMW glutenins in transgenic wheat expressing extra HMW subunits. Theor Appl Genet 100:319–327
Ayers AR, Ebel J, Valent B, Albersheim P (1976) Host–pathogen interactions: X. Fractionation and biological activity of an elicitor isolated from the mycelial walls of Phytophthora megasperma var. sojae. Plant Physiol 57:760–765
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Broekaert WF, Terras FRG, Cammue BPA, Osborn RW (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108:1353–1358
Chhikara S, Chaudhury D, Dhankher OP, Jaiwal PK (2012) Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell Tissue Organ Cult 108:83–89
Cho HS, Cao J, Ren JP, Earle ED (2001) Control of Lepidopteran insect pests in transgenic Chinese cabbage (Brassica rapa ssp. pekinensis) transformed with a synthetic Bacillus thuringiensis cry1C gene. Plant Cell Rep 20:1–7
Das B, Goswami L, Ray S, Ghosh S, Bhattacharyya S, Das S, Majumder AL (2006) Agrobacterium-mediated transformation of Brassica juncea with a cyanobacterial (Synechocystis PCC6803) delta-6 desaturase gene leads to production of gamma-linolenic acid. Plant Cell Tissue Org Cult 86:219–231
De Buck S, Windels P, De Loose M, Depicker A (2004) Single-copy T-DNAs integrated at different positions in the Arabidopsis genome display uniform and comparable beta-glucuronidase accumulation levels. Cell Mol Life Sci 61:2632–2645
de las Mercedes Dana M, Pintor-Toro JA, Cubero B (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 142:722–730
Demeke T, Hucl P, Båga M, Caswell K, Leung N, Chibbar RN (1999) Transgene inheritance and silencing in hexaploid spring wheat. Theor Appl Genet 99:947–953
Finnegan J, McElroy D (1994) Transgene inactivation: plants fight back! Nat Biotechnol 12:883–888
Ganesan M, Bhanumathi P, Ganesh Kumari K, Lakshmi Prabha A, Song PS, Jayabalan N (2009) Transgenic Indian cotton (Gossypium hirsutum) harboring rice chitinase gene (Chi II) confers resistance to two fungal pathogens. Am J Biochem Biotechnol 5:63–74
Graham LS, Sticklen MB (1994) Plant chitinases. Can J Bot 72:1057–1083
Hardt M, Laine RA (2004) Mutation of active site residues in the chitin-binding domain ChBDChiA1 from chitinase A1 of Bacillus circulans alters substrate specificity: use of a green fluorescent protein binding assay. Arch Biochem Biophys 426:286–297
Huang X, Wang J, Du Z, Zhang C, Li L, Xu Z (2013) Enhanced resistance to stripe rust disease in transgenic wheat expressing the rice chitinase gene RC24. Transgenic Res 22:939–947
Jha S, Chattoo BB (2010) Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transgenic Res 19:373–384
Jha S, Tank HG, Prasad BD, Chattoo BB (2009) Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res 18:59–69
Khan RS, Sjahril R, Nakamura I, Mii M (2008) Production of transgenic potato exhibiting enhanced resistance to fungal infections and herbicide applications. Plant Biotechnol Rep 2:13–20
Khan RS, Darwish NA, Khattak B, Ntui VO, Kong K, Shimomae K, Nakamura I, Mii M (2014) Retransformation of marker-free potato for enhanced resistance against fungal pathogens by pyramiding chitinase and wasabi defensin genes. Mol Biotechnol 56:814–823
Kim JK, Jang IC, Wu R, Zuo WN, Boston RS, Lee YH, Ahn IP, Nahm BH (2003) Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res 12:475–484
Kirkegaard JA, Robertson MJ, Hamblin P, Sprague SJ (2006) Effect of blackleg and sclerotinia stem rot on canola yield in the high rainfall zone of southern New South Wales, Australia. Aust J Agric Res 57:201–212
Kramer KJ, Muthukrishnan S (1997) Insect chitinases: molecular biology and potential use as biopesticides. Insect Biochem Mol Biol 27:887–900
Kumar V, Parkhi V, Kenerley CM, Rathore KS (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230:277–291
Li Z, Zhou M, Zhang Z, Ren L, Du L, Zhang B, Xu H, Xin Z (2011) Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct Integr Genomics 11:63–70
Limón MC, Lora JM, García I, de la Cruz J, Llobell A, Benítez T, Pintor-Toro JA (1995) Primary structure and expression pattern of the 33-kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum. Curr Genet 28:478–483
Limón MC, Chacón MR, Mejías R, Delgado-Jarana J, Rincón AM, Codón AC, Benítez T (2004) Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl Microbiol Biotechnol 64:675–685
Liu M, Sun ZX, Zhu J, Xu T, Harman GE, Lorito M (2004) Enhancing rice resistance to fungal pathogens by transformation with cell wall degrading enzyme genes from Trichoderma atroviride. J Zhejiang Univ Sci 5:133–136
Liu H, Guo X, Naeem MS, Liu D, Xu L, Zhang W, Tang G, Zhou W (2011) Transgenic Brassica napus L. lines carrying a two gene construct demonstrate enhanced resistance against Plutella xylostella and Sclerotinia sclerotiorum. Plant Cell Tissue Organ Cult 106:143–151
Lorito M, Woo SL, Garcia I, Colucci G, Harman GE, Pintor-Toro JA, Filippone E, Muccifora S, Lawrence CB, Zoina A, Tuzun S, Scala F (1998) Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc Natl Acad Sci U S A 95:7860–7865
Matsuzaki K, Yoneyama S, Fujii N, Miyajima K, Yamada K, Kirino Y, Anzai K (1997) Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry 36:9799–9806
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Ntui VO, Azadi P, Thirukkumaran G, Khan RS, Chin DP, Nakamura I, Mii M (2011) Increased resistance to fusarium wilt in transgenic tobacco lines co-expressing chitinase and wasabi defensin genes. Plant Pathol 60:221–231
Olli S, Kirti PB (2006) Cloning, characterization and antifungal activity of defensin Tfgd1 from Trigonella foenum-graecum L. J Biochem Mol Biol 39:278–283
Picart P, Pirttilä AM, Raventos D, Kristensen HH, Sahl HG (2012) Identification of defensin-encoding genes of Picea glauca: characterization of PgD5, a conserved spruce defensin with strong antifungal activity. BMC Plant Biol 12:180
Portieles R, Ayra C, Gonzalez E, Gallo A, Rodriguez R, Chacón O, López Y, Rodriguez M, Castillo J, Pujol M, Enriquez G, Borroto C, Trujillo L, Thomma BP, Borrás-Hidalgo O (2010) NmDef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high-level pathogen resistance under greenhouse and field conditions. Plant Biotechnol J 8:678–690
Sagaram US, Pandurangi R, Kaur J, Smith TJ, Shah DM (2011) Structure–activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS One 6(4):e18550
Saitoh H, Kiba A, Nishihara M, Yamamura S, Suzuki K, Terauchi R (2001) Production of antimicrobial defensin in Nicotiana benthamiana with a potato virus X vector. Mol Plant Microbe Interact 14:111–115
Schaefer SC, Gasic K, Cammue B, Broekaert W, van Damme EJ, Peumans WJ, Korban SS (2005) Enhanced resistance to early blight in transgenic tomato lines expressing heterologous plant defense genes. Planta 222:858–866
Srivastava V, Vasil V, Vasil IK (1996) Molecular characterization of the fate of transgenes in transformed wheat (Triticum aestivum L.). Theor Appl Genet 92:1031–1037
Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7:573–588
Thomma BPJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216:193–202
Tohidfar M, Mohammadi M, Ghareyazie B (2005) Agrobacterium-mediated transformation of cotton (Gossypium hirsutum) using a heterologous bean chitinase gene. Plant Cell Tissue Organ Cult 83:83–96
van der Biezen EA (2001) Quest for antimicrobial genes to engineer disease-resistant crops. Trends Plant Sci 6:89–91
van der Weerden NL, Lay FT, Anderson MA (2008) The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J Biol Chem 283:14445–14452
Wang J, Chen Z, Du J, Sun Y, Liang A (2005) Novel insect resistance in Brassica napus developed by transformation of chitinase and scorpion toxin genes. Plant Cell Rep 24:549–555
Wong JH, Ng TB (2005) Sesquin, a potent defensin-like antimicrobial peptide from ground beans with inhibitory activities toward tumor cells and HIV-1 reverse transcriptase. Peptides 26:1120–1126
Zeilinger S, Galhaup C, Payer K, Woo SL, Mach RL, Fekete C, Lorito M, Kubicek CP (1999) Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet Biol 26:131–140
Acknowledgments
This research was financially supported by the National Institute of Genetic Engineering and Biotechnology (NIGEB) of I. R. Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest.
Additional information
Communicated by: Andrzej Górny
Rights and permissions
About this article
Cite this article
Zarinpanjeh, N., Motallebi, M., Zamani, M.R. et al. Enhanced resistance to Sclerotinia sclerotiorum in Brassica napus by co-expression of defensin and chimeric chitinase genes. J Appl Genetics 57, 417–425 (2016). https://doi.org/10.1007/s13353-016-0340-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-016-0340-y