Abstract
Alternaria leaf spot caused by Alternaria brassicae, or A. brassicola, is one of the major fungal diseases of Brassica juncea (Indian mustard). To develop resistance against this fungal disease, the barley antifungal genes class II chitinase (AAA56786) and type I ribosome inactivating protein (RIP; AAA32951) were coexpressed in Indian mustard via Agrobacterium-mediated transformation. The stable integration and expression of transgenes in T0 plants were confirmed by Southern blot and Western analysis. The transgenic lines showing inheritance in Mendalian fashion (3:1) were further evaluated by in vitro studies and under greenhouse conditions for resistance to the A. brassicae fungal pathogen. The transgenic plants showed up to 44% reduction in A. brassicae hyphal growth in in vitro antifungal assays. In green house screening, the transgenic plants sprayed with A. brassicae spores showed resistance through delayed onset of the disease and restricted number, size, and expansion of lesions as compared to wild type plants. These results indicate that the expression of chitinase and RIP from a heterologous source in B. juncea provide subsequent protection against Alternaria leaf spot disease and can be helpful in increasing the production of Indian mustard.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica juncea (Indian mustard) is a major oilseed crop of the Brassicaceae family. It is cultivated in over 6 million hectares of land on the Indian subcontinent as well as in many other countries around the world. Mustard alone fulfils nearly 27.1% of vegetable oil requirements of India and is estimated to meet 41% of the demand by 2020 (Yadav and Singh 1999). Due to increased demand for edible oil and biofuels, the cultivation of oilseeds crops has gained significant importance (Kumar 1999). Unfortunately, the productivity of this crop has been hampered by several biotic and abiotic factors (Grover and Pental 2003). One of the major constraints affecting the productivity of mustard is the Alternaria leaf spot disease caused by Alternaria brassicae and A. brassicicola (Singh 1990; Meena et al. 2010). The estimated yield loss has been reported up to be as high as 46% (Kolte et al. 1987). This fungus produces symptoms throughout all parts of the plant, i.e. on cotyledons, leaves, leaf petiole, stem, inflorescence, siliquae and seeds. The disease causes the formation of dark colored necrotic lesions with concentric rings on leaves and pods. In severe cases, the vitality of the crop is badly affected and the siliquae fails to set healthy seeds. Resistance to Alternaria blight is not available in the cultivated Brassica species (Sharma et al. 2002). Wild crucifers such as Sinapis alba (Kolte et al. 1991), Camelina sativa, Capsella bursa-pastoris (Conn et al. 1988), and Neslia paniculata (Tewari and Conn 1993) have shown a high degree of resistance to A. brassicae, however transfer of disease resistance from wild sources through conventional breeding is very difficult due to sexual incompatibility with cultigens and limited gene exchange in hybrids involving resistant wild species. Consequently, conventional breeding programs have not been successful in terms of conveying fungal resistance (Yadav and Singh 1999). In light of these difficulties, a genetic engineering approach was taken to transfer antifungal proteins from different plant species to B. juncea.
Pathogenesis-related (PR) proteins are well known to provide resistance to fungal infection in various cases. Hydrolytic enzymes, such as chitinases and β-1,3-glucanases have been shown to inhibit fungal growth by degrading structural polysaccharides in fungal cell walls (Broglie et al. 1991; Leah et al. 1991; Zhu et al. 1994; Jach et al. 1995). Ribosome inactivating proteins (RIP) are a group of plant proteins that possess 28S rRNA N-glycosidase activity, which leads to the inactivation of foreign ribosomes in distantly related species and in other eukaryotes, including fungi (Stirpe et al. 1992). The constitutive expression of a single antifungal gene has been shown to enhance resistance against fungal pathogens in several crops such as tobacco, wheat, maize, rice, and B. juncea (Logemann et al. 1992; Jach et al. 1995; Bliffeld et al. 1999; Datta et al. 2001; Bieri et al. 2003; Mondal et al. 2003; Shin et al. 2008; Girhepuje and Shinde 2011). However, if enzymes from two gene families are combined, they can act synergistically with improved effect on fungus infection. When a class II chitinase and a type 1 RIP from barley were coexpressed in tobacco, increased protection against Rhizoctonia solani was demonstrated in vivo (Jach et al. 1995). The combined gene expression also significantly enhanced protection against A. alternata and Botrytis cineria. Furthermore, the combined expression of modified maize RIP and rice basic chitinase in rice significantly enhanced protection against the sheath blight pathogen R. solani (Kim et al. 2003).
In this study, to check the synergistic effect of antifungal genes, barley class II chitinase (AAA56786) and RIP (AAA32951) were used in combination to engineer B. juncea var. Pusa Jaikisan for enhanced protection against the fungus A. brassicae.
Materials and methods
Plant material, bacterial strains and vectors
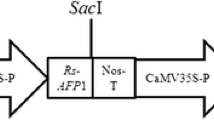
Seeds of a commercially grown cultivar of B. juncea, Pusa Jaikisan, were obtained from the Department of Plant Breeding, CCS Haryana Agricultural University, Hisar, India. The disarmed Agrobacterium tumefaciens strain EHA105 harboring a binary vector pGJ42 (Fig. 1) was used for transformation studies. The plasmid GJ42 contains two antifungal genes, barley chitinase (AAA56786) and RIP (AAA32951) under the control of CaMV35S promoter and a selectable marker gene, neomycin phosphotransferase (nptII) driven by nos promoter. The Agrobacterium culture was grown overnight in liquid YEM (Yeast extract, 1.0 g l−1; mannitol, 10 g l−1; NaCl, 0.1 g l−1; MgSO4·7H2O, 0.2 g l−1; K2HPO4, 0.5 g l−1; pH 7.2) containing 50 mg l−1 kanamycin and 20 mg l−1 rifampicin at 28°C on a rotatory shaker at 200 rpm.
T-DNA region of binary vector pGJ42 used for Agrobacterium-mediated transformation. Abbreviations: RB right border, LB left border, 35S 35Sp promoter and 35St terminator, NOS nopaline synthase promoter or terminator, nptII neomycin phosphotransferase, RIP ribosome inactivating protein, CHI chitinase
Agrobacterium-mediated transformation of B. juncea
The Brassica transformation was done as described by Prasad et al. (2000). Hypocotyls of B. juncea cv. Pusa Jaikisan were used as explants. The explants were pre-cultured on filter paper moistened with liquid MS basal medium (Murashige and Skoog 1962) containing 0.5 μM BA for 36 h. The pre-cultured explants were infected and co-cultivated with A. tumefaciens on MS medium with 2 mg l−1 BA and 2 mg l−1 NAA for 1 day. The transformed explants were selected on medium with 250 mg l−1 cefotaxime and 30 mg l−1 kanamycin (this concentration was chosen based on kanamycin toxicity tests from our preliminary experiments). The shoots were regenerated in two 2 weeks cycles. In the first cycle, the regeneration medium contained 250 mg l−1 cefotaxime and 30 mg l−1 kanamycin was used. In the second cycle, the kanamycin concentration was reduced to 20 mg l−1 for efficient shoot regeneration. The rooted, transformed shoots were established in pots in a greenhouse at 25–28°C with 16 h light and 65% humidity and grown to maturity to collect T1 seeds.
Molecular analysis of transformants
PCR and Southern hybridization
Total genomic DNA was extracted from fresh leaves of putative transformants (T0) and wild type plants using the CTAB (Cetyl trimethyl ammonium bromide) method (Rogers and Bendich 1988). Putative transformants were analyzed by PCR for the presence of the nptII gene. The 0.54 kb coding region of nptII was amplified using 20 bp oligonucleotide primers (Forward 5′-CCACCATGATATTCGGCAAC-3′ and Reverse 5′-GTGGAGGCTATTCGGCTA-3′). To ensure that reagents were not contaminated, DNA from wild type plant was included in the experiments. The amplified products were separated by electrophoresis on 1% (w/v) agarose gel and visualized by staining with ethidium bromide (Sambrook et al. 1989). Five to ten micrograms of DNA isolated from transformed and wild type plants were digested with HindIII, separated on 0.9% (w/v) agarose gel, and blotted on positively charged nylon membrane (Roche Molecular Biochemical). It was then hybridized with dCT-[P32]-labeled probe (PCR amplified nptII gene fragment of 0.75 kb) and checked with autoradiography. The probe was made using random labeling method and Southern hybridization was performed according to Sambrook et al. (1989).
Western analysis
Total protein was extracted from fresh leaves of transformed and wild type plants in a phosphate buffer (pH 7.0) as described by Jach et al. (1995) and estimated using the Bradford method (Bradford 1976). A 20 μg of protein was resolved on 12% SDS–PAGE and transferred to Hybond PVDF membrane (Biometra) using an electro-transfer system (Bangalore Genei, India). The membrane was blocked in Phosphate buffer saline (PBS) with 5% non-fat dried milk (Amresco) at 37°C for 2 h. After blocking, the membrane was incubated with primary antibodies (barley chitinase and RIP, 1:500 dilutions) for 2 h at room temperature followed by three washes with PBST (PBS with 0.01% (v/v) Tween 20) for 10 min each. Next, alkaline phosphatase conjugated secondary antibodies (Goat anti-rabbit IgG, 1:10,000 dilution, Banglore Genei) were added for 1 h. The membrane was again washed with PBST four times for 10 min each at room temperature. The proteins were detected after adding NBT-BCIP (Nitroblue tetrazolium-Bromochloroindolylphosphate) substrate and developing in the dark.
Segregation analysis
To determine the pattern of inheritance of chitinase and RIP genes, T1 seeds from each self-pollinated transformed line were germinated on MS medium containing 30 mg l−1 kanamycin for 2 weeks. The green seedlings were considered as resistant whereas non-germinated/brown seedlings as sensitive. The kanamycin resistant plants of each transgenic line were transferred to soil and further analyzed by semi-quantitative RT-PCR (Qiagen one step RT-PCR kit) in T2 plants.
Evaluation of transgenic plants for resistance to A. brassicae
In vitro antifungal assay
The in vitro inhibitory effect of soluble protein extracted from leaves of kanamycin resistant B. juncea transgenic plants on A. brassicae hyphal growth was assayed as described by Mondal et al. (2003). Wells made on antibiotic assay agar plates (Himedia) were first filled with 40 μl of spore suspension (104 spores ml−1) and incubated overnight at 30°C. After incubation, each well was differently treated with 40 μl each of transgenic leaf extract (100 μg protein/40 μl), autoclaved transgenic leaf extract, protein extraction buffer, and commercial chitinase (1.5 μg/40 μl; Sigma) solution. The plates were incubated at 25°C. The protein from wild type plants (100 μg total protein) and protein extraction buffer (Phosphate buffer, pH-7.0) served as controls. The radial growth of A. brassicae was recorded every 24 h for 5 days. The percent inhibition of hyphal growth was calculated as the ratio of the diameter of the fungal colony in treatment to the diameter of the fungal colony in wild type plants, multiplied by 100.
Greenhouse screening
A. brassicae spore culture (Accession no-ITCC 5095) was obtained from Indian Type culture collection (ITCC), Division of Plant Pathology, IARI, New Delhi. This culture was used as a test organism. The fungal culture was maintained on potato dextrose agar (PDA) medium at 22 ± 1°C. The spore suspension was prepared as described by Kanrar et al. (2002). Four plants (T1) from each of four transgenic lines and wild type were assayed for fungus infection. Two to three month old plants were inoculated with A. brassicae spore suspension adjusted to 104 spores ml−1 using a haemocytometer and 200 ml of spore suspension was sprayed on each plant using an atomizer. The plants were kept in the greenhouse at 25°C with 75–85% relative humidity. The data on lesions per leaf (average of four leaves per plant) and individual lesion diameter (in mm) were recorded 10 days after inoculation. Percent disease protection was calculated as the number of lesions per leaf in control plants minus the number of lesions per leaf in transgenic plants divided by the number of lesions per leaf in control plants, multiplied by 100.
The different disease parameters such as incubation period, latent period, necrotic lesions, lesions size and lesions per leaf were taken to compare the disease reaction of control and transgenic plants. The incubation period was considered to be from the inoculation time to the appearance of pin head/small lesions, whereas the latent period was considered to be up to the appearance of concentric rings in the lesion. Observations were recorded daily for 15 days after inoculation.
Statistical analysis
The experiments were repeated twice. Mean ± SE of diameter of lesions and percentage disease protection were calculated using Microsoft Excel software version 2003. The disease protection in in vitro inhibition and greenhouse assays were analyzed by student T-test. The segregation analysis in the progeny plants was validated by χ2 test.
Results and discussion
Generation of transgenic plants
About 36% pre-cultured explants showed regeneration on selection medium with kanamycin. A total of 15 putative transgenic plants were transferred to soil in the green house, out of which ten survived and grew to maturity as independent lines.
Molecular analysis of transformants
PCR and Southern blot analysis
The putative transgenic plants were initially screened by PCR for the presence nptII gene. 70% of putative transformants of B. juncea showed amplification of the 0.54 kb nptII gene. However, no amplification was detected in wild type plants (Fig. 2a). In order to check the integration and copy number of transgenes in the Brassica genome, seven PCR positive plants were analyzed by Southern blotting. The genomic DNA was digested with HindIII and hybridized with a 0.75 kb nptII probe. Because HindIII cuts the entire 35S-RIP/35S-CHI region of pGJ42 cassette as a 4.0 kb internal fragment, the integration will be confirmed only if the detected band sizes are higher than 4.0 kb. Out of seven plants, five putative transgenic plants showed the presence of one to two bands of size more than 4.0 kb, suggesting the integration of nptII gene at one to two independent positions in the plant genome (Fig. 2b). This showed that these transgenic plants were derived from independent transformation events. The hybridization pattern showed that four plants had a single copy and one had two copies of the T-DNA in their genome. DNA isolated from wild type plants did not hybridize with the nptII probe.
Analysis of transgenic plants by PCR, and Southern and Western blotting. a PCR analysis of control and transgenic plants with nptII marker gene: P plasmid DNA; C DNA from wild type plant; lane 3–8 DNA from transgenic lines. b Southern blot analysis of HindIII digested genomic DNA from independent transgenic lines and probed with the PCR amplified nptII fragment (0.75 kb): C wild type plant DNA digested with HindIII; lane 2–9 DNA from transgenic lines digested with HindIII; B blank lane. c Western analysis of independent transgenic lines with chitinase and RIP specific antibodies: C protein from untransformed plant; lanes 1, 4, 7, 17 transgenic lines protein
Western blotting
The expression of chitinase and RIP proteins in transgenic plants was confirmed by Western blotting (Fig. 2c). The total soluble proteins from four Southern positive transgenic lines showing a single copy T-DNA insertion and wild type plants were immunoblotted and detected using antibodies specific for barley chitinase and RIP. All four transgenic lines showed the expression of both chitinase (28 kD) and RIP (30 kD) proteins. Transgenic line 1 showed the highest expression, whereas, lines 4, 7, and 17 had lower level of chitinase and RIP expression. No expression of the barley chitinase and RIP was detected in the wild type plants.
Segregation analysis
The inheritance of transgenes to the next generation is shown in Table 1. Out of five transgenic lines, four segregated in a 3:1 ratio on kanamycin, while line 9 showed a 2:1 segregation pattern, which may be due to abnormal insertion of transgenes in the plant genome. RT-PCR analysis showed mRNA expression of the nptII gene in two randomly selected kanamycin resistant plants from each of three transgenic lines 1, 7, and 17 (Fig. 3).
Evaluation of transgenic plants for resistance to A. brassicae
In vitro antifungal assay
The crude protein extracted from three independent transgenic mustard lines (1, 7, and 17) were evaluated for their effect on the fungal hyphal growth in vitro. The total water-soluble proteins (100 μg/40 μl) from three independent transgenic plants restricted the fungal hyphal growth ranging from 36 to 44% over the wild type plants (Fig. 4a). Among the three independent lines tested, line 1 showed the maximum suppression of hyphal growth. The inhibition of fungal growth by different transgenic plants was in correlation to the levels of chitinase and RIP expression as revealed by western analysis. There was no such inhibition of fungal growth in the presence of either boiled extract or only extraction buffer. The observed inhibition of hyphal growth of A. brassicae may be due to degradation of chitin, a major structural polysaccharide of growing hyphae, by chitinase and inactivation of fungal ribosomes by RIP (Logemann et al. 1992; Leah et al. 1991).
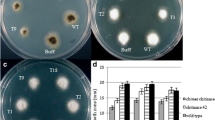
Analysis of inhibition in hyphal growth of A. brassicae in transgenic lines. a Percent inhibition in hyphal growth of A. brassicae by protein extracts of B. juncea trangenic lines overexpressing barley chitinase and RIP proteins and commercial chitinase at 1.5 μg/40 μl (CHI). b The percent protection from A. brassicae by kanamycin resistant T1 plants of transgenic B. juncea under greenhouse bioassay. Data is mean ± SE. Columns with different letters are significantly different at P < 0.05
Greenhouse screening of transgenics
In greenhouse screening, the characteristic lesions formed after spraying A. brassicae spores (104 spores/ml of H2O) were less in number on leaves of transgenic plants (<10 lesions per leaf) compared to wild type plants (>15 lesions per leaf). Individually, transgenic plants showed variability in disease protection (Fig. 4b). The lesions produced by A. brassicae in transgenic plants were restricted to 2.5–4.0 mm in diameter without formation of concentric rings, whereas in non-transgenic plants the average lesion size varied between 10 and 16 mm in diameter with concentric rings (Fig. 5). In transgenic plants, combined antifungal activities of chitinase and RIP proteins prevented the formation of this kind of concentric ring pattern by dissolution of growing hyphae and inhibition of the spread of fungus across the tissue layers. Different host-pathogen systems have shown resistance in the form of reduced lesion size due to restricted growth of fungal pathogens (Bliffeld et al. 1999; Broglie et al. 1991; Jach et al. 1995).
Other parameters such as incubation period (from time of inoculation to appearance of pinhead/small size lesions), latent period (from time of inoculation to appearance of concentric rings in the lesions), and symptoms such as leaf tip drying and necrotic lesions are shown in Table 2 to demonstrate the effect of A. brassicae on the whole plant. The incubation period of A. brassicae was delayed by 3 days in transgenic plants compared to wild type plants. The maximum number of small size lesions reached the concentric ring stage 10 days after inoculation in transgenic plants as compared to wild type plants where it took 4–5 days to reach the concentric ring stage. The latent period was also longer (8 days) in transgenic plants than wild type plants (4 days). It has been shown under field conditions in conventional breeding programs that susceptible rapeseed mustard variety ‘Parkash’ has shorter incubation and latent periods than resistant ‘Towar’ cultivar (Saharan and Kadian 1983). In the present study, transgenic plants showed a significant delay in disease development due to longer incubation and latent periods. Similarly, Kanrar et al. (2002) reported that transgenic B. juncea plants expressing the rubber tree lactin, have exhibited resistance against A. brassicae. A longer incubation period of 120 h in transgenic plants rather than 60 h in control plants was the major factor for delay in disease progression in transgenic plants. The latent period was also longer in transgenic plants as compared to wild type plants under greenhouse conditions. The delay in onset of disease is attributed to the antifungal activity in the transgenic plants. Previously, the constitutive expression of chitinase in B. juncea has shown resistance to Alternaria blight by delaying the onset of disease and reducing the number of lesions (Mondal et al. 2003, 2007). However, the amount of transgenic leaf protein used in in vitro fungal growth inhibition was eight fold higher (400 μg/20 μl) (Mondal et al. 2007) than those used in the present study (100 μg/40 μl). Similarly, Broglie et al. (1991) showed a delay in infection of R. solani in chitinase expressing transgenic tobacco. Since the hydrolytic chitinase enzyme will degrade the fungal cell wall and RIP’s 28S rRNA N-glycosidase activity leads to the inactivation of foreign ribosomes of distantly related species and of the other eukaryotes including fungi, combined expression of chitinase and RIP is expected to even further increase resistance. Jach et al. (1995) reported that combined expression of barley chitinase and RIP in tobacco significantly enhanced protection against the fungal pathogen R. solani compared to the protection level obtained by the expression of only a single barley transgene. In rice, the coexpression of modified RIP and rice basic chitinase provided tolerance to R. solani but failed to provide tolerance to Bipolaris oryzae and Magnaporthe grisea (Kim et al. 2003). Our results are consistent with those reported in previous studies. However, in the present study, without a direct comparison of plants with single transgenes, it is difficult to ascertain whether the combined effect of chitinase and RIP genes is better than either gene alone.
In conclusion, the constitutive co-expression of chitinase and RIP in B. juncea confers protection against Alternaria leaf spot by arresting the growth of the pathogen A. brassicae, and thereby decreases disease severity in the transgenic plants compared to control plants. However, further experiments are needed to evaluate fungal resistance of transgenic lines under field conditions and to test for resistance to other pathogens.
Abbreviations
- BA:
-
6-Benzylaminopurine
- NAA:
-
α-Naphthaleneacetic acid
- MS:
-
Murashige and Skoog
References
Bieri S, Potrykus I, Futterer J (2003) Effects of combined expression of antifungal barley seed proteins in transgenic wheat on powdery mildew infection. Mol Breed 11:37–48
Bliffeld M, Mundy J, Potrykus I, Futterer J (1999) Genetic engineering of wheat for increased resistance to powdery mildew disease. Theor Appl Genet 98:1079–1086
Bradford MM (1976) A rapid and sensitive method for quantitation of proteins using the principle of protein-dye binding. Anal Biochem 72:248–254
Broglie K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais CJ, Broglie R (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194–1197
Conn KL, Tewari JP, Dahiya JS (1988) Resistance to Alternaria brassicae and phytoalexins-elicitation in rapeseed and other crucifers. Plant Sci 56:21–25
Datta K, Tu J, Oliva N, Ona I, Velazhahan R, Mew TW et al (2001) Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci 160:405–414
Girhepuje PV, Shinde GB (2011) Transgenic tomato plants expressing a wheat endochitinase gene demonstrate enhanced resistance to Fusarium oxysporum f. sp. lycopersici. Plant Cell Tissue Org Cult 105:243–251
Grover A, Pental D (2003) Breeding objectives and requirements for producing transgenics for major field crops of India. Curr Sci 84:310–320
Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C (1995) Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J 8:97–109
Kanrar S, Venkateswari JC, Kirti PB, Chopra VL (2002) Transgenic expression of hevein, the rubber tree lectin, in Indian mustard confers protection against Alternaria brassicae. Plant Sci 162:441–448
Kim JK, Jang IC, Wu R, Zuo WN, Boston RS, Lee YH, Ahn P, Nahm BH (2003) Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Trans Res 12:475–484
Kolte SJ, Awasthi RP, Vishwanath (1987) Assessment of yield losses due to Alternaria blight in rapeseed and mustard. Ind Phytopathol 40:209–211
Kolte SJ, Bardoloi DK, Awasthi RP (1991) The search of resistance to major diseases of rapeseed-mustard in India. In: Proceedings of GCJRC 8th international rapeseed congress, Saskatoon, Canada, pp 219–225
Kumar PR (1999) Rapeseed mustard research in India: 21st century strategies. 10th International Rapeseed Congress. Canberra, Australia. http://www.regional.org.au/gcirc/6/634.htm-34k
Leah R, Tommerup H, Svendsen I, Mundy J (1991) Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem 266:1564–1573
Logemann J, Jach G, Tommerup H, Mundy J, Schell J (1992) Expression of a barley ribosome-inactivating protein leads to increased fungal protection in transgenic tobacco plants. Bio/Technology 10:305–308
Meena PD, Awasthi RP, Chattopadhyay C, Kolte SJ, Kumar A (2010) Alternaria blight: a chronic disease in rapeseed-mustard. J Oilseed Brass 1:1–11
Mondal KK, Chatterjee SC, Viswakarma N, Bhattacharya RC, Grover A (2003) Chitinase-mediated inhibitory activity of Brassica transgenic on growth of Alternaria brassicae. Curr Microbiol 47:171–173
Mondal KK, Bhattacharya RC, Koundal KR, Chatterjee SC (2007) Transgenic Indian mustard (Brassica junicea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep 26:247–252
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant 15:473–497
Prasad KVSK, Sharmila P, Kumar PA, Saradhi PP (2000) Transformation of Brassica juncea (L.) Czen. with bacterial codA gene enhances its tolerance to salt stress. Mol Breed 6:489–499
Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoot RA (eds) Plant mol biol manual. Kluwer, Dordrecht, pp 1–11
Saharan GS, Kadian AK (1983) Analysis of components of horizontal resistance in rapeseed and mustard cultivars against Alternaria brassicae. Indian Phytopathol 36:503–507
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sharma G, Kumar VD, Haque A, Bhat SR, Prakash S, Chopra VA (2002) Brassica coenospecies: a rich reservoir for genetic resistance to leaf spot caused by Alternaria brassicae. Euphytica 125:411–417
Shin S, Mackintosh CA, Lewis J, Heinen SJ, Radmer L, Dill-Macky R, Baldridge GD, Zeyen RJ, Muehlbauer GJ (2008) Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J Exp Bot 59:2371–2378
Singh RS (1990) Plant diseases, 6th edn. Oxford and IBH, New Delhi
Stirpe F, Barbieri L, Battelli MG, Soria M, Lappi DA (1992) Ribosome-inactivation proteins from plants: present status and future prospects. Bio/Technology 10:405–412
Tewari JP, Conn KL (1993) Reactions of some wild crucifers to Alternaria brassicae. Bull OILS SROP 16:53–58
Yadav JS, Singh NB (1999) In: Proceedings of the 10th international rapeseed congress, Canberra, Australia
Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ (1994) Enhanced protection against fungal attack by constitutive coexpression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology 12:807–812
Acknowledgments
Authors are grateful to the Max Planck Institute fur Zuchtungsforschung, Germany and Prof. S. Muthukrishnan, USA for providing the gene construct and the barley anti-chitinase and anti-RIP antibodies, respectively. SC is thankful to M. D. University, Rohtak for University Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chhikara, S., Chaudhury, D., Dhankher, O.P. et al. Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae . Plant Cell Tiss Organ Cult 108, 83–89 (2012). https://doi.org/10.1007/s11240-011-0015-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-011-0015-7