Abstract
Transgenic rice (Oryza sativa L. cv. Pusa basmati 1), overexpressing the Rs-AFP2 defensin gene from the Raphanus sativus was generated by Agrobacterium tumefaciens-mediated transformation. Expression levels of Rs-AFP2 ranged from 0.45 to 0.53% of total soluble protein in transgenic plants. It was observed that constitutive expression of Rs-AFP2 suppresses the growth of Magnaporthe oryzae and Rhizoctonia solani by 77 and 45%, respectively. No effect on plant morphology was observed in the Rs-AFP2 expressing rice lines. The inhibitory activity of protein extracts prepared from leaves of Rs-AFP2 plants on the in vitro growth of M. oryzae indicated that the Rs-AFP2 protein produced by transgenic rice plants was biologically active. Transgene expression of Rs-AFP2 was not accompanied by an induction of pathogenesis-related (PR) gene expression, suggesting that the expression of Rs-AFP2 directly inhibits the pathogens. Here, we demonstrate that transgenic rice plants expressing the Rs-AFP2 gene show enhanced resistance to M. oryzae and R. solani, two of the most important pathogens of rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is the staple food crop of over 50% of the world population. Rice blast, caused by Magnaporthe oryzae, is one of the most devastating diseases of rice. M. oryzae is an Ascomycete fungus, produces specialised infection structures known as appressoria that infect aerial tissues and hyphopodia that can infect root tissues (Sesma and Osbourn 2004). The fungus is highly variable and hence, durable resistance through breeding has attained limited success. Sheath blight caused by Rhizoctonia solani, which is a Basidiomycete that reproduces asexually, exists primarily as vegetative mycelium and/or sclerotia, and has a broad host range (Anderson 1982). Sheath blight is another fungal disease to which rice is highly vulnerable (Ou 1985). Attempts to control sheath blight with resistant cultivars have not been successful because good source of resistance are not available. Control through chemical methods significantly increases production cost as well as environmental burden. Thus, transgenic rice with enhanced resistance against rice blast and sheath blight would have a significant advantage in reducing crop losses.
Plants defend against antagonistic microorganisms using an array of antimicrobial compounds (Broekaert et al. 1995). Defensins are a class of evolutionarily and structurally related small, highly basic, cysteine-rich peptides, displaying a broad-spectrum in vitro antifungal activity (Osborn et al. 1995; Thomma et al. 2002). Rs-AFP2 (5.1 kD) a plant defensin from the seeds of Raphanus sativus, interacts with glucopyranosylceramide (GlcCer) present in the plasma membrane of fungal hyphae, leads to increased K+ efflux and Ca2+ influx, membrane potential changes, and exerts antifungal activity against a broad spectrum of plant pathogenic filamentous fungi by causing hyperbranching and growth reduction of the hyphal tips (Thevissen et al. 2004). The IC50 (Inhibitory concentration) of Rs-AFP2 for R. solani was found to be 20 μM (unpublished data). Rs-AFP2 inhibited the growth in vitro of M. oryzae and other phytopathogens at micromolar concentration (Terras et al. 1992). We, therefore, sought to produce transgenic rice plants expressing Rs-AFP2, and evaluate their response to rice blast and sheath blight, two of the most important pathogens of rice.
Materials and methods
Plasmid construction and rice transformation
Rs-AFP2 gene was amplified from the plasmid pFAJ3105 (Francois et al. 2002) and cloned at BamHI site of pAHC17 under maize Ubiquitin (Ub1) promoter (Christensen and Quail 1996), to obtain pUb1-Rs. Plasmid pUb1-Rs was digested and cloned at HindIII site in pCAMBIA-1305.2 (Cambia, Australia) to obtain pUb1-Rs-T. The binary vector (pUb1-Rs-T) was mobilized into Agrobacterium tumefaciens strain LBA4404 harbouring pSB1 (a plasmid that carries virB, virG and virC; Komari et al. 1996). The scutellum-derived rice calli were transformed with Agrobacterium tumefaciens strain LBA4404 (pSB1, pUb1-Rs-T) as described (Hiei et al. 1994). Total DNA from various independent transgenic lines was extracted (Dellaporta et al. 1983) and screened by PCR (using Rs-AFP2 specific primers). The PCR profile was 94°C, 30 s; 50°C, 30 s; 72°C, 1 min, for 30 cycles in a Minicycler (MJ Research, Waltham, MA, USA).
Southern blot analysis
The genomic DNA (10 μg) from non-transformed and T2 transgenic plants was digested with restriction enzymes KpnI/SacI as recommend by the supplier; electrophoresed on 0.8% agarose gels and blotted onto Hybond N+ nylon membrane (Amersham, Buckinghamshire, England). DNA was fixed to the membranes by UV cross-linking, 12 × 104 μJ cm−2 (Spectrolinker™ XL-1000, Spectronics Corporation, USA). Hybridisation probe (283 bp Rs-AFP2 gene) was labeled non-radioactively using Gene Images AlkPhos Direct Labelling and Detection System (Amersham Biosciences, Hong Kong). Hybridization was performed as recommended by supplier (Amersham Biosciences, Hong Kong).
Northern blot analysis
Total RNA was isolated from the leaves of transgenic and control plants as described by Logeman et al. (1987). RNAs (20 μg/sample) were subjected to formaldehyde-containing agarose gel electrophoresis, and transferred to Hybond N+ nylon membrane (Hybond-N, Amersham, Buckinghamshire, England). Hybridisation probe was labeled non-radioactively using Gene Images AlkPhos Direct Labelling and Detection System (Amersham Biosciences, Hong Kong). Hybridization was performed as recommended by supplier (Amersham Biosciences, Hong Kong).
Raising antibodies against Rs-AFP2
Rs-AFP2 coding sequence was translationally fused to Glutathione-S-Transferase (GST) gene in pEG(KT) vector (Mitchell et al. 1993) and expressed in S. cerevisiae strain, s288C by ‘one-step transformation’ method (Chen et al. 1992). Four percent galactose was used to induce the expression of GST-Rs-AFP2 fusion protein in S. cerevisiae transformants. Purification of the fusion protein was carried out as described by the supplier (Amersham, Buckinghamshire, England). The purified fusion protein was treated with factor Xa to get purified Rs-AFP2, which was then used to raise polyclonal antibodies in rabbit. Unfractionated rabbit blood serum was used as anti-Rs-AFP2 for all the analysis.
Western blot analysis
Total soluble proteins extracted from 100 mg of seed/leaf/root tissues of four Ub1-Rs-T transgenic lines (Ub1-Rs-T31, Ub1-Rs-T26, Ub1-Rs-T24 and Ub1-Rs-T42; hereafter referred as “transgenic lines”) were homogenized in 0.1 M PBS supplemented with 1 mM PMSF and estimated by Bradford (1976) method. Around 20 μg of total soluble proteins from each sample were heated for 10 min at 85°C and heat-labile denatured proteins were removed by centrifugation (12,000g), and were separated on a 15% SDS-PAGE at constant voltage (200 V). Purified Rs-AFP2 (50 ng) was used as a positive control. Immunoblots were developed with antibody raised against Rs-AFP2, using the alkaline phosphatase detection method (Bangalore Genei, Bangalore, India).
Estimation of Rs-AFP2 in leaf tissue of transgenic plants
Approximately, 20 μg of protein extracted from 21-days-old leaf tissues of transgenic lines (Ub1-Rs-T31, Ub1-Rs-T26, Ub1-Rs-T24 and Ub1-Rs-T42) was taken as the antigen for indirect enzyme-linked immunosorbent analysis (ELISA), using antibodies against Rs-AFP2. Known concentrations of purified Rs-AFP2 (2–100 ng) were used as a standard to plot absorbance (405 nm) versus concentration of purified Rs-AFP2 (ng per assay system), to estimate the levels of Rs-AFP2 in transgenic rice lines. Four plants of each line were used for further characterization. Average values were used for comparing different transgenic lines unless mentioned. All the samples in 96-well ELISA plate were incubated for 1–2 h followed by five times wash with 1× PBS containing 0.2% Tween 20. After blocking the wells with 3% BSA for 2 h, another wash of 1× PBS was given. Primary antibody (Rabbit anti-Rs-AFP2 antibody) was diluted to 1:500 in blocking solution, added to the wells, and incubated for 1 h, followed by 5 times washes with 1× PBS. Secondary antibody (HRP conjugated goat anti-rabbit IgG antibody; Bangalore Genei, Bangalore) was diluted to 1:1,000, added to the wells, and incubated for 1 h. To detect the secondary antibody bound molecules, 50 μl of 1× substrate tetramethyl benzidine (TMB)/hydrogen peroxide (H2O2) was added to the wells and the reaction was stopped by adding 1 N H2SO4. Final reaction system was diluted to 1 ml and abosorbance was measured at 405 nm. Absorbance values obtained in the samples from transgenics were used to estimate amount of Rs-AFP2 with the help of the standard graph.
Immunohistochemical procedure
As measured by ELISA, Ub1-Rs-T42 showed the highest expression of Rs-AFP2. Thus, it was used for immunohistochemical localization studies. Leaf and root tissues from 21-days-old Ub1-Rs-T42 and control plants were fixed with 10% formaldehyde, 5% acetic acid, and 85% ethanol for 30 min at room temperature. The tissue was then dehydrated through a graded ethanol series (30 min each in 30, 50, 70, 95, and 100% EtOH). Dehydrated tissue was gradually embedded in paraffin wax. A rotary microtome (Weswox Optik, model-TR HI 66, Ambala Cantt, India) and a freshly sharpened knife blade were used for sectioning. Tissue ribbons with 8–10 μm transverse sections were fixed on clean glass slides. Slides were then deparaffinised in xylene for 10–15 min, and rehydrated through a graded ethanol series (10 min each in 100, 95, 70, 50, and 30% EtOH). In order to quench most of the tissue autofluorescence from chlorophyll and cell walls, sections were incubated in 0.1% Toluidine blue for overnight. The sections were then washed in PBS/BSA (20 mM Sodium phosphate buffer, pH 7.0, 150 mM NaCl, and 10 μg/ml BSA); followed by transfer to the blocking solution (100 μl of 10% BSA (USB, Cleveland, Ohio, USA) in 1 ml PBS), incubation for 1 h in a moist chamber and washes with PBS/BSA. The sections were incubated for 90 min with anti-Rs-AFP2 antibody diluted to 1:100 in blocking solution. Following incubation, the sections were washed thrice for 15 min each in PBST (PBS with 0.1% Tween 20). Washed sections were then stained with Fluorescein isothiocyanate (FITC) conjugated anti-rabbit IgG antibodies raised in goat (Sigma Chemical Co, St Louis, MO, USA) diluted to 1:20 in PBS for 30 min. Three washes each of 15 min were given with PBST to the stained sections and mounted in water. The slides were observed under an epifluorescence microscope (Nikon, Eclipse 80i, Kanagawa, Japan) with a CCD camera attached. Control slides were treated without anti-Rs-AFP2 antibody but with FITC-conjugated goat anti-rabbit IgG antibody. Control slides were treated without anti-Rs-AFP2 antibody but with FITC-conjugated goat anti-rabbit IgG antibody.
Fungal spores were germinated overnight on glass slide. Growing fungal mycelia was treated with purified Rs-AFP2 (3 μM) for 30 min at 28°C and fixed with 10% formaldehyde, 5% acetic acid, and 85% ethanol for 30 min at room temperature and incubated in 0.1% toluidine blue for overnight. The samples were incubated for 90 min with anti-Rs-AFP2 antibody (diluted to 1:100) in blocking solution and then stained with FITC-conjugated anti-rabbit IgGs raised in goat (Sigma Chemical Co, St Louis, MO, USA), diluted to 1:20 in PBS for 30 min. The slides were observed under an epifluorescence microscope at 1000× magnification. For all the imunolocalization studies.
Rice blast assay
Twenty one-day-old transgenic lines (4 plants of each line) and non-transformed control plants were transferred into pots in an inoculation chamber; each pot was inoculated by spraying 1 × 105 spores per ml of M. oryzae (strain B157 corresponding to international race IC9) and incubated in a growth chamber maintained at 28°C, 90% relative humidity, 200 μmol/m2s1 light intensity and under 14 h photoperiods. Blast disease development was monitored as % diseased leaf area (%DLA = Diseased leaf area × 100/Total leaf area), over a period of 10 days post inoculation (dpi) (Bonman and Mackill 1988).
Sheath blight assay
R. solani from stock cultures maintained on sorghum seeds, was cultivated in 1 l sterile potato-dextrose broth (PDB) (Hi-Media, India) for 4–6 days at 28–30°C at 100 rpm. Mycelia were harvested and homogenised by vortexing in fresh sterile PDB medium; 0.1 g fresh weight per ml mycelial suspension was mixed with rice hull-rice grain mixture and incubated at room temperature (28–30°C) for 7–10 days. About 25 seeds, each from non-transformed plants and four transgenic lines were de-husked, surface sterilized and placed on solid Murashige and Skoog (MS) medium containing 3% sucrose. Seedlings were incubated in a growth chamber maintained at 28°C and 90% relative humidity under 16 h light and 8 h dark for 3–4 weeks and then transferred to un-inoculated soil for a week. Five-gram aliquots of inoculated rice-hull substrate were placed in the middle of each plant at its maximum tillering stage and infection was monitored (Jach et al. 1995).
In vitro antifungal activity assay and microscopic studies
The fungal mycelia were treated with 20 μg of total protein from leaf tissue of transgenic plants or 5 μg of purified Rs-AFP2, for all microscopic studies unless otherwise stated. Approximately, 100 μl of M. oryzae (isolate B157) suspension (103 spores) were germinated on a glass slide at 28°C in the presence or absence of Rs-AFP2, mounted with appropriate concentration of glycerol to maintain constant relative humidity in a sealed humidity chamber for 24 h. Growth inhibition was assayed after 24 h, in terms of percent germination or altered morphology of hyphae as compared to the control samples. The treated and untreated samples were air dried, fixed with 10% formaldehyde, 5% acetic acid, and 85% ethanol for 30 min at room temperature, and washed thoroughly with water. The fixed samples were stained with 0.3 g/l calcofluor white (Sigma-Aldrich, USA) for 30 min at room temperature. Samples were washed with 0.1 M phosphate buffered saline (pH 7.0) before resuspending in the mounting medium. Glycerol was used as a mounting agent for fluorescence microscopy. All samples were examined at 400× magnification under epifluorescence microscope (Eclipse 80i, 40X, Nikon, Japan), equipped with 100-W high-pressure mercury lamp (Osram HBO 103 W/2 N), and filter set for calcofluor white (UV-2A, Nikon, Japan).
Results
Generation and characterization of Rs-AFP2 transformed plants
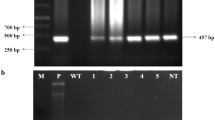
Transgenic rice plants expressing Rs-AFP2 were generated via Agrobacterium-mediated transformation, using hygromycin phosphotransferase gene (hpt) as a selectable marker (Fig. 1b). Rs-AFP2 protein folds into a small and compact β-barrel, which is stabilized by four internal disulfide bridges, and disulfide bridge formation takes place in Endoplasmic Reticulum; the ER signal peptide, therefore, is required. Therefore, the predicted expression product had a leader peptide (28 amino acids) at its amino-terminus derived from the Dm-AMP1 precursor. Twenty-eight UbI-Rs-T primary plants (T0) were obtained. A 283 bp PCR amplified product corresponding to the size of Rs-AFP2 was observed in all the 28 putative transformed plants and positive control (pUbI-Rs-T plasmid), which was absent in DNA of the non-transformed plants (Fig. 1a; data of three representative lines are presented, data for other lines are not shown). Mature plant leaves of homozygous T2 generation transgenic lines were harvested and checked for transgene integration by using Southern blot analysis (Fig. 1c). Single bands were detected in four lines (Rs-T-31, Rs-T-26, Rs-T-24 and Rs-T-42), which were further analysed for the expression of transgene. As expected, the protein of Rs-AFP2 was expressed constitutively in the transgenic lines but not in control plants (Fig. 2). Leaves (Fig. 2a), roots (Fig. 2b) and seeds (Fig. 2c) showed the presence of transgene product. The amount of Rs-AFP2 in leaves from a series of T2 rice plants transformed with pUbI-Rs-T was determined using indirect ELISA. Expression levels ranged from 4.5 ± 0.72 to 5.29 ± 0.61 μg/mg of total soluble proteins, in the leaf extracts of transgenic lines (Fig. 3). None of the Rs-AFP2 lines showed any obvious phenotypic changes compared with control plants throughout the life cycle (Data not shown).
a Total genomic DNA extracted from transgenic plants was amplified by PCR using specific primers for Rs-AFP2 coding region. Lane 1 100 bp marker; lane 2 positive control (pUbI-Rs-T plasmid) lane 3–5: Rs-AFP2 transformed plants; lane 6 negative control (Non-transformed Pusa basmati 1). b Plant expression vector pUb-R-T. pUb1 Ubiquitin promoter, SP signal peptide, tNos 3′ untranslated terminator region of the Agrobacterium tumefaciens nopaline synthase gene, p35S CaMV35S promoter, hpt hygromycin phosphotransferase. c Southern blot analysis of KpnI-digested genomic DNA from non-transformed and representative Rs-AFP2-transformed transgenic plants in T2 generation. Genomic DNA (10 μg) was digested with restriction enzymes and blotted onto nitrocellulose membranes after electrophoresis in a 0.8% agarose gel. The membrane was hybridized with 283 bp Rs-AFP2 coding sequence; lane 1 ladder, lane 2 Rs-T-14, lane 3 Rs-T-26, lane 4 Rs-T-17, lane 5 Rs-T-42, lane 6 Rs-T-31, lane 7 Rs-T-24, lane 8 negative control (non-transformed rice plant)
Immunodetection of Rs-AFP2 in transgenic rice lines. Total leaf protein extracts from T2-progeny plants were subjected to immunoblot analysis with polyclonal anti-Rs-AFP2. About 20 μg of total soluble proteins from each sample (T2-transformed and non-transformed rice plants) were separated on a 15% SDS–PAGE. GST-Rs-AFP2 was cleaved with factor Xa to get purified Rs-AFP2 and 50 ng of purified Rs-AFP2 was used as positive control. a Leaves; lane 1 marker (CPM1; Cat no: 105305; Bangalore genie Pvt Ltd, Banglore, India), lane 2 Rs-T-31, lane 3 Rs-T-26, lane 4 Rs-T-24, lane 5 Rs-T-42, lane 6 positive control (purified Rs-AFP2), lane 7 negative control (non-transformed Pusa basmati 1); b root; lane 1 positive control, lane 2 Rs-T-31, lane 3 Rs-T-26, lane 4 Rs-T-24, lane 5 Rs-T-42, lane 6 negative control (non-transformed Pusa basmati 1); c seed; lane 1 positive control, lane 2 Rs-T-31, lane 3 Rs-T-26, lane 4 Rs-T-24, lane 5 Rs-T-42, lane 6: negative control (non-transformed Pusa basmati 1)
ELISA of T2-transgenic rice plants expressing Rs-AFP2. Levels of Rs-AFP2 in different T2-lines were estimated by indirect ELISA. Concentration of Rs-AFP2 in the protein extracts from transgenic leaf tissues was estimated by plotting the absorbance values on standard graph prepared using known concentrations of purified Rs-AFP2. The data are presented as means ± SEM from three independent estimations. The value obtained for the non-transformed plant was subtracted from the values for the Rs-AFP2-lines to eliminate the background levels
The expression of OsPR1a (gi: 9801265) type genes has been widely used as an indicator for induction of defense responses against rice blast (Mittler et al. 1995). To explore possible activation of the plant defense system in the Rs-AFP2 transgenic rice lines, expression of the endogenous OsPR-1a gene was analyzed at the RNA level, without challenging with the pathogen. The transcripts for OsPR-1a were not detected in Rs-AFP2 transgenic lines (Fig. 4). As expected, OsPR-1a transcripts were detected in M. oryzae-infected control Pusa basmati 1 (Fig. 4, lane 1).
Northern-blot analysis of Rs-AFP2-expressing lines for the expression of rice OsPR-1a gene. About 20 μg/sample of total RNA were separated in a formaldehyde/agarose gel, transferred onto nylon membranes and hybridized to a non-radioactively labeled PR1a probe; lane 1 M. oryzae-infected wild-type plants, lane 2 control, non-infected wild type plants, lane 3 Rs-T-26, lane 4 Rs-T-24, lane 5 Rs-T-42; b rRNA bands (28 and 18S) from an ethidium bromide-stained gel of the corresponding RNA samples indicate equivalent loading of RNA
Localization of the Rs-AFP2 protein in rice tissues and on M. oryzae
Sub-cellular localization of Rs-AFP2 was determined by treating the transgenic leaf and root sample with anti-Rs-AFP2 followed by FITC-conjugated anti-rabbit IgG. Detectable levels of Rs-AFP2 were observed in the leaf (Fig. 5a) and root apoplast (Fig. 5b) of transformed rice, while cytoplasmic content remained unlabelled.
a Immunolocalization of Rs-AFP2 in transgenic rice leaf. Transverse sections were prepared from leaf tissues of Rs-AFP2 transgenic lines and control non-transformed plants; A transverse sections of transgenic rice leaf, B control non-transformed leaf tissue. Sections ‘A’ and ‘B’ were treated with anti-Rs-AFP2 and FITC-conjugated goat anti-rabbit IgG. Bar corresponds to 10 μm. b Localization of Rs-AFP2 in transgenic rice root. Transverse sections were prepared from root tissues of Rs-AFP2 transgenic lines and control non-transformed plants; A transverse sections of transgenic root, B control (non-transformed root tissue). Sections ‘A’ and ‘B’ were treated with anti-Rs-AFP2 and FITC-conjugated goat anti-rabbit IgG
Further, the localization of Rs-AFP2 was studied by treating M. oryzae mycelia with Rs-AFP2 and anti-Rs-AFP2 antibody followed by FITC-conjugated goat anti-rabbit IgG. Distinct fluorescence was observed in the fungal membrane (data not shown).
Disease tolerance of transgenic rice
We examined the effects of the overexpression of Rs-AFP2 on resistance of transgenic plants to fungal pathogens. Disease resistance was evaluated on T2 transgenic lines; Ub1-Rs-T31, Ub1-Rs-T26, Ub1-Rs-T24 and Ub1-Rs-T42 and the control line, with M. oryzae and R. solani. All the four Rs-AFP2 lines showed significant reduction in disease severity to blast infection (Fig. 6a). We compared antifungal activities of leaf protein extracts from transgenic plants and non-transformed control plants in terms of percent germination of M. oryzae spores; in vitro antifungal activities in the leaf protein extracts of transgenic plants were prominent and showed a dose dependent inhibition (Fig. 7c). When transgenic plants were challenged with M. oryzae, there was a remarkable difference in diseased leaf area (% DLA) between non-transformed (69.68 ± 2.52) and transgenic lines (11.01 ± 1.74 to 25.64 ± 1.50) at 99% confidence interval (CI) (Fig. 6a). To determine whether Rs-AFP2 lines show a broad-spectrum resistance, plants were infected with R. solani. Compared with the controls, Rs-AFP2 lines exhibited moderately increased resistance against R. solani, as indicated by reduced % infected plants. The % infected plants in transgenic lines ranged from 36.38 ± 4.82 to 48.85 ± 6.88; whereas, the value was 66.33 ± 3.55 in non-transformed plants at 99% CI (Fig. 6b).
ain planta bioassay of M. oryzae (105 spores/ml) on transgenic lines. A Whole plant infection assay of non-transformed control and transgenic plants, B Blast infected leaves of Rs-AFP2 lines and non-transformed control rice plants. Infected leaf area was measured as % DLA (Diseased leaf area) at 10 dpi. The data are presented as means ± SEM from three independent estimations. bin planta bioassay of Rs-AFP2transgenic lines with R. solani. The number of plants showing disease symptoms was used as disease scoring system. A Whole plant infection assay of non-transformed control and transgenic lines, B response of transgenic lines and non-transformed plants to sheath blight. Twenty seeds, each from non-transformed plants and the transgenic lines were used for sheath blight assay. The data are presented as means ± SEM from three independent estimations
a Disease response of transformed and non-transformed rice leaves challenged with M. oryzae spores. Twenty-one day old Rs-T-42 transgenic lines and non-transformed control plants were inoculated with M. oryzae spores. Transverse sections of Rs-AFP2 transgenic lines and non-transformed control were prepared. The samples were stained with calcofluor white. A Transverse sections of transgenic leaves with its corresponding bright field image, B non-transformed control with its corresponding bright field image. Bar corresponds to 10 μm. bM. oryzae mycelia when treated with total protein from leaf tissue of transgenic plants Rs-T-42 showed abnormal morphology. Approximately, 103 spores of M. oryzae (100 μl) suspension were germinated on a glass slide with 20 μg of total protein from leaf tissue of transgenic plants, Growth inhibition was assayed after 24 h, in terms of percent germination or altered morphology of hyphae as compared to the control samples. The treated and untreated samples were fixed and stained with 0.3 g/l calcofluor white. AM. oryzae spore germination in presence of total protein from leaf tissue of transgenic plants, BM. oryzae spore germination in presence of total protein from leaf tissue of non-transformed control plants. Bar corresponds to 10 μm. c Antifungal activities of leaf protein extracts from transgenic plants to non-transformed plants in terms of percent germination of M. oryzae. About 105 spores/ml of the M. oryzae was germinated in sterile yeast extract glucose-broth with various amounts of Rs-AFP2, at 25°C for 12 h. % spore germination was calculated on YEG agar plate. Percentage spore germination in presence of Rs-AFP2 was estimated compared to 100% germination in medium without Rs-AFP2. The data are presented as average from three independent estimations. \( {\frac{{\% \,{\text{Spore}}\,{\text{germination = Spore}}\,{\text{germination}}\,{\text{in}}\,{\text{presence}}\,{\text{of}}\,{\text{Rs}} - {\text{AFP}}2 \times 100}}{{{\text{Spore}}\,{\text{germination}}\,{\text{in}}\,{\text{medium}}\,{\text{with}}\,{\text{buffer}}\,({\text{control}})}}} \)
Microscopic analysis was carried out to follow the disease symptom development on inoculated leaves and roots. Tissue sections of inoculated leaves were cleaned with 6% KOH, stained with calcofluor white, and examined under epifluorescence microscope. In the control leaves infected with M. oryzae spores, extensive fungal colonisation was observed (Fig. 7). In contrast, reduced fungal colonization and growth inhibition was prominent in leaves of the Rs-AFP2 lines under the same experimental conditions (Fig. 7a). Hyphae showing abnormal morphology could be detected, when M. oryzae was treated with total soluble protein from transgenic lines (Fig. 7b).
Discussion
Success in the development of cultivars with enhanced resistance to pathogens involving transgenic expression of antifungal genes depends on the nature of the recipient plant, the specific pathogen, and the source of transgene. In this study, transgenic rice plants constitutively expressing Rs-AFP2 were generated by Agrobacterium-mediated transformation. This gene was chosen in view of earlier reports that Rs-AFP2 shows in vitro antifungal activity against several agronomically important fungal pathogens (Terras et al. 1995). The elite indica rice cultivar Pusa basmati 1, known to be susceptible to the M. oryzae and R. solani, was used in this study. Results presented here indicated that constitutive expression of the Rs-AFP2 gene in rice results in enhanced resistance against M. oryzae and R. solani.
Expression level of Rs-AFP2 was analysed by indirect ELISA, from leaf protein extracts of transgenic plants. Expression level ranged between 0.45 and 0.53% of total soluble protein, which was approximately 2 times higher than the report of tobacco and tomato plants transformed with Ca35S driven Rs-AFP2 gene constructs cloned from Radish (Terras et al. 1995; Koike et al. 2002). Further, the levels of Rs-AFP2 in rice plants were sufficient to impart resistance against M. oryzae and R. solani. Constitutive expression of the radish defensin (Rs-AFP2) enhanced resistance of tobacco plants to the foliar pathogen Alternaria longipes (Terras et al. 1995) and similarly in tomato to A. solani (Koike et al. 2002). In addition, transgenic rice constitutively expressing a plant defensin (wasabi and Dm-AMP1) provided a robust resistance against the agronomically important fungus M. oryzae (Kanzaki et al. 2002; Jha et al. 2008).
The important prerequisite to control the pathogen in the transgenic plant is the presence of antifungal proteins at the site of interaction with the pathogen. During the early onset of the infection process, interaction between plant and pathogen occurs in the extracellular space at various sites, like leaves, roots or stem. We have used Rs-AFP2 driven by strong and constitutive maize ubiquitin promoter and the signal peptide derived from the Dm-AMP1. The Dm-AMP1 signal peptide had successfully targeted the Dm-AMP1 to apoplast in transgenic rice (Jha et al. 2008). The expression of transgene in different plant tissues was ascertained by western blot analysis. Extracellular location of Rs-AFP2 in transgenic plants was confirmed by immunohistochemical localization studies in the sections prepared from leaves and roots of the transgenic plants. The immunohistological observations revealed the apoplastic location of Rs-AFP2 and confirmed that the N-terminal signal sequence of the Dahlia Dm-AMP1 protein directs secretion of Rs-AFP2 to the intercellular spaces.
Another important aspect of the plant-produced Rs-AFP2 was its biological activity in transgenic plants. The present study showed that the Rs-AFP2 protein is being correctly processed and/or folded in the rice secretory pathway. The expression level of the Rs-AFP2 gene was sufficient to suppress the growth and development of M. oryzae (77%) and R. solani (45%) as compared to non-transformed plants. Constitutively expressed Rs-AFP2 gene influences the proliferation of both the hemi-biotrophic fungi; this was observed when plants were challenged with M. oryzae and R. solani. Small necrotic lesions were observed, which did not spread out to form the large lesions as seen on the control plants. We also observed a consistent difference in the severity of tissue damage between control and Rs-AFP2 transgenic plants, when monitored for the development of disease symptoms through microscopic analysis. Strong evidence of enhanced resistance, due to a direct effect of the overexpressed Rs-AFP2, comes from growth anomalies of the fungal hyphae on the transgenic lines. The concentration of purified Rs-AFP2 completely inhibiting spore germination of M. oryzae was 3 μM and no fungal growth was seen above this concentration. Significant changes in the hyphal morphology were also observed at concentrations that only partially inhibited the germination of conidia (<IC50) (Fig. 7a–c). These include abnormal hyperbranching of germinated conidia, compared to well-extended mycelial growth in the controls. Leaves from non-transformed control plants infected with M. oryzae spores showed extensive fungal colonisation in contrast to reduced fungal colonization and growth inhibition in leaves of the Rs-AFP2 lines under the same experimental conditions (Fig. 7a). Hyphae showing abnormal morphology could be detected, when M. oryzae was treated with total soluble protein from transgenic lines (Fig. 7b, c). Bulbous structure at the hyphal tip and hyperbranching of the hyphae and an apparent collapse of the plasma membrane in transgenic lines was observed. Similar effects on fungal growth were reported for fungicides (Robson et al. 1989; Wiebe et al. 1990), plant defensins (Terras et al. 1995), plant knottins (Prasad et al. 2008) and transgenic plants overexpressing Ace-AMP1 (Patkar and Chattoo 2006). Furthermore, transgene expression of Rs-AFP2 gene was not accompanied by an induction of pathogenesis-related (PR) gene expression supporting that the transgene product itself is active against M. oryzae.
Interestingly, Rs-AFP2 is localized to the plasma membrane (data not shown). GlcCer mutants of both S. cerevisiae, C. albicans and Pichia pastoris are resistant against Rs-AFP2 strongly suggesting the role of GlcCer in Rs-AFP2-mediated growth inhibition (Thevissen et al. 2004). Inhibition of germination and hyphal growth of A. nidulans and A. fumigatus by blocking the biosynthesis of GlcCer has also been reported (Levery et al. 2002). Further, GlcCer plays a key role in the pathogenicity of C. neoformans (Rittershaus et al. 2006). Moreover, the RsAFP2-induced generation of reactive oxygen species in susceptible fungi was reported, pointing to an induction of apoptosis (Aerts et al. 2006). Thus, resistance of the transgenic rice plants expressing Rs-AFP2 against M. oryzae was due to interaction of transgene product with the fungal membrane, and its inhibition directly by Rs-AFP2. Consequently, the present report expands the phylogenetic range over which Rs-AFP2 has been shown to be effective in conferring disease resistance at the level of greenhouse tests.
References
Aerts AM, Francois IEJA, Meert EMK, Li Q, Cammue BPA, Thevissen K (2006) The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. FEBS Lett 580:1903–1907
Anderson NA (1982) The genetics and pathology of Rhizoctonia solani. Annu Rev Phytopathol 20:329–347
Bonman JM, Mackill DJ (1988) Durable resistance to rice blast disease. Oryza 25:103–110
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Broekaert WF, Terras FR, Cammue BP, Osborn RW (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108:1353–1358
Chen DC, Yang BC, Kuo TT (1992) One-step transformation of yeast in stationary phase. Curr Genet 21:83–84
Christensen AH, Quail P (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version 2. Plant Mol Biol Rep 1:19–22
Francois IE, De Bolle MF, Dwyer G, Goderis IJ, Woutors PF, Verhaert PD, Proost P, Schaaper WM, Cammue BP, Broekaert WF (2002) Transgenic expression in Arabidopsis of a polyprotein construct leading to production of two different antimicrobial proteins. Plant Physiol 128:1346–1358
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6(2):271–282
Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C (1995) Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J 8(1):97–109
Jha S, Tank HG, Prasad BD, Chattoo BB (2008) Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Res 18:59–69
Kanzaki H, Nirasawa S, Saitoh H, Ito M, Nishihara M, Terauchi R, Nakamura I (2002) Overexpression of the wasabi defensin gene confers enhanced resistance to blast fungus (Magnaporthe oryzae) in transgenic rice. Theor Appl Genet 105:809–814
Koike M, Okamoto T, Tsuda S, Imai R (2002) A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem Biophys Res Commun 298:46–53
Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T (1996) Vectors carrying two separate T-DNAs for cotransformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J 10:165–174
Levery M, Momany R, Lindsey MS, Toledo J, Shayman M, Fuller K, Brooks RL, Doong AH (2002) Straus and H.K. Takahashi. FEBS Lett 525:59–64
Logeman J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Ann Biochem 163:16–20
Mitchell DA, Marshall TK, Deschenes RJ (1993) Vectors for the inducible expression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715–723
Mittler R, Shulaev V, Lam E (1995) Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7:29–42
Osborn RW, De Samblanx GW, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees SB, Broekaert WF (1995) Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett 368:257–262
Ou SH (1985) Rice diseases, 2nd edn. Commonwealth Mycological Institute Publication, Kew, pp 280–282
Patkar RN, Chattoo BB (2006) Transgenic indica rice expressing ns-LTP like protein shows enhanced resistance to both fungal and bacterial pathogens. Mol Breeding 17:159–171
Prasad BD, Jha S, Chattoo BB (2008) Transgenic indica rice expressing Mirabilis jalapa antimicrobial protein (Mj-AMP2) shows enhanced resistance to the rice blast fungus Magnaporthe oryzae. Plant Sci 175:364–371
Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Poeta MD (2006) Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest 116:1651–1659
Robson GD, Kuhn PJ, Trinci APJ (1989) Effect of validamycin A on the inositol content and branching of Rhizoctonia cerealis and other fungi. J Gen Microbiol 135:739–750
Sesma A, Osbourn AE (2004) The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431:582–586
Terras FRG, Schoofs HME, De Bolle MFC, Van Leuven F, Rees SB, Vanderleyden J, Cammue BPA, Broekaert WF (1992) Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem 267:15301–15309
Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, van Leuven FV, Vanderleyden J, Cammue BPA, Broekaert WF (1995) Small cysteine-rich antifungal proteins from radish: their role in host defence. Plant Cell 7:573–588
Thevissen K, Warnecke DC, François IEJA, Leipelt M, Heinz E, Ott C, Zähringer U, Thomma BPHJ, Ferket KKA, Cammue BPA (2004) Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem 279:3900–3905
Thomma BPHJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216:193–202
Wiebe MG, Robson GD, Trinci APJ (1990) Edifenphos (Hinosan) reduces hyphal extension, hyphal growth unit length and phosphatidylcholine content of Fusarium graminearum A3/5, but has no effect on specific growth rate. J Gen Microbiol 136:979–984
Acknowledgments
We thank Dr. B. Cammue, University of Leuven, Belgium, for providing the plasmid pFAJ3105. This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jha, S., Chattoo, B.B. Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transgenic Res 19, 373–384 (2010). https://doi.org/10.1007/s11248-009-9315-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-009-9315-7