Abstract
Methane is classified as the second major greenhouse gas with a global warming potential 25 times higher than carbon dioxide. Wastewater treatment plants (WWTPs) are considered as one of the main anthropogenic sources for global methane emissions. Utilizing the anaerobic digestion driven biogas, methanotrophs can offer a prominent solution for coupling methane mitigation with value-added resources recovery. Hence, methanotrophs can play a pivotal role in the paradigm shift to consider wastewater streams as proactive energy and value-added material resource instead of waste requiring further treatment. This review is destined to summarize the recent accomplishments in three methanotrophic-based biotechnological applications which are methanol, biopolymers production and biological nitrogen removal processes. Moreover, methanotrophs taxonomy, metabolism, and growth conditions are reviewed. In addition, the possibility to link the aforementioned applications within the operation of existing WWTPs in order to transform “energy-consuming treatment processes” into “energy-saving and energy-positive systems” is discussed.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Methane (CH4) is considered as the second major greenhouse gas (GHG) (Francisco José Fernández 2005). Methane concentration in the atmosphere has reached 1.75 ppm and is expected to reach 4 ppm in the year 2050 (Hanson and Hanson 1996). Compared to the major GHG (i.e., CO2), methane molecule can absorb 30 times more heat with a lifetime span of approximately 12 years and contributes to more than 25% of the global warming phenomena (GWP) (Scheutz et al. 2009). Hence, the focus on CH4 emissions mitigation will have 20–60 times greater effect on GWP than the CO2 emissions reduction (Hanson and Hanson 1996). Particularly, anthropogenic methane sources are estimated to contribute for 63% of the global methane emissions including landfills, fossil fuels burning, rice cultivation, coal mining, oil recovery and waste management (Strong et al. 2015). Recently, significant efforts focus on developing new technologies for methane mitigation and the recovery of value-added products through different biotechnologies such as fuel production, electricity generation, and biopolymers production (Nikiema et al. 2007).
In order to activate the methane molecule, the bond between carbon and hydrogen (C–H bond) must be broken which is, unfortunately, one of the most inactive hydrocarbon bond requiring 438.8 kJ/mol (Park and Lee 2013). Consequently, various expensive and energy intensive thermochemical techniques were introduced to break the C–H bond and utilize the methane including chemical catalysts, high temperatures, and high pressure (Fei et al. 2014). On the other hand, methane can be utilized biologically under ambient temperature and atmospheric pressure (Conrado and Gonzalez 2014). Moreover, the biocatalysts responsible for methane utilization are found in diverse environments (e.g., pH, temperature, oxygen concentrations, pollutants existence and substrate availability), which reflect its adaptability to different operational conditions. Collectively, it can be concluded that biological methane utilization is more efficient, simpler, and cheaper in comparison to thermochemical conversion.
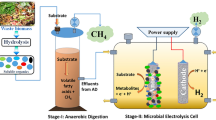
Biologically, two distinct microbial clusters can utilize methane and activate its stable C–H bond (1) ammonia oxidizing bacteria (AOBs) and (2) methane oxidizing bacteria (methanotrophs). AOBs partially oxidize methane via the ammonia monooxygenase (AMO) while using ammonia as their energy source (Hanson and Hanson 1996; Taher and Chandran 2013). AOBs can only be employed in methane to methanol conversion with relatively low productivities due to the competition between methane and ammonia on the AMO (Ge et al. 2014). On the other hand, methanotrophs can be employed in various biotechnological applications including methanol, biopolymers, single-cell protein (SCP), and ectoine production (Strong et al. 2015). Figure 1 illustrates the potential of biogas utilization by methanotrophs and AOBs. Hence, methanotrophs are an active, advantageous, and prominent research area, especially, in the enhancement of its productivity and overcoming the challenges from the perspective of genetic engineering, enzymology, biotechnology and bioreactor engineering.
This review work is destined to focus on the potential and the sustainability of mitigating the biomethane and recovering value-added resources using methanotrophs including; methanol production and biopolymers accumulation. In addition, methanotrophs taxonomy and metabolism are comprehensively reviewed. Lastly, an integration vision of the methanotrophic bioreactor into wastewater treatment plants and their incorporation in the nitrogen removal processes is demonstrated.
2 Methanotrophs
Methanotrophs are a unique cluster of microorganisms that have the ability to utilize methane as their sole carbon and energy source (Anthony 1982). Naturally, methane oxidation is carried out by methane oxidizing bacteria and anaerobic methane oxidizing archaea (Cui et al. 2015). Methanotrophic archaea can anaerobically couple methane oxidation, via the reverse methanogensis pathway, with the reduction of multiple types of electron acceptors; i.e., sulfate, nitrate, and metal ions (Mn4+ and Fe3+) (Ge et al. 2014). Whereas, the gram-negative methane oxidizing bacteria is a descending cluster from the methylotrophic bacteria (Semrau et al. 2010). In methane oxidizing bacteria, methane oxidation into methanol (CH3OH) is catalyzed by the methane monooxygenase (MMO) enzyme. Methanol is converted subsequently into formaldehyde (CHOH). Instantly, formaldehyde is oxidized into carbon dioxide (CO2) with formate (CHOOH) as an intermediate (Anthony 1982). Furthermore, formaldehyde, formate, and carbon dioxide can be utilized for cell synthesis requirements via the ribulose monophosphate (RuMP), serine, and Calvin-Benson-Bassaham (CBB) cycles, respectively (Chistoserdova and Lidstrom 2013a). It is noteworthy that the ability to produce methanol and biopolymers from methane is exclusive to the methane oxidizing bacteria. Hence, even though the nomenclature methanotrophs includes both the methane oxidizing bacteria and the anaerobic methane oxidizing archaea, this review cover only the methane oxidizing bacteria.
2.1 Methanotrophs taxonomy
Aerobic methanotrophs are phylogenetically located in the Verrucomicrobia phylum and the Gamma and Alpha subdivisions of Proteobacteria phylum (Murrell 2010; Sun et al. 2013). As shown in Fig. 2, aerobic methanotrophs are clustered into three main types; type I methanotrophs or Gamma-Proteobacteria methanotrophs forming the Methylococcaceae and Methylothermaceae families, type II methanotrophs or Alpha-Proteobacteria methanotrophs lying in the Methylocystaceae and Beijerinckiaceae families, and type III or Verrucomicrobia methanotrophs descending from the Methylacidiphilaceae family. Lastly, Candidatus Methylomirabilis oxyfera (M. oxyfera) is the only known anaerobic methanotrophic bacteria. It belongs to the gram-negative NC10 phylum which all of its members have not been isolated in pure culture yet (Shen et al. 2015).
2.1.1 Type I methanotrophs
Type I methanotrophs can be found in many environments, including freshwaters and sediments, marine environments, rice fields, hot springs, soils, landfills, coal-mine surface, drainage water (Bowman 2006, 2014; Semrau et al. 2010), denitrification reactors, silage and manure wastes (Trotsenko et al. 2009), sewage and activated sludge, and anaerobic digesters (Ho et al. 2013a, b; Kits et al. 2015b). Type I methanotrophs possess a typical well developed intracytoplasmic membrane (ICM) throughout the cell, which appears as stacks of vesicular discs. Therefore, the expression of particulate methane monooxygenase (pMMO) is conferred due to its location inside the ICM (Semrau et al. 2010). Moreover, some genera including Methylomagnum, Methaylovulum, and few strains within Methylomonas and Methylocaldum can express soluble methane monooxygenase (sMMO) as well as pMMO (Iguchi et al. 2011; Kalyuzhnaya 1999; Khalifa et al. 2015). Even though type I methanotrophs assimilate carbon via the RuMP pathway at the level of formaldehyde, Methylocaldum genus, Methylomagnum ishizawai, and Methylococcus capsulatus species possess the serine pathway enzymes (Bodrossy et al. 1997; Khalifa et al. 2015; Takeuchi et al. 2014). Despite it cannot grow autotrophically, Methylococcus strains can assimilate carbon dioxide in association with methane as the cellular carbon through a partially functional CBB cycle (Bowman et al. 1993).
2.1.2 Type II methanotrophs
Type II methanotrophs widely exist in different environments especially at low oxygen and high methane concentrations (Amaral and Knowles 1995). These environments include soil and freshwater sediments, rice fields, coal-mine drainage water, landfills, acidic wetlands, forest soils, groundwater aquifers and sewage sludge (Bowman et al. 1993; Knief 2015; Marín and Arahal 2014). Methylocystis and Methylosinus genera possess an ICM aligned to the cell periphery, while the ICM for the Methylocapsa genus appears as stacks of membrane vesicles packed in parallel on only one side of the cell membrane. Accordingly, the three of them express pMMO. Moreover, Methylosinus and some Methylocystis strains can possess sMMO (Bowman et al. 1993; Marín and Arahal 2014; Whittenbury et al. 1970). In contrast, Methylocella and Methyloferula genera miss the extensive ICM system and only express sMMO. These two genera develop a vesicular membrane system composed of spherical or ovoid-shaped membrane vesicles located on the periphery of the cytoplasm (Semrau et al. 2011; Vorobev et al. 2011). In order to increase their methane oxidation surface area, methanotrophs form more ICM in the presence of methane. It is noteworthy that a better ICM is developed while growing on methane rather than methanol (Bowman 2006). All type II methanotrophs can accumulate poly-β-hydroxybutyrate (PHB) as a survival mechanism under nutrients unbalanced conditions. Carbon, in the form of formate, is assimilated via the serine pathway while all other pathways are not found (Bowman 2006; Marín and Arahal 2014).
2.1.3 Type III methanotrophs
Type III methanotrophs were firstly isolated in 2007 from hot acidic habitats including acidic hot springs, volcanic mud, geothermal mud areas (Knief 2015; van Teeseling et al. 2014). Type III methanotrophs do not possess the typical proteobacterial ICM except for Methylacidimicrobium fagopyrum which possess a type I-like ICM. However, Methylacidiphilum strains have an ICM of carboxysome-like structures or vesicular membranes (Op den Camp et al. 2009). Whereas, no ICM system was observed in both Methylacidimicrobium tartarophylax and Methylacidimicrobium cyclopophantes (van Teeseling et al. 2014). All methanotrophic Verrucomicrobia possess pMMO only which raises the question about the location of pMMO in strains that do not have an ICM (Erikstad and Birkeland 2015; van Teeseling et al. 2014). Carbon, in the form of carbon dioxide (CO2), is assimilated via the CBB cycle, while the complete RuMP and serine cycle enzymes are not found (Erikstad and Birkeland 2015; van Teeseling et al. 2014). Therefore, unlike proteobacterial methanotrophs, CO2 presence stimulate the bacterial growth, but, no growth on CO2 only was reported (Op den Camp et al. 2009; van Teeseling et al. 2014).
2.1.4 Anaerobic methanotrophs
Anaerobic methanotrophs were discovered in 2006 with the ability of coupling anaerobic methane oxidation with nitrite reduction (Raghoebarsing et al. 2006) which attracted several researchers to develop the nitrite-dependent anaerobic methane oxidation (N-DAMO) process to be coupled with anaerobic ammonium oxidation (ANAMMOX) for nitrogen removal in WWTPs (Wang et al. 2017b). They were enriched from freshwater environments, sewage sludge, and wetland soils. However, they have not been isolated in pure culture yet. Moreover, M. oxyfera is the only bacterial type that can grow anaerobically on methane (Wang et al. 2017a). Even though no ICM system was observed in M. oxyfera, pMMO is the initiator of methane oxidation which is located on the cytoplasmic membrane surface. The genomic analysis of M. oxyfera revealed the existence of incomplete RuMP and serine pathways and the full CBB cycle (Wu et al. 2011). Later, it was confirmed that carbon is fixed in the level of CO2 via the CBB cycle. Whereas, it is suggested that the incomplete serine pathway is used to detoxify the formaldehyde (Rasigraf et al. 2014; Shen et al. 2015).
2.2 Methane metabolism in methanotrophs
All methanotrophs oxidize methane terminally to carbon dioxide through a series of linked reactions to fulfill their energy and cell replication requirements with the aid of their secreted enzymes as illustrated in Fig. 3 (Hanson and Hanson 1996).
As shown in Eqs. (1)–(3), methane is oxidized to methanol in a reaction catalyzed by MMO which splits the oxygen molecule into two atoms. One oxygen atom is incorporated in methane hydroxylation while the other atom is reduced to H2O (Madigan et al. 2015). As the latter reaction requires reducing equivalents, methane hydroxylation is considered as an energy consuming reaction unlike the remaining reactions which are energy producing (Chistoserdova and Lidstrom 2013a).
*A is a reducing equivalent providing 2e− and 2H+; NAD(P)H in sMMO; not confirmed yet in pMMO.
The multi-function oxidase MMO is found in two forms; the cytoplasmic soluble form “sMMO” and the copper containing particulate form “pMMO” located in the ICM (Semrau et al. 2010). sMMO utilizes nicotinamide adenine dinucleotide NAD(P)H generated from formaldehyde and formate oxidation as reducing equivalents. On the other hand, the electron donor for pMMO has not been identified yet (Karthikeyan et al. 2015). It was assumed that ubiquinol (Q8H2) could be the most probable pMMO electron donor relying on pMMO similarities with AMO. However, the mechanism of ubiquinone reduction to ubiquinol is still not clear (Kalyuzhnaya et al. 2015). One hypothesis is that electrons transfer between the methanol oxidizing enzyme methanol dehydrogenase (MDH) and pMMO owing to their close locations (Culpepper and Rosenzweig 2014). Alternatively, NAD(P)H from formaldehyde and formate oxidation may be the responsible for ubiquinone reduction. This hypothesis is supported by the observed enhancement in methane oxidation and methanol production with external formate addition (Trotsenko and Murrell 2008). Hence, it was suggested that pMMO might be utilizing various electron sources according to the growth conditions (Kalyuzhnaya et al. 2015).
pMMO-expressing cells oxidize methane more efficiently than sMMO as they have higher methane affinity and growth yields (Kalyuzhnaya et al. 2015). On the other hand, sMMO have a broader substrate range than pMMO which makes it more attractive for several biotechnological processes (Smith et al. 2010). The expression of both sMMO and pMMO enzymes is controlled by copper concentration. pMMO is expressed in copper concentrations above 1 µmol/g (dry weight) of cells, while sMMO is expressed in concentration below 1 µmol/g (dry weight) of cells (Hanson and Hanson 1996). Both enzymes properties are summarized in Table 1.
The produced methanol is further oxidized to formaldehyde via the quinoprotein methanol dehydrogenase (MDH) located in the periplasm, as expressed in Eq. (4) (Chistoserdova and Lidstrom 2013a). Methanol oxidation to formaldehyde is accompanied with the reduction of pyrroloquinoline quinone (PQQ) to PQQH2. Afterwards, PPQH2 is oxidized and transfer electrons (2 electrons) either to the terminal oxidase with cytochromes-c and other carriers as intermediates or to regenerate the reducing equivalents needed for methane hydroxylation as previously described (Smith et al. 2010). Generally, methanol oxidation is an energy-conserving step regardless of the electrons deviation.
*pQQH2 is further oxidized and transfer 2e− and 2H+ to terminal oxidase or pMMO.
Formaldehyde plays a pivotal role in methanotrophs metabolism as the central intermediate. Part of the formed formaldehyde is terminally oxidized to CO2 for energy generation. The other part is incorporated in the carbon assimilation pathways for cell replication; RuMP pathway for type I and serine pathway for type II. Formaldehyde is rapidly directed to either cycles due to its toxic effect on methanotrophs (Karthikeyan et al. 2015).
Two systems have been suggested for formaldehyde oxidation to formate expressed in Eq. (5). Firstly, oxidation is catalyzed by formaldehyde dehydrogenase (FaDH) which is either NAD-linked or PQQ-containing and cytochrome-linked enzyme (Chistoserdova and Lidstrom 2013a). The quinoprotien FaDH was confirmed to be the major formaldehyde oxidizing enzyme in Methylococcus capsulatus while expressing pMMO only (Zahn et al. 2001). This finding supports the hypothesis that the electron source for pMMO can be linked to formaldehyde oxidation. The second suggestion is the tetrahydromethanopterin (H4MPT)-linked formaldehyde oxidation pathway. Within H4MPT pathway, MtdB enzyme is the responsible of NAD(P)+ reduction to NAD(P)H (Chistoserdova et al. 2009). Nonetheless, different formaldehyde oxidation systems were found in methylotrophs and might be existing in methanotrophs only as a formaldehyde detoxification strategy (Chistoserdova and Lidstrom 2013a). Finally, formate is oxidized to carbon dioxide, as demonstrated in Eq. (6). This reaction is catalyzed by the NAD dependent enzyme formate dehydrogenase (FDH) which functions as sMMO electron source (Smith et al. 2010).
NAD(P)H or PQQH2 is produced according to the pathway.
The last three oxidation reactions, Eqs. (4)–(6), produce 6 electrons (2e− each). Two of them travel back to methane oxidation in the form of reducing equivalents. The remaining electrons (4e−) traverse the electron transport chain (ETC) generating energy by producing adenosine triphosphate (ATP) through the proton motive force. Finally, the four electrons reduced the terminal electron acceptor O2 to H2O, as shown in Eq. (7). Accordingly, the overall reaction can be written as expressed in Eq. (8).
The overall reaction.
As mentioned before, the nitrite dependent anaerobic methane oxidizing bacteria M. oxyfera lacks the ICM. However, the same pathway took place with two preliminary reactions, shown in Eqs. (9) and (10). Nitrite (NO2−) is converted to NO via the periplasmic nitrite reductase (cd1 Nir). Subsequently, two NO molecules are converted to N2 and O2. The produced O2 is further utilized in the methane oxidation pathway and acts as the terminal electron acceptor (Shen et al. 2015; Wu et al. 2011).
The oxygen produced is further involved in methane oxidation pathway.
The overall reaction.
For the methane assimilation pathways, as previously mentioned, type I undergoes the RuMP pathway and type II uses the serine pathway. Whereas, type III and the anaerobic methanotrophs assimilate carbon via CBB cycle. Each cycle has different reactions and enzymes involved in their methane assimilation pathway for energy production and cell replication (Chistoserdova and Lidstrom 2013a; Wu et al. 2011).
The RuMP pathway is initiated by the reaction between formaldehyde and ribulose-5-phosphate yielding fructose-6-phosphate. Two specific enzymes mediate this reaction; hexulosephosphate synthase (HPS) and hexulosephosphate isomerase (HPI) found only in type I methanotrophs. Fructose-6-phosphate follows a series of reactions producing pyruvate and glyceraldehyde-3 phosphate. Glyceraldehyde is then utilized to regenerate ribulose-5-phosphate and complete the cycle for biomass synthesis, while the pyruvate is incorporated in an incomplete TCA cycle for CO2 production. The main intermediates of the RuMP cycle are found in the form of sugar phosphates (Hanson and Hanson 1996; Kalyuzhnaya et al. 2015; Khmelenina et al. 2015).
In the serine cycle, formaldehyde is converted to methyl-H4MPT then to methylene-H4F mediating the reaction of formaldehyde with glycine to activate the serine cycle (Kalyuzhnaya et al. 2015; Karthikeyan et al. 2015). Furthermore, the cycle produce acetyl-CoA which is the key entry for the complete TCA cycle required for energy generation and biomass synthesis (Chidambarampadmavathy et al. 2015). The main intermediates in the serine cycle are found in the form of amino acids and CoA derivatives (Kalyuzhnaya et al. 2015).
In general, type I methanotrophs have higher growth rate than type II (Kalyuzhnaya et al. 2015). Moreover, type I is more energy efficient as it requires 1 ATP for assimilation of 3 formaldehyde molecules compared to 3 ATP and 2 NADH to assimilate 2 formaldehyde molecules and 1 CO2 molecule in type II (Karthikeyan et al. 2015). Up to 50% of produced CO2 by type II can be incorporated in cell synthesis compared to 15% in type I, what makes the biogas produced from anaerobic digesters consisting of about 40% CO2 suitable for type II (Kalyuzhnaya et al. 2015). One more advantage for type II is their ability to accumulate biopolymers under nutrient limitation conditions (Karthikeyan et al. 2015).
2.3 Factors affecting methanotrophs growth
The specific growth rates for pure methanotrophic cultures using methane as carbon and energy source are illustrated in Table 2. Type I methanotrophs have the highest growth rates ranging from 0.2 to 0.3 h−1 in some strains (Hirayama et al. 2011, 2014; Whittenbury et al. 1970; Wise et al. 2001). The highest growth rate reported in type II methanotrophs is 0.15 to 0.18 h−1 for Methylocystis strains (Dedysh et al. 2007; Whittenbury et al. 1970). Type III methanotrophs are slower than both types with growth rates ranging from 0.013 to 0.07 h−1 (Op den Camp et al. 2009; van Teeseling et al. 2014). Anaerobic M. oxyfera is the slowest growing methanotrophs with a doubling time that can reach up to 2 months (Shen et al. 2015). However, no specific type dominates in methane sufficient conditions. This can be referred to the fact that each type has different favorable growth conditions such as pH, temperature, nutrients, methane and oxygen availability. Furthermore, factors like methane solubility and bioreactor characteristics can notably affect the methanotrophic microbial yield and biomass density. Unfortunately, various factors affecting methanotrophs growth are still unclear and need to be reviewed and investigated either in natural habitats or bioreactors.
2.3.1 Temperature
More than 70% of methanotrophs grow optimally in a temperature ranging from 20 to 35 °C, as shown in Table 2. However, high temperatures (55–60 °C) are preferable for the type I Methylothermus genus and type III Methylacidiphilum genus (Bowman 2014; Op den Camp et al. 2009). Within type I, Methylococcus and Methylocaldum genera [which form type X a subset of type I methanotrophs (Bowman 2006)] grow optimally at temperature range of 42–55 °C (Trotsenko et al. 2009). In contrast, Methylosphaera genus grows at lower temperature ranging from 10 to 15 °C (Bowman 2006). Furthermore, all type II methanotrophs (except some strains within Methylocystis) and some type I methanotrophs (Methylosphaera, Methyloprofundus, Methyloglobulus, Methylovulum, and some species within Methylobacter, Methylosarcina, Methylomonas, Methylomicrobium) are able to survive at lower temperatures (4–10 °C) (Bowman 2014; Marín and Arahal 2014; Tavormina et al. 2015). Moreover, it was observed that type I methanotrophs predominated at lower temperatures (3–10 °C) in a sample enriched from landfill cover soils while both types grew normally at 20 °C (Börjesson et al. 2004).
2.3.2 pH and salinity
As presented in Table 2, more than 90% methanotrophs prefer to grow in pH ranges from 5.5 to 8. However, verrucomicrobial methanotrophs are more acidophilic with optimum growth at pH ranging from 1.5 to 3.5. Methylacidiphilum fumariolicum (strain SoIV) and Methylacidimicrobium tartarophylax sp. are the most acidophilic methanotrophs growing at pH between 0.5 and 0.8 (Op den Camp et al. 2009; van Teeseling et al. 2014). In contrast, Methylomicrobim species grow better in alkaline mediums (pH 8–10) (Bowman 2006; Kalyuzhnaya et al. 2008).
For the salinity, most of methanotrophs do not require NaCl for their growth with wide tolerance ranging from 0.2 up to 10% NaCl (w/v) (Bowman 2014; Semrau et al. 2010). However, Methylosoma genus is intolerant to NaCl (Rahalkar et al. 2007). Furthermore, a study on mixed culture showed that methane uptake and growth rate remained constant with salinity up to 7 mg/L, while a noticeable decline occurred after increasing the salinity above this level (van der Ha et al. 2010).
2.3.3 Substrates
Generally, all methanotrophs prefer methane as their carbon and energy source. However, in the absence of methane, methanotrophs except most of the Methylocaldum species, Methylobacter tundripaldum sp., and Methylocystis rosea sp. can grow on methanol (Bowman 2014; Lidstrom 2006; Marín and Arahal 2014). However, at relatively high methanol concentrations, growth is fully or partially inhibited due the excessive accumulation of the toxic formaldehyde resulting from methanol oxidation (Graham et al. 1993; Whittenbury et al. 1970). Despite its inhibitory effect, Methylocella tundrae prefers methanol than methane (Dedysh et al. 2004). On the other hand, some strains within Methylocapsa and Methylocella genera have the ability to grow on other C1 compounds like formate, and methylamines (Dedysh et al. 2004; Dunfield et al. 2003, 2010).
For a long time, it was believed that methanotrophs grow only on C1 compounds. However, it was discovered that some type II strains can grow on multi-carbon substrates (Semrau et al. 2011). Methylocella species can grow on acetate, ethanol, malate, succinate, and pyruvate without losing its vitality. Methylocella silvestris BL2T grew faster on acetate than on methane and interestingly methane consumption rate increased after growing on acetate. Moreover, acetate was preferred when both substrates were available (Dedysh et al. 2005). Contrarily, Methylocapsa aurea can grow on acetate at the expense of the growth rate (Dunfield et al. 2010). In addition, many strains within Methylocystis genus can grow on acetate, whereas Methylocystis bryophila can poorly grow on pyruvate and ethanol (Belova et al. 2011, 2013; Im et al. 2011). Recently, it was reported that the strain Methylocystis hirsuta has the ability to grow on different multi carbon substrates including acetic, propionic, butyric and valeric acids (López et al. 2018a). Interestingly, Methylocystis hirsuta reached a higher biomass density when supplemented with both methane and any of the aforementioned multi carbon organic acids at adequate concentrations.
2.3.4 Methane and oxygen concentrations
In conditions where methane concentration is above 1%, type II methanotrophs form stable but slow growing communities in which the fluctuation in CH4 and O2 concentrations do not affect the community structure or their abundance (Chi et al. 2012a, b; Henckel et al. 2000; Semrau et al. 2010). Moreover, type II dominates in very low methane concentration, below 0.06%, due to their unique pMMO2 which is found in most Methylocystis sp., Mehtylosinus sporium, some of Methylosinus trichoporium strains (Baani and Liesack 2008; Knief and Dunfield 2005). It was reported in several studies that high methane to oxygen ratios, low oxygen concentration, are more preferable for type II growth (López et al. 2018b; Semrau et al. 2010). Unlike type II, type I methanotrophs grow faster with communities more sensitive to fluctuating methane and oxygen concentrations. As a result, the microbial structure of type I-dominated cultures change with the variation in the concentration of either methane or oxygen (Chi et al. 2012a, b; Henckel et al. 2000; López et al. 2014). Thus, type I mostly is expected to dominate in the first stage of any enrichment process. Moreover, it was reported that type I outcompete at methane concentrations between 0.06 and 1% where type II can grow but with lower abundance (Cantera et al. 2016; López et al. 2014; Semrau et al. 2010). This finding is supported by the fact that pMMO, unlike sMMO, has higher affinity to methane (Kalyuzhnaya et al. 2015). However, it is noteworthy that methane and oxygen mixing ratio or concentrations are not reliable selection parameter between type I and type II, unless other factors were manipulated, i.e., copper concentration or nitrogen source.
Generally, the increase in methane and oxygen concentrations is usually associated with remarkable enhancement in microbial growth and methane uptake rates of both types regardless of their relative abundance (Chi et al. 2012a, b; Li et al. 2014; López et al. 2014). However, some studies reported a decline in methane oxidation occurred at high oxygen concentration. This hypothesis was based on two observations; (1) the possession of oxygen sensitive nitrogenase, the enzyme responsible for fixing atmospheric nitrogen, which can be revealed by adding other nitrogen sources such as nitrate or ammonia, (2) the excessive accumulation of the toxic formaldehyde (Amaral and Knowles 1995; Pfluger et al. 2011).
2.3.5 Methane solubility
One of the major obstacles facing methanotrophs enrichments is the energy intensive agitation required to overcome methane low water solubility effects (dimensionless Henry’s law constant equals 30 at 25 °C) (Ordaz et al. 2014). Furthermore, attached growth systems usually exhibit very low yields and growth rates due to the limited methane transfer rate (Fennell et al. 1992; Pfluger 2010).
In an approach to increase methane solubility and its delivery to the bacteria, a pressure bioreactor was employed. Even though high biomass densities ranged from 18 to 65 g/L were achieved, pressure bioreactors are cost intensive and less secured than reactors working under atmospheric pressure (Helm et al. 2008; Wendlandt et al. 2001, 2005). Furthermore, loop bioreactors were also used for the enhancement of methane delivery to methanotrophs due to the advantage of their circular flow. Such advantage can be accomplished in fluidized systems by a propeller or a jet drive and optimum for gases having low solubility in water. Methylocystis hirsuta strain was cultivated in two loop bioreactors with different configurations; bubble column bioreactor and forced-liquid vertical loop bioreactor. Biomass density up to 2.9 g/L was successfully obtained (Rahnama et al. 2012). Another system was tested for methane solubility enhancement which is the two-phase partitioning bioreactor (TPPB) in which a non-aqueous phase (NAP) with higher affinity towards methane than water is employed. When 10% v/v of silicon oil was added, growth rate increased by 330% in the TPPB enriched with the Methylosinus sporium (DSMZ 17706) strain (Ordaz et al. 2014). Relying on the same concept, the addition of 5% v/v paraffin oil to the growth medium including Methylosinus trichosporium OB3b in a 5-l fermenter resulted in increasing the biomass density from 1 to 6 g/L after 160 h. Furthermore, the biomass density reached 14 g/L in the medium supplemented with paraffin oil after 240 h (Han et al. 2009). In addition, multiple agents were evaluated to enhance methane limited mass transfer including polymers, nanoparticles and electrolytes. However, bioreactor configuration modification found to be more efficient. Moreover, the addition of the aforementioned agents is associated with some challenges such as the downstream processing and their effect on the microbial community (Stone et al. 2017).
2.3.6 Nitrogen sources
All type II methanotrophs and few type I methanotrophs (Methylococcus, Methylosoma, Methyloglobulus, Methyloprofundus, and some strains within Methyomonas, and Methylobacter) have the ability to fix atmospheric nitrogen via the oxygen sensitive nitrogenase. Whereas, Methylosphaera has more oxygen tolerance to nitrogen fixation (Bowman 2006; Bowman et al. 1997). Thus, type II methanotrophs dominate under N-limiting conditions or high carbon to nitrogen (C/N) ratios. Whereas, type I methanotrophs require high nitrogen content or lower C/N ratios for better growth (He et al. 2011; López et al. 2013; Zhang et al. 2014). This distinguishing ability was manipulated as a selection tool in the selection of type II methanotrophs from mixed cultures in low dissolved oxygen concentrations (Pfluger et al. 2011).
On the other hand, methanotrophs prefer to grow on nitrate and ammonium as the cellular nitrogen source (Bowman 2006; López et al. 2013). Ammonium presence can partially inhibit the methanotrophic growth because of the competition between ammonium on MMO or the accumulation of excessive toxic hydroxylamine or nitrite. Whereas, nitrate supports higher methanotrophic growth rates than ammonium for both type I and type II methanotrophs (Karthikeyan et al. 2016; Sundstrom and Criddle 2015). Interestingly, it was found that higher moisture content can relieve ammonium toxicity influence. Whereas, substrate competition effects can be eliminated under higher methane to ammonium ratios (He et al. 2011; Wang et al. 2011; Zhang et al. 2014).
Under methane sufficient conditions, type II methanotrophs, unlike type I, can build more stable communities in the presence of ammonium with minor inhibitory effects (Mohanty et al. 2006; Visscher et al. 2001; Zhang et al. 2014). Supportively, it was reported that ammonium had a minor inhibitory effect on the type II Methylocystis sp. in comparison with type I cultures including Methylomicrobium album, Methylosinus sporium, and Methylomonas methanica (Nyerges and Stein 2009). It was suggested that Methylocystis sp. might possess a multiple enzymes system that can detoxify hydroxylamine (Nyerges and Stein 2009). Furthermore, Methylocystis dominated an activated sludge culture after 24 days of incubation under continuous ammonium feeding conditions (Myung et al. 2015a). Interestingly, a recent study reported that the sMMO expressing Methylosinus trichosporium OB3b has a slightly higher growth yield and methane oxidation rate when growing on ammonium than nitrate (Zhang et al. 2017b). Collectivity, it can be concluded that using nitrate as nitrogen source results in higher growth and methane oxidation rates with type I dominant cultures. Whereas, ammonium continues presence or N-limiting conditions result in type II dominant cultures with slower growth rates.
In contrast to the findings mentioned above, some reported batch experiments revealed that the addition of ammonium to different N-limiting soils was accompanied with an increase in methane oxidation rates and type I abundance (He et al. 2011; Lee et al. 2009; Zhang et al. 2014). However, this can be referred to type I faster response for the nutrients sufficiency conditions not type I preference of ammonium as the nitrogen source (Semrau et al. 2010). Supporting this hypothesis, a notable decline in methane oxidation rates was noticed in the same cultures. Furthermore, the addition of nitrate to N-limiting cultures was found to have greater stimulatory effect than the ammonium (Karthikeyan et al. 2016; Mohanty et al. 2006; Zhang et al. 2017b).
2.3.7 Copper
As mentioned before, copper concentrations regulate the expression of MMO enzyme (Semrau et al. 2010). pMMO is well developed in concentrations above 1 µM, while sMMO is expressed below this level (Hanson and Hanson 1996). However, several studies discovered that copper concentration is not a reliable parameter in the selection of type I or type II as most of methanotrophs can express pMMO (Cantera et al. 2016; Pieja et al. 2011a). Moreover, some sMMO lacking methanotrophs can survive and grow under very low copper concentration like Methylomicrobium and Methylobacter. This can be referred to their possession of unique copper uptake mechanisms like the methanobactin one (Cantera et al. 2016; Semrau et al. 2010). In addition, some type I like Methylomonas possess both sMMO and pMMO and dominate in cultures having high copper concentrations (Cantera et al. 2016; van der Ha et al. 2012a, b). In mixed cultures, some studies mentioned that copper addition significantly increase methane uptake and growth rate especially between 1 and 4.31 µM (Cantera et al. 2016; Ho et al. 2013a, b; López et al. 2013). Conversely, other studies noticed either no or minor increase (van der Ha et al. 2012a, b; Karthikeyan et al. 2016). Whereas, higher copper concentrations was reported to have an inhibitory effects due to its toxicity (Ho et al. 2013a, b; Lee et al. 2009). These contradicted reports suggest that copper concentration is not the most decisive factor on the microbial activity which depends on other factors such as initial community structure, methane concentration, and nitrogen source. For instance, if the dominant genus is Methylomonas, no or minor increase will be associated with copper increase, while in the case of type II existence an increase will be observed (Graham et al. 1993; van der Ha et al. 2010, 2012a, b). Moreover, as mentioned in the previous sections, ammonium presence in the growth medium with relatively high concentrations may result pMMO enzyme inhibition and better expression of sMMO. Whereas, lower methane concentrations stimulate pMMO enzyme expression due to its higher methane affinity (Karthikeyan et al. 2016).
3 Value-added resources recovery using methanotrophs in wastewater
WWTPs represent 4% of the yearly global methane production which make them a valuable biogas source (Ho et al. 2013a, b). In 1986, Corder and his team were the first to utilize methanotrophs enriched from anaerobic digester sludge confirming their potential of accumulating methanol from methane (Corder et al. 1986). Recently, numerous studies isolated or reported the existence of different types of methanotrophs from different locations within WWTPs such as waste activated sludge (WAS), and anaerobically digested sludge (Myung et al. 2015a; Reyes et al. 2015; Siniscalchi et al. 2015). Moreover, methanotrophs successfully dominated mixed cultures seeded from the aforementioned types of sewage sludge in different studies (Cantera et al. 2016; Jewell et al. 1992; Kampman et al. 2014).
In this section, three potential biotechnological applications for methanotrophs are reviewed to be employed in an existing WWTPs for value-added recovery.
3.1 Methanol production using methanotrophs
In addition to its negative effect on the environment, multiple obstacles limit the direct energy generation from biogas such as the existence of impurities and moisture, its low handling and collecting capabilities, and lack of convenient infrastructure for gas distribution. Moreover, combined heat and energy technologies show low electricity efficiency (η ≈ 25–45%) (Bachmann et al. 2015; Ge et al. 2014). Alternatively, methanol can be used as a fuel either standalone or blended with gasoline. Methanol, as a fuel, is more feasible than methane due to its higher transportability, and security (AlSayed et al. 2018). Furthermore, more energy can be derived from methanol (15.8 MJ/L) compared with methane (38.1 × 10−3 MJ/L) (Hwang et al. 2014). Methanol has lower NOx and SO2 emission than natural gas (Murray and Furlonge 2009). In addition, methanol is considered as a substitute carbon source for sugar used in biochemical industry (Schrader et al. 2009). Additionally, methanol is commonly used as an external carbon source used to enhance denitrification process in wastewater treatment (Ginige et al. 2008). Collectively, methanol is considered as a multiple use commodity with a prominent role as an efficient, sustainable substitute for methane produced from WWTPs.
3.1.1 Bio-methanol production metabolism
Bio-methanol can be produced from methane either via utilizing the MMO enzyme or the whole methanotrophic cell as biocatalysts. The former process has major drawbacks such as the high cost and enzymes instability (Park and Lee 2013). Hence, biological methanol production using methanotrophs can be more feasible especially using the anaerobic digestion driven biogas (Sheets et al. 2016).
As expressed before in Eqs. (1)–(3), methanotrophs hydroxylate methane in an energy requiring step which is catalyzed by MMO. The produced methanol is rapidly oxidized to carbon dioxide with formaldehyde and formate as intermediates. Oxidations are catalyzed by methanol dehydrogenase (MDH), formaldehyde dehydrogenase (FaDH), formate dehydrogenase (FDH), respectively. Accordingly, it is essential to inhibit MDH activity for extracellular accumulation of methanol which, subsequently, inhibiting the following oxidation steps (Mehta et al. 1987). Unfortunately, the latter three steps are electrons producing step which provide the cells with the energy needed to govern simultaneous methane uptake. Furthermore, cellular carbon (used for cell synthesis and replication) is assimilated in the level formaldehyde, formate, and carbon dioxide. Thus, the inhibition of methanol oxidation would be associated with two major challenges (1) notable decline in bacterial growth, and (2) lack of energy needed for methane uptake to be, subsequently, converted into methanol. Therefore, an additional source of electrons, usually formate, is added to maintain cell vitality and for continuous methane uptake (Bjorck et al. 2018). Figure 4 illustrates the methanol production pathway.
3.1.2 Factors affecting bio-methanol production
Even though intensive researches have been conducted on bio-methanol production from methane using methanotrophs, several challenges still hinder the process upscaling. To the moment, no studies have successfully developed a feasible and stable methanol production process. Most of the reported studies were performed in batch scale and short term. Methanol production periods ranged from 8 to 24 h. Whereas, longer production durations were associated with relatively lower productivities (Bjorck et al. 2018; Ge et al. 2014). This can be referred to the bacterial decay associated with methanol production caused by the metabolic reactions inhibition. In order to make methanol production process industrially feasible, multiple factors, (i.e., pH, Temperature, cell densities, and gases mixing ratio) need be optimized as well as electrons supply and MDH inhibition strategies.
-
Biomass culture
Most of the previous studies were performed using pure cultures resulting in various methanol productivities (Bjorck et al. 2018). The most common strain used for methanol production is Methylosinus trichosporium (Ge et al. 2014). The highest methanol productivity obtained using M. trichosporium was equal 4101 mgmethanol/L/day (Mehta et al. 1991). Whereas, the highest reported methanol productivity using methanotrophs was attained using a novel type I Methylomonas sp. DH-1 isolated from brewery waste sludge which was equal to 7968 mgmethanol/L/day (Hur et al. 2017). Isolated from solid state anaerobic digester, Methyloculdum sp. 14B strain has shown relatively high methanol productivity of 1090 mgmethanol/L/day (Sheets et al. 2016). In addition, multiple strains within type II methanotrophs have been used including Methylosinus sporium, Methylocella tundrae, Methylocystis bryophila, Methylocella silvestris, Methyloferula stellate, and Methylomonas methanica. However, the resulted methanol concentrations were relatively low (lower than 200 mgmethanol/L) (Mardina et al. 2016; Patel et al. 2016c, 2017).
Han et al. was the first to accumulate methanol using methanotrophic mixed culture. A consortium of strains (Methylosinus trichosporium OB3b, Methylococcus capsulatus, and Methylosinus sporium) isolated from landfill soil resulted in methanol concentration of 220 mgmethanol/L (Han et al. 2013). Recently, a mixed culture dominated by type I methanotrophic was enriched from waste activated sludge and used for methanol production. The attained methanol productivity was equal to 2110 mg/L/day which is comparable to the pure cultures (AlSayed et al. 2018). It is noteworthy to mention that the highest methanol productivities in pure and mixed cultures were obtained using type I methanotrophs (AlSayed et al. 2018; Hur et al. 2017). This can be elucidated by type I methanotrophs higher growth rates and methane uptake rates, as discussed before.
-
Nutrients
As mentioned before, copper is a crucial nutrient for pMMO and sMMO expression regulation. Therefore, methanol productivity increased with copper addition. It was reported that concentration ranging from 1 to 5 µM Cu2+ would notably increase methanol production. However, it was found that copper concentrations higher than 10 µM inhibited both the methanol production and bacterial growth. Also, addition of 10 µM iron in the culturing medium resulted in an enhanced methanol production due to its positive effect on MMO activity (Furuto et al. 1999; Pen et al. 2014; Sheets et al. 2016). Interestingly, it was found that the addition of both 10 µM Fe and 5 µM Cu doubled methanol production of M. sporium (Patel et al. 2016c). Furthermore, it was found that methanol concentrations increased by 60% after nutrients addition which can be referred to their effect on the cellular activity and enzymes expression (AlSayed et al. 2018).
-
Headspace composition
Theoretically, 1 mol of methane and 1 mol of oxygen are required to produce 1 mol of methanol. However, the reported values for methane to oxygen ratio in the headspace always deviates from the theoretical ratio (Ge et al. 2014). This deviation can be referred to the limitation of oxygen and methane mass transfer and/or further methanol oxidation (Hur et al. 2017). Methane concentrations used in the previous studies varied from 20 to 80% (Bjorck et al. 2018; Hwang et al. 2014). It was reported that increasing methane concentration up to 50% leads to higher methanol production, especially, at incubation times longer than 24 h (Patel et al. 2016c, 2017). A recent study showed that further increase in methane, up to 80%, leads to higher methanol productivity and methane uptake rate (AlSayed et al. 2018).
In WWTPs, anaerobic digesters (ADs) are the methane factories which generate methane along with other gases such as CO2 and H2S forming the biogas. Typically, biogas consists of 50–80% of methane, 20–50% of carbon dioxide, up to 2% of H2S, and other trace impurities such as NH3 and siloxanes (Lebrero and Chandran 2017). Interestingly, raw biogas from commercial scale AD system was efficiently used for methanotrophic microbial growth and methanol production (Sheets et al. 2016). In agreement, Patel et al. found that the presence of carbon dioxide with concentrations up to 20% resulted in increasing methanol concentrations from 3.86 to 4.35 mM in comparison with pure methane (Patel et al. 2016a). Elucidating this increase, Xin et al. has demonstrated that carbon dioxide with concentrations up to 40% can inhibit the MDH and trace methanol concentrations was accumulated (Xin et al. 2004a). Methyloculdum sp. SAD2 strain isolated from commercial scale AD tolerates up to 500 ppm of H2S and convert methane into methanol with conversion ratios comparable to the pure methane (Zhang et al. 2016). Collectively, it can be concluded that the main components of biogas—other than the methane—either have limited influence or enhance methanotrophs growth or methanol production.
The presence the hydrogen gas (H2) in the headspace was reported to enhance methanol production. Patel et al. reported that the use of biohythane, a mixture of CH4 60–70% (v/v) and H2 10–15% (v/v), enhanced methanol production by 1.9-fold in comparison with pure methane as a feed. The mechanism of H2 positive effect has not been confirmed yet, however, it was proposed that methanotrophs utilize it as source of reducing power for NADH generation (Patel et al. 2016a, 2017).
-
MDH inhibition
Methanol oxidation is catalyzed by the PQQ linked MDH, as shown in Eq. (4), in which cytochromes-c are utilized as the electron carriers to the terminal oxidase (Chistoserdova and Lidstrom 2013a). Only trace methanol concentrations can be observed unless methanol oxidation is inhibited. The most common strategy used for MDH inhibition is the addition of chemicals inhibitors including; phosphate, NaCl, cyclopropanol, EDTA, MgCl2, and NH4Cl (Hwang et al. 2014). Such inhibitors were used either separately or combined in order to inhibit MDH activity.
Methanol accumulation not only depends on MDH inhibition but also MMO activity which controls the methane oxidation rate. For instance, NH4Cl was found to inhibit both MMO and MDH activity resulting in lower methanol yields, even though high methane to methanol conversion ratio was attained. This low methanol yield is due to the decline in the methane uptake rate caused by MMO partial inhibition (Han et al. 2013; Yoo et al. 2015). As discussed before, it is suggested that MDH supplies MMO with the electrons needed for methane hydroxylation (Kalyuzhnaya et al. 2015). Hence, it was reported that the optimum MDH relative activity should be in the range of 50% (Takeguchi et al. 1997). Further MDH inhibition would be associated with a decline in MMO activity, and by consequence, limited methane hydroxylation (Takeguchi et al. 1997). Furthermore, full MDH inhibition would fully eliminate any cellular carbon assimilation and energy generation.
Cyclopropanol with an optimum concentration of 0.67 µM was reported as the most efficient MDH inhibitor. At this concentration, MDH activity is inhibited by 50% while decreasing the MMO activity by 5.2% only, as shown in Table 3 (Furuto et al. 1999; Takeguchi et al. 1997). However, cyclopropanol is not commonly used due to its instability under aerobic conditions and difficulty in preparation. Moreover, it is an irreversible inhibitor as it produces a stable compound from the interaction with the free and MDH-linked PQQ (Ge et al. 2014; Kim et al. 2010).
Furthermore, high salts concentrations (i.e., NaCl, NH4Cl and phosphate) can disrupt electrons transport between PQQ and cytochromes-c decreasing the MDH activity (Ge et al. 2014). NH4Cl is not commonly used due to its inhibitory effect on MMO activity, as shown in Table 3 (Kim et al. 2010; Yoo et al. 2015). On the other hand, NaCl is an advantageous inhibitor due to its stability, abundance and low cost. It was reported that 200 mM of NaCl is the optimum concentration for methanol production (Lee et al. 2004). However, it was observed that NaCl concentrations higher than 100 mM distorts the cell morphology. Thus, it is recommended to use combination of NaCl and other inhibitors such as EDTA to eliminate the cell distortion effect (Lee et al. 2004). Interestingly, EDTA addition resulted in approximately 1.8 times higher methanol concentration in comparison with using NaCl only (Kim et al. 2010; Pen et al. 2014).
Phosphate is the most commonly used MDH inhibitor with concentrations ranging from 40 to 100 mM. It is considered to be uncompetitive and reversible (Bjorck et al. 2018; Lebrero and Chandran 2017). As presented in Table 3, phosphate is usually used in combination with other inhibitors such as EDTA and MgCl2 due to the notable decline in MMO activity at phosphate concentrations above 100 mM (Mardina et al. 2016; Takeguchi et al. 1997). Even though MgCl2 inhibitory mechanism is still unknown, various studies observed that the addition of MgCl2 with concentrations ranging from 5 to 20 mM to phosphate buffer significantly enhance methanol yields (Duan et al. 2011; Ge et al. 2014). On the other hand, the addition of 50 µM–1 mM of EDTA to phosphate buffer yields higher methanol concentrations. It decreases the MDH activity by blocking the electron transfer to the cytochrome-c as it binds to the cytochrome-binding area in the MDH (Ge et al. 2014; Han et al. 2013). As shown in Table 3, EDTA had higher inhibitory effect on the MDH activity. However, it was reported that MgCl2 addition to phosphate results in higher methanol concentrations in comparison with EDTA. This can be elucidated by the fact that EDTA has negative effect on MMO due to its chelation effect on the MMO metal ions. Whereas, Mg ions support MMO activity and the methanotrophic cell growth (Mardina et al. 2016; Patel et al. 2016c; Sheets et al. 2016).
As a cost effective MDH inhibition alternative, Xin et al. reported that the presence of high carbon dioxide concentrations can partially inhibit MDH activity while the other electrons producing steps still occurring. However, the maximum achieved methanol production rate was too low (0.1 mg/L/h) (Xin et al. 2004a).
-
Electron donor
As previously illustrated, methanol accumulation should be associated with external addition of a reducing power source. Theoretically, two electrons (1 mol of NADH) are needed to produce 1 mol of methanol. Both formaldehyde and formate can be added as reducing power source. However, formaldehyde is toxic and has inhibitory effects on the methanotrophic activity. Hence, most of studies were adding formate for simultaneous methane uptake and methanol production (Bjorck et al. 2018). Wide range of formate concentrations (from 14.3 to 120 mM of sodium formate) were reported as the optimum concentration for methanol production (Bjorck et al. 2018; Ge et al. 2014). Interestingly, Sheets et al. mentioned that excessive formate addition resulted in methanol accumulation with productivities up to (500–1000 mg/L/day) without adding any MDH inhibitors. It was referred to the higher activity of FDH than MDH resulting in a higher production rate of reducing power (Sheets et al. 2016). On the other hand, methanol was successfully accumulated for about 24 h without formate addition. However, the production rate was low and a sharp decline was noticed after 24 h (Han et al. 2013; Yoo et al. 2015). These reports suggest that methanotrophs can utilize other electron donors such as lipids.
Unfortunately, the addition of sodium formate is not economically feasible at the commercial scale. Therefore, alternative strategies should be investigated such as generating electrons electrochemically or using facultative methanotrophs. Interestingly, Xin et al. used PHB as a reducing power reserve while converting carbon dioxide (not methane) into methanol. After 144 h, cells with 38.6% of PHB produced 3 mg/L methanol without losing its vitality (Xin et al. 2007). Even though low methanol yield was observed, employing PHB as an electron donor is shown to be viable. The low methanol yield can be resolved by the use of methane instead of carbon dioxide and by applying more efficient MDH inhibition strategy.
-
Cell density
Biomass density has contradictive results on its effect on methanol accumulation (Ge et al. 2014). Some studies reported that the optimum cell density of M. trichosporium is 3000 mg/L, while a decline in methanol accumulation was noticed with further increase in the cell density (Mehta et al. 1991; Xin et al. 2007, 2004b). In contrast, Takeguchi et al. (1997) found that the maximum methanol accumulation occurred at cell density as low as 35 mg/L. Whereas, Lee et al. (2004) reported the peak accumulation at a cell density of 600 mg/L. Other studies using M. tundrae (Mardina et al. 2016) and M. bryophila (Patel et al. 2016b) reported an increase by 1.35 and 1.5 times in methanol concentrations while increasing the cell density from 1.5 to 9 and 3 to 18 g/L, respectively. The aforementioned studies were performed using different bioreactor configurations, gases delivery techniques, and shaking speeds which can elucidate the contradicted results. At high cell density, Duan et al. used 5% paraffin oil to eliminate the mass transfer negative effect and higher concentration of MDH inhibitors. Methanol concentration as high as 1130 mg/L was achieved by increasing the M. trichosporium at cell density of 17,000 mg/L (Duan et al. 2011). These results suggest that increasing cell density should be accompanied with an increase in methane concentration, MDH inhibitors, and overcoming mass transfer limitation. However, it is noteworthy that, in all reports, the specific methanol productivity decreases with the biomass increase (Bjorck et al. 2018). Therefore, it can be concluded that enhancements in the bioreactor engineering are still needed to achieve higher methanol concentrations at higher biomass densities.
Accordingly, the suggested conditions for methanol production using methanotrophs are illustrated in Table 4.
3.2 PHB production by methanotrophs
Methanotrophs can convert biogas to a biodegradable polymer PHB from the PHA family. Therefore, these microorganisms can combine the need for biogas emissions remediation and a cheaper feedstock to produce bioplastics which is one of the main dragging forces to the mass production of biopolymers and contribute to 30–50% of the production cost (Khosravi-Darani et al. 2013). Methanotrophs can convert methane aerobically to PHB under unbalanced growth conditions, deficiency or limitation of essential nutrients i.e., nitrogen and phosphorus prevent their cellular growth and force them to store PHB as intracellular granules (Karthikeyan et al. 2015). Figure 5 shows a suggested schematic diagram for the cycle of PHB production using biogas. After extracting the accumulated PHB inside the bacterial cell, it can be either used as it is or combined with other polymers to get a desired product.
3.2.1 Mechanism of PHB production in methanotrophs
Under balanced conditions when all the essential nutrients are available, methanotrophs proceed to TCA cycle to obtain their energy needs. While under unbalanced conditions when one or more nutrients are not available, they switch to the PHB cycle to provide the energy required for the cell maintenance i.e., survival in this case. Acetyl-CoA plays the intermediate role under limited conditions; it is the first entry to the PHB cycle with the aid of some specific enzymes. Firstly, Acetyl-CoA is converted to Acetoacetyl-CoA through the enzyme β-ketothiolase encoded by PhaA gene that is then reduced to β-hydroxybutryl-CoA by the enzyme Acetoacetyl-CoA reductase encoded by PhaB gene. PHB synthetase enzyme encoded by PhaB gene, which is a polymerase, starts to form PHB from β-hydroxybutryl-CoA. The second part of the cycle includes the enzyme PHB depolymerase encoded by PhaZ gene that depolymerizes PHB granules formed to hydroxybutrate monomers. Secondly, acetoacetate is formed with the aid of β-hydroxybutyrate dehydrogenase which is then converted to Acetoacetyl-CoA by Acetoacetate succinyl-CoA transferase to complete the cycle (Chidambarampadmavathy et al. 2015; Zhang et al. 2009). Figure 6 illustrates the pathway for the PHB cycle in methanotrophs.
As described above, Acetyl-CoA is crucial for the PHB cycle and most probably can be produced only through the serine pathway (Babel 1992). Moreover, the RuMP pathway does not proceed a complete TCA cycle to obtain energy from Acetyl-CoA (Karthikeyan et al. 2015) which supports the hypothesis that PHB accumulation is exclusive to type II methanotrophs. Moreover, the consumption of the accumulated PHB generates the reducing equivalent NAD(P)H+ which is utilized in methane oxidation as a survival mechanism. As previously mentioned, sMMO is mainly found in type II methanotrophs and only use the NAD(P)H+ as reducing equivalents for the methane oxidation step. Accordingly, several strains from type I and type II methanotrophs were tested for the existence of PhaC “PHB polymerase”. Only type II strains possess this gene confirming the conclusion of their exclusiveness for PHB accumulation (Pieja et al. 2011a).
While some mixed cultures can grow using the stored biopolymers when other carbon sources were missing (Ciğgin et al. 2007; Dircks et al. 2001; Majone et al. 1998), PHB was assumed to function as a carbon source for methanotrophs growth under the limited conditions (Chistoserdova and Lidstrom 2013b; Sipkema et al. 2000). However, experimental studies revealed that PHB is not produced at methane absence, showing that PHB cannot be used as a sole carbon source. Thus, PHB can be used only as a source of reducing equivalents for the methane uptake and its conversion to methanol (Pieja et al. 2011b).
3.2.2 Optimization of growth and PHB accumulation in type II methanotrophs
Since PHB accumulation is likely limited to type II methanotrophs, previous studies focused mainly on two targets, the factors affecting type II growth and conditions enhancing their PHB accumulation capability. Accordingly, pure culture studies were conducted to understand the effect of different nutrients concentrations on the growth of these bacteria and their role in type II metabolism while fewer studies considered mixed cultures. In the following section, we will discuss some of the reported factors affecting the growth and PHB accumulation of different type II methanotrophic strains.
-
Nitrogen source
Nitrogen limitation was one of the most discussed conditions for PHB accumulation by methanotrophs. Moreover, it was concluded that the nitrogen limitation choice can be an effective parameter for long-term productivity of PHB in mixed cultures (Myung et al. 2015a). However, contradictive data are available for the optimum choice of nitrogen source to maximize the bacterial growth and enhance their PHB accumulation capacity.
Type II strains have different responses to the nitrogen source available in the growing medium. The strain Methylosinus trichosporium OB3b accumulated 50% PHB after nitrogen depletion in a nitrate salt medium with an initial concentration of 20 mmol (Shah et al. 1996). In another study, the same strain accumulated 38% PHB in nitrogen limited conditions after growing in a medium with initial nitrate concentration of 10 mmol (Pieja et al. 2011a). Using ammonium as a nitrogen source with an initial concentration of 10 mmol resulted in only 13% PHB accumulation, while switching to nitrogen gas increased the PHB accumulation to 45% (Rostkowski et al. 2013). When both nitrate and ammonium were used with concentrations of 10 and 8 mmol, respectively, PHB accumulation reached 30% (Doronina et al. 2011).
On the other hand, Methylocystis parvus OBBP accumulated 60% PHB after growing on ammonium and transferred to nitrogen limited conditions (Rostkowski et al. 2013) compared to 36% accumulation when it was growing using nitrate as nitrogen source (Pieja et al. 2011a). The strain Methylosinus trichosporium IMV3011 accumulated 47% PHB after growing on both ammonium and nitrate as nitrogen sources with initial concentration of 16 and 10 mmol respectively (Song et al. 2012). In addition, Methylocystis hirsuta accumulated up to 51% PHB after growing on 13 mmol ammonium then transferred to deficiency conditions (Rahnama et al. 2012).
In mixed culture consortium, an enrichment dominated by the Methylocystis GB25 strain accumulated 51% PHB under ammonium deficiency conditions (Wendlandt et al. 2001). Another enrichment growing on nitrogen gas prior to nitrogen limitation accumulated 43% PHB and this enrichment was dominated by the Methylocystis and Methylosinus genus (Pieja et al. 2011a). Also, an enrichment dominated by Methylocystis genus and growing on 13.5 mmol ammonium accumulated 39% PHB in nitrogen limitation conditions (Myung et al. 2015a). Recently, a new nitrogen source was introduced to a mixed consortium of PHB producing methanotrophs, which is urea. Urea is gradually converted to ammonium decreasing its toxic effect and then can be utilized by methanotrophs resulting in 39% PHB accumulation under deficiency conditions (Sundstrom and Criddle 2015).
All of the previous studies especially on mixed cultures revealed that when nitrate was chosen as nitrogen source biomass density was higher. However, the PHB production was less due to the invasion of the non-producing PHB type I methanotrophs. Contrarily, growth on ammonium produced higher level of PHB at the expense of the biomass (Criddle et al. 2015a). This can be referred to the toxicity produced by the hydroxylamine resulting from the co-oxidation of ammonium. Interestingly, type II methanotrophs have higher resistance to ammonium toxicity. Based on these data, a new strategy was developed on a mixed culture from an activated sludge where the mixed culture firstly grew on ammonium as a selection for the PHB producing microorganisms and then transferred to grow on nitrate to increase their biomass density. These microorganisms mainly composed of type II methanotrophs accumulated about 40% PHB. This strategy appears to be successful, however, some modification can be applied to increase the PHB productivity including the optimization of nitrogen concentrations (Criddle et al. 2015b).
-
Phosphorus
Phosphorus concentration affects both type II growth and their PHB accumulation. A concentration of 2–25 mmol is needed to maintain the sMMO activity (Bowman and Sayler 1994). Whereas, concentrations above 40 mmol completely inhibited the growth of Methylosinus trichosporium OB3b (Park et al. 1991). On the other hand, phosphorus deficiency resulted in higher PHB accumulation values. Methylocystis parvus OBBP accumulated 31% PHB in a phosphorus deficient modified medium containing 0.12 mmol compared to 18% PHB in a control medium with phosphorus concentration of 2.9 mmol (Sundstrom and Criddle 2015). In addition, under phosphorus deficiency conditions, PHB accumulation reached 46% in an enrichment dominated by the Methylocystis GB25 strain (Wendlandt et al. 2001). Moreover, decreasing phosphorus concentrations from 7.3 to 5.7 mmol increased PHB accumulation from 16 to 26.5% by Methylosinus trichosporium IMV3011 strain (Zhang et al. 2008).
-
Copper
Similar to nitrogen, contradictive data is reported for the effect of copper on the PHB accumulation capacity for different strains. In a study on Methylosinus trichosporium OB3b, bacteria accumulated 42% PHB in a copper free medium while accumulating 50% PHB in a medium supplemented with 10 µm copper (Shah et al. 1996). On the other hand, PHB accumulation for Methylocystis parvus OBBP increased from 18 to 49% when copper concentration was decreased from 15 to 5 µm (Sundstrom and Criddle 2015). In a study on the combined effect of nitrogen source and copper on the growth rates and PHB accumulation of the Methylosinus trichosporium OB3b strain, it was found that the highest growth rate was achieved while the bacteria was growing on ammonium in a copper free medium and accumulated 25% PHB under limited conditions. On the other hand, switching to nitrate with 5 µm copper relatively decreased the growth rate but the PHB accumulation reached 51% (Zhang et al. 2017b).
-
Other nutrients
Most of the used media for the cultivation of methanotrophs had iron concentrations ranging from 6 to 20 µm (López et al. 2014; Sundstrom and Criddle 2015; Zhang et al. 2008). In addition, it was found that relatively higher iron concentrations ranging from 40 to 80 µm supports sMMO activity (Park et al. 1991). However, increasing iron concentration from 4.6 to 60 µm during nitrogen limited conditions declined the PHB accumulation of Methylocystis hirsuta from 28 to 19% due to copper existence in the culturing medium (García-Pérez et al. 2018). Moreover, iron deficiency did not result in high PHB accumulation (11%) in an enrichment dominated by the Methylocystis GB25 strain while sulphur deficiency resulted in 33% PHB accumulation (Helm et al. 2008).
On the other hand, magnesium and potassium deficiency resulted in a PHB accumulation of 28 and 34% respectively (Wendlandt et al. 2005). Decreasing potassium concentration from 0.58 to 0.09 mmol increased PHB accumulation from 18 to 28% in Methylocystis parvus OBBP strain while Methylocystis hirsuta accumulated 12.5% PHB under potassium limited conditions (García-Pérez et al. 2018). In addition, when calcium concentration was decreased to 7.2 µm PHB accumulation was doubled to reach 39% in Methylocystis parvus OBBP strain (Sundstrom and Criddle 2015). Moreover, sMMO activity was significantly affected when 1 mmol of zinc was added to the medium and completely stopped by 0.01 mmol of mercury (Grosse et al. 1999).
-
Temperature and pH
Most of the studies were carried out in temperature ranging from 20 to 40 °C and rarely discussed the effect of temperature on biomass yields and PHB accumulation. However, pMMO activity declined sharply after 45 °C in a study on the Methylosinus trichosporium OB3b strain (Takeguchi and Okura 2000) while sMMO activity decreased at temperatures above 30 °C (Park et al. 1991).
Regarding the pH, most of the studies used culturing medium having a pH ranging from 6 to 7. However, it was reported that increasing the medium acidity was one of the conditions that favored the PHB accumulation of type II methanotrophs. The acidic medium increased the dissolution of CO2 in the culturing medium which can be employed as an input for the serine cycle (Pieja et al. 2011a).
-
Methane and oxygen
At high oxygen levels the rate of methanol to formaldehyde may increase leading to the inhibition of the whole metabolic reaction (Costa et al. 2001). In a study on the effect of oxygen partial pressure on biomass growth and PHB accumulation, the maximum PHB accumulation for Methylosinus trichosporium OB3b was 45% at 0.2 atm., while Methylocystis parvus OBBP accumulated 60% PHB at 0.3 atm. On the other hand, The maximum biomass yield was observed at 0.4 atm. (Rostkowski et al. 2013). While in another study on the combined effect of the oxygen partial pressure and the methane to oxygen ratio in a methanotrophic mixed culture, the maximum PHB content for cultures grown at 0.1 atm was achieved when the methane to oxygen ratio was 1:1. Changing this ratio had a negative effect on the amount of PHB accumulated by almost 25%. However, for cultures grown at an oxygen partial pressure of 0.2 atm, increasing the methane to oxygen ratio up to 3:1 did not have a significant effect and the PHB accumulation reached 45%. Contrarily, decreasing the methane to oxygen ratio to 1:3 declined the PHB accumulation to 37% (Zhang et al. 2017a). Therefore, the suggested growth conditions for targeting type II methanotrophs are illustrated in Table 5.
3.2.3 Molecular weight of extracted biopolymers
The molecular weight of the PHB accumulated in type II methanotrophs is mainly regulated by the combined effect of the PHB synthetase and depolymerase enzymes activity (Song et al. 2012). In addition, under nutrient deficiency conditions the activity of these enzymes increases while increasing the molecular weight of the PHB accumulated (Wendlandt et al. 2001; Xin et al. 2013). Addition of inhibitors to the TCA cycle induced the PHB cycle and resulted in higher PHB accumulation with higher molecular weight during the nutrient deficiency conditions (Song et al. 2012; Zhang et al. 2008). Table 6 shows the molecular weight of accumulated PHB corresponding to different nutrient deficiency conditions or addition of some organic acids (inhibitors for the TCA cycle).
3.2.4 Biomass and PHB yields
Most of the studies reported the PHB yields under nitrogen limitation condition only. However, other studies reported biomass yields and PHB yields under different nutrients deficiency conditions rather than nitrogen. Table 7 shows different values for the PHB yields reported in different studies for some type II strains. On the other hand, fewer studies reported the biomass yields for their experiments, generally, biomass yield ranged from 0.4 to 0.8 gDCW/gCH4 (Myung et al. 2015a; Pfluger et al. 2011; Pieja et al. 2012; van der Ha et al. 2012a, b).
3.2.5 Growth rates
As all other parameters, methanotrophs growth rate is affected by the cultivation conditions as pH, copper and nitrogen source. When the pH of the copper free growing medium was increased from 6 to 8.5 the specific growth rate of Methylosinus trichosporium OB3b decreased from 0.087 to 0.039 h−1 (Park et al. 1991). On the other hand, the specific growth rate for the same strain was 0.07, 0.095 and 0.0083 h−1 at copper concentrations of 0.21, 1.25 and 20 µmol, respectively (Takeguchi and Okura 2000). For the strain Methylocystis parvus OBBP reducing copper concentration from 15 to 5 µmol increased the specific growth rate from 0.065 to 0.08 h−1 (Sundstrom and Criddle 2015). Compared to other nitrogen sources, nitrogen gas resulted in the slowest growth rates for type II methanotrophs. In a study on the Methylosinus trichosporium OB3b strain, the growth rate was 0.015 h−1 during nitrogen gas growth compared to 0.11 h−1 while growing on ammonium under the same conditions. However, in terms of PHB accumulation, the bacteria grown on both nitrogen sources could accumulate up to 40% PHB under nutrient limited conditions (Zhang et al. 2017b). This slow growth rate can be contributed to the oxygen sensitivity of the nitrogen fixing genes for type II methanotrophs as increasing the oxygen partial pressure from 0.1 to 0.2 atm. decreased the growth rate of Methylosinus trichosporium OB3b from 0.24 to 0.06 h−1 (Rostkowski et al. 2013). Table 8 illustrates some of the recorded growth rates for different type II strains in correspondence to the nitrogen source.
3.2.6 Bioreactor configuration
An effective bioreactor configuration for scaling up PHB production from methanotrophs has some challenges to overcome. First, mixing two flammable gases oxygen and methane safely without affecting the overall performance of the bioreactor. Secondly, enhance the methane delivery to the microorganisms due to the low methane solubility which has been discussed earlier in the methane solubility section. Thirdly, minimizing the power requirements and operational costs. Lastly, achieving a sustainable capability of maintaining the growth of type II methanotrophs with a stable PHB accumulation. Production of PHB through methanotrophs occurs in two stages, a growth phase and a PHB accumulation phase. These two phases can take place in the same reactor or separately where PHB accumulation is expected to take place at the end of the logarithmic growth phase and the beginning of the stationary phase.
Bioreactors with different configurations tested the ability of type II methanotrophs for PHB accumulation while predicting the optimum mode of operation. For instance, Methylosinus trichosporium OB3b accumulated 45% PHB after 160 h in a copper free medium. While, a PHB content of 50% was achieved in a medium supplemented with 10 µmol copper after 120 h when the growth and PHB accumulation phases were simultaneously occurring in a stirred tank reactor (Shah et al. 1996). Moreover, PHB accumulation reached 51% after 24 h under deficiency conditions in a pressure bioreactor operated in a continuous mode during the growth phase and in a batch mode in the PHB phase using a methane-utilizing mixed culture (Wendlandt et al. 2001). In an attempt to deduce the optimum operating mode for PHB producing methanotrophs in a sequencing batch reactor having a 24 h cycle duration, an increase in the PHB production trend was observed over time when cycles started with 8 h of methane limitation (Pieja et al. 2012).
On the other hand, Methylocystis hirsuta accumulated 42.5% in a bubble column reactor under nitrogen limited conditions when the growth phase and PHB accumulation phase were separated using natural gas (Rahnama et al. 2012). Upon modifying the bubble column reactor configuration was with internal gas recirculation, the same strain accumulated up to 35% PHB during simultaneous growth and nitrogen limited cycles while treating air emissions polluted with 4% (v/v) methane (García-Pérez et al. 2018). Table 9 summarizes the results obtained for bioreactors studies under different conditions.
3.2.7 Co-polymers production by methanotrophs
While scaling up the biopolymers production process using methanotrophs is still under research, thermal and mechanical properties of the accumulated biopolymers are taken into consideration for a wider application. Unfortunately, PHB application can be limited due to its low thermal stability as the melting temperature for PHB (≈ 180 °C) is close to the degradation temperature (≈ 200 °C) making it harder in processing (Cal et al. 2016). Moreover, the stiffness, brittleness and high crystallinity are considered from the main drawbacks of PHB. PHB co-polymers can provide a possible solution for the aforementioned drawbacks. The incorporation of hydroxyvalerate (HV) units to PHB results in the formation of the co-polymer poly hydroxybutyrate-co-valerate PHBV which has a lower melting temperature, crystallinity, water permeability and enhanced mechanical properties (Strong et al. 2016). Properties of the produced co-polymer PHBV mainly depends on the HV fraction. For example, increasing the HV fraction from 3 to 25 mol% decreased the melting temperature from 170 to 137 °C (Lee 1996).
During the PHB accumulation phase, under nutrient limited condition, a co-substrate is introduced to the culturing medium such as citrate, propionate or valerate yielding ketovaleryl-CoA which is then converted into hydroxyvaleryl-CoA. The formed hydroxyvaleryl-CoA combines with hydroxybutyrate to form the PHBV polymers as illustrated in Fig. 7. The accumulation of PHBV is linked to the methane oxidation rate, moreover, it is prohibited in the absence of methane. Methane oxidation rate increased when valerate is added to the growing medium and increased the energy requirements for methane oxidation. The electron equivalent conversion fraction for energy (fe) increased from 0.25 without valerate to 0.35 and 0.45 after addition of 100 and 400 mg/L valerate respectively (Myung et al. 2015a) which can be linked to the ATP required for the valerate uptake (Myung et al. 2016a), as shown in Fig. 7.
Different co-substrates were tested for PHBV production. Valerate achieved the highest results, on the other hand, pentanol resulted in a decreased methane uptake and PHBV yield (Cal et al. 2016). In addition, valerate outcompeted propionate in the incorporation of HV monomers and total co-polymer production.
Valerate concentrations above 0.7% v/v showed an inhibitory effect on HV incorporation and on PHBV accumulation in general for the strain Methylocystis sp. WRRC1. At high valerate concentrations PHB was accumulated with 15% content only compared to 30% PHB in the absence of valerate. On the other hand, at 0.34% v/v valerate, the PHBV accumulated was 60% with 50% incorporated HV monomers. Furthermore, eliminating copper from the PHBV accumulation phase resulted in increasing biopolymer accumulation to 78% with an HV content of 58% (Cal et al. 2016). However, the inhibitory level of the added co-substrate seems to be strain specific where the total amount the biopolymer accumulated decreased after concentration of 100 mg/L for both the strain Methylocystis parvus OBBP (Myung et al. 2016a) and 115 mg/L for Methylocystis hirsuta (López et al. 2018a). Table 10 shows results for PHBV production using different methanotrophic strains.
Interestingly, Methylocystis parvus OBBP was able to produce different biopolymers from the PHA family when organic acids (co-substrates) including 3HB, butyrate, valerate, hexanoate, and octanoate were added during nitrogen limited conditions (Myung et al. 2017). The products were not only limited to PHB and PHBV but also included P(3HB-co-4HB), P(3HB-co-5HV-co-3HV) and P(3HB-co-6HHx-co-4HB). The composition of the final PHA was controlled by the number of carbon atoms and the existence and location of hydroxyl group in the organic acid. This finding opens a new insight towards the methanotrophic-based production of biopolymers which can be devoted towards different applications.
3.3 Nitrification and denitrification pathways for methanotrophs
One of the interesting characteristics for methanotrophs is their flexible response towards different nitrogen sources under different oxygen conditions. Methanotrophs have the ability to nitrify and denitrify. In this section, we will discuss the nitrification and denitrification capability of methanotrophs as well as the responsible enzymes discovered that facilitate these processes.
3.3.1 Ammonia oxidation by methanotrophs
Under aerobic conditions methanotrophs share similar properties with AOB, more specifically, the methane monooxygenase enzyme “MMO” share some properties with the ammonia monooxygenase enzyme “AMO” in the reducing equivalent, inhibitor and active sites, though ammonia can compete with methane for the pMMO oxidation step (Stein and Klotz 2011; Zhu et al. 2016). Ammonia oxidation results in the formation of the toxic hydroxylamine. Both types of bacteria oxidize the formed hydroxylamine rapidly to nitrite, however unlike methanotrophs, AOBw have the ability to provide their energy and cellular growth needs from the hydroxylamine oxidation step (Nyerges and Stein 2009; Soliman and Eldyasti 2016).
Although ammonia is mainly used by methanotrophs as a nutrient, ammonia oxidation by methanotrophs can inhibit their growth in two ways; first through the competition with methane for pMMO, secondly through the accumulation of the toxic hydroxylamine and nitrite which inhibits the FDH enzyme (Kits et al. 2015a). However, excess supplementation of methane can mitigate the competition effect of ammonia. More interestingly, some methanotrophs developed detoxification strategies to overcome the inhibitory effects of hydroxylamine and nitrite. However, genes expressed and enzymes encoded are strain specific and do not follow the known classification for methanotrophs i.e., type I and type II.
Enzymes involved in ammonia oxidation pathway in methanotrophs includes the hydroxylamine oxidoreductase enzyme (HAO) encoded by the haoAB genes which oxidizes hydroxylamine to nitrite, another enzyme having the same function which is P460 enzyme (encoded by cyp(cytL)) was also discovered in methanotrophs. In addition, ammonia oxidation results in an increased activity for the cytS gene encoding cytochrome-c which converts nitric oxide to nitrous oxide.
Detoxification strategies is different between methanotrophic strains. Hydroxylamine can be converted to nitric oxide through the hydroxylamine oxidoreductase enzyme that is furtherly converted to nitrous oxide using the cytochrome-c. On the other hand, some strains are capable of reducing hydroxylamine back to ammonia as a detoxification mechanism probably by a hydroxylamine reductase enzyme (Dam et al. 2013). For nitrite detoxification, it can be either converted to nitric oxide using NirK, NirS and HaoA nitrite reductases and other unknown enzymes or reduced back to ammonia through the NirB assimilatory nitrite reductase (Campbell et al. 2011; Stein and Klotz 2011).
Although some methanotrophs can resist the hydroxylamine and nitrite formed through ammonia oxidation, at elevated levels of ammonia they would not be able to handle it and the methane oxidation will be inhibited. In a study on the inhibitory effects of ammonium and nitrite on methane oxidation, two type I methanotrophs (M. album and M. methanica) were compared to another two type II methanotrophs (Methylocystis sp. and M. sporium) at different methane concentrations. All the strains were capable of producing nitrite from ammonia except for M. methanica. At low methane concentrations, ammonia had a higher inhibitory effect on methane oxidation than nitrite while at higher methane concentration the inhibitory effects for both ammonium and nitrite were somehow the same. Rates of methane oxidation were equally inhibited at higher ammonium levels at all methane concentration for the four strains tested (Nyerges and Stein 2009). Despite M. methanica could not produce nitrite from ammonia, ammonia had similar inhibitory effects on its methane oxidation confirming the ability of some methanotrophs to oxidize hydroxylamine directly to nitric and nitrous oxides.
On the other hand, 16 different methanotrophic strains were tested to determine the inhibitory concentration of ammonium, hydroxylamine and nitrite on the growth rates. Most of the cells from different strains could tolerate up to 40 mM ammonium without inhibiting their growth. Hydroxylamine and nitrite had inhibitory effects at concentrations of 1 and 2 mM, respectively. Moreover, all the type I tested strains produced nitrous oxide through the oxidation of hydroxylamine while type II produced nitrous oxide from nitrite when they were grown on ammonium (Hoefman et al. 2014b).
3.3.2 Denitrification activity of methanotrophs
The cost of adding an external carbon source in wastewater treatment plants with low organic loadings is one of the main drawbacks for the conventional denitrification process. Methane can be considered as a cheap and non-toxic external donor that can be produced from the existing anaerobic digesters in any WWTP. Many studies showed the contributions of methanotrophs to the nitrogen removal process directly or indirectly. Anaerobic methane oxidation coupled to denitrification gained a lot of interest in the past few years and the advantages, challenges, mechanism and potential to be incorporated in WWTPs has been deeply investigated (Wang et al. 2017a). Briefly, M. oxyfera was widely studied to develop the nitrite dependent anaerobic methane oxidation (DAMO) process (Wang et al. 2017a). In addition, nitrate-DAMO is supported by the existence of the methane-oxidizing archaea M. nitroreducens in the microbial community where M. nitroreducens convert nitrate into nitrite which can be then reduced by M. oxyfera (Hu et al. 2011). However, it is noteworthy to mention that nitrite-DAMO can still occur if other microorganisms support nitrate reduction rather than M. nitroreducens and biogas containing 0.5% H2S supported the synergistic activity between nitrite-DAMO bacteria, nitrate-reducing and sulfide-oxidizing bacteria (López et al. 2017). Moreover, the possibility of co-culturing DAMO with anaerobic ammonium oxidation (ANAMMOX) microorganisms was demonstrated in several lab-scale studies for the combined removal of methane and nitrogen from wastewater streams (Cai et al. 2015; Ding et al. 2014). However, the major challenge facing this process is the stability and performance when subjected to real operational conditions to fulfil WWTPs requirements and to maintain the parameters required for the long-term dominance of both slow-growing microorganisms (van Kessel et al. 2018).
One of the widely discussed methods for methane dependent nitrogen removal is the aerobic methane oxidation coupled to denitrification (AME-D) in which methane is oxidized by the aerobic methanotrophs releasing organic compounds that are then utilized by the existing denitrifying bacteria as their electron donor under anoxic conditions (Modin et al. 2007). These organic compounds include methanol, acetate, citrate and proteins. During the AME-D process, the dissolved oxygen level is kept at a minimum level to support the growth for the aerobic methanotrophs while preventing any inhibition on the anoxic denitrifiers. Another method is the simultaneous nitrification and denitrification process (SNR) with the co-existence of aerobic methanotrophs, autotrophic nitrifiers and heterotrophic denitifiers in the same reactor under the same operating conditions (Lee et al. 2001). Lastly, the newly discovered capability of aerobic methanotrophs to denitrify by themselves under anoxic conditions.
Elder studies on AME-D process proposed that denitrification only occurred through the activity of the heterotrophic denitrifying bacteria in the anaerobic regions of the bioreactors i.e., central parts of the formed flocs or biofilms or by aerobic denitrifiers (Modin et al. 2007). Moreover, many studies could not differentiate between the nitrate used for assimilation by the methanotrophs and the nitrate utilized by the heterotrophs. Methanotrophs were excluded from the denitrification activity happening. However, recent studies revealed the capability of aerobic methanotrophs to denitrify by themselves under anoxic conditions (Dam et al. 2013; Kits et al. 2015a). Most of the denitrifying genes were sequenced in the studied methanotrophic strains with increased activity in micro aerobic environments (Zhu et al. 2016). This discovery will give a new perspective to the AME-D process with better understanding of the bacterial activity occurring which will lead to proper bioreactors design. For complete denitrification to take place, number of genes encoding denitrifying enzymes should exist in the bacteria. Main enzymes with their corresponding genes are shown in Table 11. Denitrification by aerobic methanotrophs can be described as a strategy for energy conservation to sustain their respiration in the absence of oxygen (Kits et al. 2015a). Switching the terminal electron acceptor from oxygen to nitrate or nitrite will decrease their oxygen requirements and the low levels of available oxygen levels can be used only for the methane oxidation step (Kits et al. 2015b). However, as previously mentioned the existence and the activity of the denitrification genes is strain specific and not all the prescribed enzymes exist in all methanotrophs. For example, two type I methanotrophs can result in partial denitrification through different paths; M. denitrificans FJG1 can reduce nitrate to nitrous oxide under anoxic conditions while M. album strain BG8 can produce nitrous oxide from the reduction of nitrite due to the lack of the nitrate reductase in its genome sequence (Zhu et al. 2016).
More precisely the denitrification ability of aerobic methanotrophs should be named as a partial denitrification process as most of the studied strains released only nitrous oxide and not nitrogen gas. To the best of our knowledge, only the strain Methylocystis sp.SC2 could complete the denitrification pathway and efficiently use the nitrous oxide reductase to convert nitrous oxide to nitrogen gas under oxygen limiting conditions (Dam et al. 2013). Table 12 illustrates the enzymes involved in the denitrification activity of some methanotrophic strains detected in previous studies.
Interestingly methanotrophs with the ability of accumulating intracellular PHB can use this biopolymer as an alternative energy source under anoxic conditions (Vecherskaya et al. 2009). The fact that both PHB degradation and denitrification take place under anoxic conditions can support the idea of their interlinking (Dam et al. 2013). From this perspective, the ability of an enriched methanotrophs from activated sludge and dominated by Methylocystis sp. for nitrous oxide production under anoxic conditions was tested (Myung et al. 2015b). After an ammonium-growing phase followed by a PHB accumulation phase, the enriched methanotrophs were exposed to anoxic conditions where nitrite was added and PHB oxidation and nitrous oxide production were monitored. According to this methodology, an ammonium removal of 99 and 70% of nitrite removal were achieved with a conversion efficiency of 70%. When ammonium and nitrite were added together the ammonium and nitrite removal were only 28 and 8% respectively. Moreover, PHB accumulation reached 40% before anoxic conditions then decreased to 33% after nitrous oxide production. The electron balance conducted indicated that PHB oxidation under anoxic conditions supported the conversion of nitrite to nitrous oxide. The maximum observed N2O production rate was 2.1 mg N2O–N g VSS−1 h−1. One other important thing this study shows is the observed nitrogen gas produced at a production rate of 1.25 mg N2 − N g VSS−1 h−1 indicating the existence of some strains in the enriched culture can complete the denitrification process.
Methanotrophs can be very flexible considering different nitrogen sources under different conditions. Moreover, they have a promising potential to take part in either nitrification or denitrification in biological nitrogen removal processes. Identifying the possible nitrogen assimilation pathways (as shown in Fig. 8) for different methanotrophic strains as well as their nitrogen production rates is key factor for a scaling up decision. However, future researches should focus on factors selecting the biomass involved the denitrification process, maximizing their nitrogen removal capability and testing their operation in larger scale bioreactors under real operational conditions.
4 Potential applications for integration of methanotrophs in WWTPs
WWTPs can be a potential source for all the requirements needed for sustainable methanotrophic cultivation. Part of the waste activated sludge can be used as the seed culture for methanotrophic enrichment while the biogas produced from the anaerobic digesters can be used as their carbon and energy source. Moreover, the three applications discussed earlier can be linked together and have the potential to be integrated with other processes existing in the WWTPs as shown in Fig. 9.
The biogas collected from the anaerobic facility can function in two routes regarding methanotrophs. Firstly, the biogas can be directly transferred to support the methanotrophic cultivation in different applications. Secondly, the biogas can be used as an external carbon source for the denitrification process in a conventional biological nitrogen removal (BNR) system. During the latter application, the biogas methane can be employed either for the AME-D process or to support the nitrate-DAMO.
After mixing the biogas with the waste activated sludge in the methanotrophs cultivation tank, the enriched bacteria can be directed towards two applications; either to the methanol production tank or biopolymers accumulation tank. After purification, the methanol produced by the methanotrophs can be stored to be used as a commodity or can be directed to the conventional BNR system as an external carbon source to sustain the denitrification activity in the anoxic tank.
When the enriched methanotrophs are transferred to nutrient deficiency conditions for PHB accumulation, the PHB rich cells can proceed to an extraction facility for biopolymers production. Another high-potential application for PHB producing methanotrophs is their participation in shortcut BNR systems. The first step in shortcut BNR systems is ammonia oxidation to nitrite followed by nitrite reduction to nitrogen gas i.e., denitritation. Methanotrophs can couple PHB degradation with nitrite reduction as the second step in shortcut systems under anoxic conditions. Moreover, methanotrophs can be employed directly for denitritation under anoxic conditions and nitrite-DAMO can be enhanced by coupling it with ANAMMOX in a single bioreactor for complete nitrogen removal.
However, it is important to mention that nitrite reduction using aerobic methanotrophs (either directly or with PHB) will mainly result in a partial denitrification process with nitrous oxide as a terminal product. Achieving a complete denitrification using PHB accumulating methanotrophs will result in a 50% increase in the biopolymer requirements (Myung et al. 2015b). On the other hand, until now it is hard to directly reach a complete denitrification using methanotrophs as most of the studied strains lack the enzyme N2OR that reduces the nitrous oxide to nitrogen gas.
Another application for PHB-rich cells is using the produced PHB in the methanol production process. The high cost of formate addition as external electron source is one the main obstacles limiting the upscaling of methanol production using methanotrophs. Interestingly, PHB provides the cell with reducing equivalent in case of methane deficiency. Thus, it is theoretically applicable that PHB-rich cells efficiently produce methanol relying on the accumulated PHB as reducing equivalent source. Noteworthy, coupling PHB degradation with methane to methanol conversion has not been tested yet. Therefore, the response of PHB-rich cells to the presence of methane and MDH inhibitors has not been investigated.
References
AlSayed A, Fergala A, Khattab S, ElSharkawy A, Eldyasti A (2018) Optimization of methane bio-hydroxylation using waste activated sludge mixed culture of type I methanotrophs as biocatalyst. Appl Energy 211:755–763. https://doi.org/10.1016/j.apenergy.2017.11.090
Amaral JA, Knowles R (1995) Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett 126:215–220. https://doi.org/10.1111/j.1574-6968.1995.tb07421.x
Anthony C (1982) The biochemistry of methylotrophs. Academic Press, London
Baani M, Liesack W (2008) Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci 105:10203–10208. https://doi.org/10.1073/pnas.0702643105
Babel W (1992) Pecularities of methylotrophs concerning overflow metabolism, especially the synthesis of polyhydroxyalkanoates. FEMS Microbiol Rev 9:141–148. https://doi.org/10.1111/j.1574-6968.1992.tb05831.x
Belova SE, Baani M, Suzina NE, Bodelier PLE, Liesack W, Dedysh SN (2011) Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ Microbiol Rep 3:36–46. https://doi.org/10.1111/j.1758-2229.2010.00180.x
Belova SE, Kulichevskaya IS, Bodelier PLE, Dedysh SN et al (2013) Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int J Syst Evol Microbiol 63:1096–1104. https://doi.org/10.1099/ijs.0.043505-0
Bjorck CE, Dobson PD, Pandhal J (2018) Biotechnological conversion of methane to methanol: evaluation of progress and potential. AIMS Bioeng 5:1–38. https://doi.org/10.3934/bioeng.2018.1.1
Bachmann N, Jansen J, Baxter D, Bochmann G, MONTPART (2015) Sustainable biogas production in municipal wastewater treatment plants. EU Science Hub—European Commission [WWW Document]. EU Sci Hub. https://ec.europa.eu/jrc/en/publication/sustainable-biogas-production-municipal-wastewater-treatment-plants. Accessed 31 Oct 2016
Boden R, Cunliffe M, Scanlan J, Moussard H, Kits KD, Klotz MG, Jetten MSM, Vuilleumier S, Han J, Peters L, Mikhailova N, Teshima H, Tapia R, Kyrpides N, Ivanova N, Pagani I, Cheng J-F, Goodwin L, Han C, Hauser L, Land ML, Lapidus A, Lucas S, Pitluck S, Woyke T, Stein L, Murrell JC (2011) Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J Bacteriol 193:7001–7002. https://doi.org/10.1128/JB.06267-11
Bodrossy L, Holmes EM, Holmes AJ, Kovács KL, Murrell JC (1997) Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs Methylocaldum gen. nov. Arch Microbiol 168:493–503
Börjesson G, Sundh I, Svensson B (2004) Microbial oxidation of CH4 at different temperatures in landfill cover soils. FEMS Microbiol Ecol 48:305–312. https://doi.org/10.1016/j.femsec.2004.02.006
Bowman J (2006) The methanotrophs—the families Methylococcaceae and Methylocystaceae. In: The prokaryotes. Springer, New York, pp 266–289
Bowman JP (2014) The family Methylococcaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin, pp 411–440
Bowman JP, Sayler GS (1994) Optimization and maintenance of soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Biodegradation 5:1–11. https://doi.org/10.1007/BF00695208
Bowman JP, Sly LI, Cox JM, Hayward AC (1990) Methylomonas fodinarum sp. nov. and Methylomonas aurantiaca sp.nov.: two closely related type I obligate methanotrophs. Syst Appl Microbiol 13:279–287. https://doi.org/10.1016/S0723-2020(11)80199-2
Bowman JP, Sly LI, Nichols PD, Hayward AC (1993) Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol 43:735–753
Bowman JP, Sly LI, Stackebrandt E (1995) The phylogenetic position of the family Methylococcaceae. Int J Syst Bacteriol 45:622. https://doi.org/10.1099/00207713-45-3-622a
Bowman JP, McCammon SA, Skerrat JH (1997) Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 143:1451–1459
Cai C, Hu S, Guo J, Shi Y, Xie G-J, Yuan Z (2015) Nitrate reduction by denitrifying anaerobic methane oxidizing microorganisms can reach a practically useful rate. Water Res 87:211–217. https://doi.org/10.1016/j.watres.2015.09.026
Cal AJ, Sikkema WD, Ponce MI, Franqui-Villanueva D, Riiff TJ, Orts WJ, Pieja AJ, Lee CC (2016) Methanotrophic production of polyhydroxybutyrate-co-hydroxyvalerate with high hydroxyvalerate content. J Biol Macromol, Int. https://doi.org/10.1016/j.ijbiomac.2016.02.056
Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG (2011) Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol Lett 322:82–89. https://doi.org/10.1111/j.1574-6968.2011.02340.x
Cantera S, Lebrero R, García-Encina PA, Muñoz R (2016) Evaluation of the influence of methane and copper concentration and methane mass transport on the community structure and biodegradation kinetics of methanotrophic cultures. J Environ Manag 171:11–20. https://doi.org/10.1016/j.jenvman.2016.02.002
Chi Z, Lu W, Wang H, Zhao Y (2012a) Diversity of methanotrophs in a simulated modified biocover reactor. J Environ Sci 24:1076–1082. https://doi.org/10.1016/S1001-0742(11)60889-9
Chi Z-F, Lu W-J, Li H, Wang H-T (2012b) Dynamics of CH4 oxidation in landfill biocover soil: effect of O2/CH4 ratio on CH4 metabolism. Environ Pollut 170:8–14. https://doi.org/10.1016/j.envpol.2012.06.005
Chidambarampadmavathy K, Karthikeyan OP, Heimann K (2015) Biopolymers made from methane in bioreactors. Eng Life Sci. https://doi.org/10.1002/elsc.201400203
Chistoserdova L, Lidstrom ME (2013a) Aerobic methylotrophic prokaryotes. In: The prokaryotes. Springer, Berlin, pp 267–285
Chistoserdova L, Lidstrom PME (2013b) Aerobic methylotrophic prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin, pp 267–285. https://doi.org/10.1007/978-3-642-30141-4_68
Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME (2009) The expanding world of methylotrophic metabolism. Annu Rev Microbiol 63:477–499. https://doi.org/10.1146/annurev.micro.091208.073600
Ciğgin AS, Karahan O, Orhon D (2007) Effect of feeding pattern on biochemical storage by activated sludge under anoxic conditions. Water Res 41:924–934. https://doi.org/10.1016/j.watres.2006.11.017
Conrado RJ, Gonzalez R (2014) Envisioning the bioconversion of methane to liquid fuels. Science 343:620–621. https://doi.org/10.1126/science.1250214
Corder RE, Johnson ER, Vega JL, Clausen EC, Gaddy JL (1986) Biological production of methanol from methane. http://www.anl.gov/PCS/acsfuel/preprint%20archive/Files/33_3_LOS%20ANGELES_09-88_0469.pdf. Accessed March 2018
Costa C, Vecherskaya M, Dijkema C, Stams AJM (2001) The effect of oxygen on methanol oxidation by an obligate methanotrophic bacterium studied by in vivo 13C nuclear magnetic resonance spectroscopy. J Ind Microbiol Biotechnol 26:9–14. https://doi.org/10.1038/sj.jim.7000075
Criddle CS, Rostkowski KH, Sundstrom ER, Leland Stanford Junior University (2015a) Process for the selection of PHB-producing methanotrophic cultures. U.S. Patent 9,062,340
Criddle CS, Sundstrom ER, Leland Stanford Junior University (2015b) Intermittent application of reduced nitrogen sources for selection of PHB producing methanotrophs. U.S. Patent Application 14/404,527
Cui M, Ma A, Qi H, Zhuang X, Zhuang G (2015) Anaerobic oxidation of methane: an “active” microbial process. MicrobiologyOpen 4:1–11. https://doi.org/10.1002/mbo3.232
Culpepper MA, Rosenzweig AC (2014) Structure and Protein–protein interactions of methanol dehydrogenase from Methylococcus capsulatus (Bath). Biochemistry (Mosc) 53:6211–6219. https://doi.org/10.1021/bi500850j
Dam B, Dam S, Blom J, Liesack W (2013) Genome analysis coupled with physiological studies reveals a diverse nitrogen metabolism in Methylocystis sp. strain SC2. PLoS ONE 8:e74767. https://doi.org/10.1371/journal.pone.0074767
Danilova OV, Kulichevskaya IS, Rozova ON, Detkova EN, Bodelier PLE, Trotsenko YA, Dedysh SN (2013) Methylomonas paludis sp. nov., the first acid-tolerant member of the genus Methylomonas, from an acidic wetland. Int J Syst Evol Microbiol 63:2282–2289. https://doi.org/10.1099/ijs.0.045658-0
Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM (2000) Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol 50:955–969
Dedysh SN, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Liesack W, Tiedje JM (2002) Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int J Syst Evol Microbiol 52:251–261. https://doi.org/10.1099/00207713-52-1-251
Dedysh SN, Berestovskaya YY, Vasylieva LV, Belova SE, Khmelenina VN, Suzina NE, Trotsenko YA, Liesack W, Zavarzin GA (2004) Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int J Syst Evol Microbiol 54:151–156. https://doi.org/10.1099/ijs.0.02805-0
Dedysh SN, Knief C, Dunfield PF (2005) Methylocella species are facultatively methanotrophic. J Bacteriol 187:4665–4670. https://doi.org/10.1128/JB.187.13.4665-4670.2005
Dedysh SN, Belova SE, Bodelier PLE, Smirnova KV, Khmelenina VN, Chidthaisong A, Trotsenko YA, Liesack W, Dunfield PF (2007) Methylocystis heyeri sp. nov., a novel type II methanotrophic bacterium possessing ‘signature’ fatty acids of type I methanotrophs. Int J Syst Evol Microbiol 57:472–479. https://doi.org/10.1099/ijs.0.64623-0
Dedysh SN, Didriksen A, Danilova OV, Belova SE, Liebner S, Svenning MM (2015) Methylocapsa palsarum sp. nov., a methanotroph isolated from a subArctic discontinuous permafrost ecosystem. Int J Syst Evol Microbiol 65:3618–3624. https://doi.org/10.1099/ijsem.0.000465
Deutzmann JS, Hoppert M, Schink B (2014) Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov. Syst Appl Microbiol 37:165–169. https://doi.org/10.1016/j.syapm.2014.02.001
Ding Z-W, Ding J, Fu L, Zhang F, Zeng RJ (2014) Simultaneous enrichment of denitrifying methanotrophs and anammox bacteria. Appl Microbiol Biotechnol 98:10211–10221. https://doi.org/10.1007/s00253-014-5936-8
Dircks K, Henze M, van Loosdrecht MC, Mosbaek H, Aspegren H (2001) Storage and degradation of poly-beta-hydroxybutyrate in activated sludge under aerobic conditions. Water Res 35:2277–2285
Doronina NV, Ezhov VA, Trotsenko YA (2011) Growth of Methylosinus trichosporium OB3b on methane and poly-β-hydroxybutyrate biosynthesis. Appl Biochem Microbiol 44:182–185. https://doi.org/10.1134/S0003683808020099
Duan C, Luo M, Xing X (2011) High-rate conversion of methane to methanol by Methylosinus trichosporium OB3b. Bioresour Technol 102:7349–7353. https://doi.org/10.1016/j.biortech.2011.04.096
Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN (2003) Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol 53:1231–1239. https://doi.org/10.1099/ijs.0.02481-0
Dunfield PF, Belova SE, Vorob’ev AV, Cornish SL, Dedysh SN (2010) Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol 60:2659–2664. https://doi.org/10.1099/ijs.0.020149-0
Erikstad H-A, Birkeland N-K (2015) Draft genome sequence of “Candidatus Methylacidiphilum kamchatkense” Strain Kam1, a thermoacidophilic methanotrophic verrucomicrobium. Genome Announc 3:e00065-15. https://doi.org/10.1128/genomeA.00065-15
Fei Q, Guarnieri MT, Tao L, Laurens LML, Dowe N, Pienkos PT (2014) Bioconversion of natural gas to liquid fuel: opportunities and challenges. Biotechnol Adv 32:596–614. https://doi.org/10.1016/j.biotechadv.2014.03.011
Fennell DE, Underhill SE, Jewell WJ (1992) Methanotrophic attached‐film reactor development and biofilm characteristics. Biotechnol Bioeng 40:1218–1232. https://doi.org/10.1002/bit.260401012
Francisco José Fernández RTA (2005) Methanogenesis and methane oxidation in wetlands. Implications in the global carbon cycle. Hidrobiológica 15:327–349
Furuto T, Takeguchi M, Okura I (1999) Semicontinuous methanol biosynthesis by Methylosinus trichosporium OB3b. J Mol Catal Chem 144:257–261. https://doi.org/10.1016/S1381-1169(99)00007-2
García-Pérez T, López JC, Passos F, Lebrero R, Revah S, Muñoz R (2018) Simultaneous methane abatement and PHB production by Methylocystis hirsuta in a novel gas-recycling bubble column bioreactor. Chem Eng J 334:691–697. https://doi.org/10.1016/j.cej.2017.10.106
Ge X, Yang L, Sheets JP, Yu Z, Li Y (2014) Biological conversion of methane to liquid fuels: status and opportunities. Biotechnol Adv 32:1460–1475. https://doi.org/10.1016/j.biotechadv.2014.09.004
Geymonat E, Ferrando L, Tarlera SE (2011) Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int J Syst Evol Microbiol 61:2568–2572. https://doi.org/10.1099/ijs.0.028274-0
Ginige MP, Bowyer JC, Foley L, Keller J, Yuan Z (2008) A comparative study of methanol as a supplementary carbon source for enhancing denitrification in primary and secondary anoxic zones. Biodegradation 20:221–234. https://doi.org/10.1007/s10532-008-9215-1
Graham DW, Chaudhary JA, Hanson RS, Arnold RG (1993) Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors. Microb Ecol 25:1–17
Grosse S, Laramee L, Wendlandt KD, McDonald IR, Miguez CB, Kleber HP (1999) Purification and characterization of the soluble methane monooxygenase of the type II methanotrophic bacterium Methylocystis sp. strain WI 14. Appl Environ Microbiol 65:3929–3935
Han B, Su T, Wu H, Gou Z, Xing X-H, Jiang H, Chen Y, Li X, Murrell JC (2009) Paraffin oil as a “methane vector” for rapid and high cell density cultivation of Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 83:669–677. https://doi.org/10.1007/s00253-009-1866-2
Han J-S, Ahn C-M, Mahanty B, Kim C-G (2013) Partial oxidative conversion of methane to methanol through selective inhibition of methanol dehydrogenase in methanotrophic consortium from landfill cover soil. Appl Biochem Biotechnol 171:1487–1499. https://doi.org/10.1007/s12010-013-0410-0
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
He P, Yang N, Fang W, Lü F, Shao L (2011) Interaction and independence on methane oxidation of landfill cover soil among three impact factors: water, oxygen and ammonium. Front Environ Sci Eng China 5:175–185. https://doi.org/10.1007/s11783-011-0320-8
Helm J, Wendlandt K-D, Jechorek M, Stottmeister U (2008) Potassium deficiency results in accumulation of ultra-high molecular weight poly-β-hydroxybutyrate in a methane-utilizing mixed culture. J Appl Microbiol 105:1054–1061. https://doi.org/10.1111/j.1365-2672.2008.03831.x
Henckel T, Roslev P, Conrad R (2000) Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ Microbiol 2:666–679. https://doi.org/10.1046/j.1462-2920.2000.00149.x
Heyer J, Berger U, Hardt M, Dunfield PF (2005) Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int J Syst Evol Microbiol 55:1817–1826. https://doi.org/10.1099/ijs.0.63213-0
Hirayama H, Suzuki Y, Abe M, Miyazaki M, Makita H, Inagaki F, Uematsu K, Takai K (2011) Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int J Syst Evol Microbiol 61:2646–2653. https://doi.org/10.1099/ijs.0.028092-0
Hirayama H, Fuse H, Abe M, Miyazaki M, Nakamura T, Nunoura T, Furushima Y, Yamamoto H, Takai K (2013) Methylomarinum vadi gen. nov., sp. nov., a methanotroph isolated from two distinct marine environments. Int J Syst Evol Microbiol 63:1073–1082. https://doi.org/10.1099/ijs.0.040568-0
Hirayama H, Abe M, Miyazaki M, Nunoura T, Furushima Y, Yamamoto H, Takai K (2014) Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int J Syst Evol Microbiol 64:989–999. https://doi.org/10.1099/ijs.0.058172-0
Ho A, Lüke C, Reim A, Frenzel P (2013a) Selective stimulation in a natural community of methane oxidizing bacteria: effects of copper on pmoA transcription and activity. Soil Biol Biochem 65:211–216. https://doi.org/10.1016/j.soilbio.2013.05.027
Ho A, Vlaeminck SE, Ettwig KF, Schneider B, Frenzel P, Boon N (2013b) Revisiting methanotrophic communities in sewage treatment plants. Appl Environ Microbiol 79:2841–2846. https://doi.org/10.1128/AEM.03426-12
Hoefman S, Heylen K, De Vos P (2014a) Methylomonas lenta sp. nov., a methanotroph isolated from manure and a denitrification tank. Int J Syst Evol Microbiol 64:1210–1217. https://doi.org/10.1099/ijs.0.057794-0
Hoefman S, van der Ha D, Boon N, Vandamme P, De Vos P, Heylen K (2014b) Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol 14:83. https://doi.org/10.1186/1471-2180-14-83
Hoefman S, van der Ha D, Iguchi H, Yurimoto H, Sakai Y, Boon N, Vandamme P, Heylen K, De Vos P (2014c) Methyloparacoccus murrellii gen. nov., sp. nov., a methanotroph isolated from pond water. Int J Syst Evol Microbiol 64:2100–2107. https://doi.org/10.1099/ijs.0.057760-0
Hu S, Zeng RJ, Keller J, Lant PA, Yuan Z (2011) Effect of nitrate and nitrite on the selection of microorganisms in the denitrifying anaerobic methane oxidation process. Environ Microbiol Rep 3:315–319. https://doi.org/10.1111/j.1758-2229.2010.00227.x
Hur DH, Na J-G, Lee EY (2017) Highly efficient bioconversion of methane to methanol using a novel type I Methylomonas sp. DH-1 newly isolated from brewery waste sludge. J Chem Technol Biotechnol 92:311–318. https://doi.org/10.1002/jctb.5007
Hwang IY, Lee SH, Choi YS, Park SJ, Na JG, Chang IS, Kim C, Kim HC, Kim YH, Lee JW, Lee EY (2014) Biocatalytic conversion of methane to methanol as a key step for development of methane-based biorefineries. J Microbiol Biotechnol 24:1597–1605. https://doi.org/10.4014/jmb.1407.07070
Hwang IY, Hur DH, Lee JH, Park C-H, Chang IS, Lee JW, Lee EY (2015) Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst. J Microbiol Biotechnol 25:375–380. https://doi.org/10.4014/jmb.1412.12007
Iguchi H, Yurimoto H, Sakai Y (2011) Methylovulum miyakonense gen. nov., sp. nov., a type I methanotroph isolated from forest soil. Int J Syst Evol Microbiol 61:810–815. https://doi.org/10.1099/ijs.0.019604-0
Im J, Lee S-W, Yoon S, DiSpirito AA, Semrau JD (2011) Characterization of a novel facultative Methylocystis species capable of growth on methane, acetate and ethanol: facultative methanotrophy in a Methylocystis sp. Environ Microbiol Rep 3:174–181. https://doi.org/10.1111/j.1758-2229.2010.00204.x
Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K (2008) Methane oxidation at 55 C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci 105:300–304. https://doi.org/10.1073/pnas.0704162105
Jewell WJ, Nelson YM, Wilson MS (1992) Methanotrophic bacteria for nutrient removal from wastewater: attached film system. Water Environ Res 64:756–765
Kalyuzhnaya MG (1999) Methylomonas scandinavica sp.nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst Appl Microbiol 22:565–572
Kalyuzhnaya MG, Stolyar SM, Auman AJ, Lara JC, Lidstrom ME, Chistoserdova L (2005) Methylosarcina lacus sp. nov., a methanotroph from Lake Washington, Seattle, USA, and emended description of the genus Methylosarcina. Int J Syst Evol Microbiol 55:2345–2350. https://doi.org/10.1099/ijs.0.63405-0
Kalyuzhnaya MG, Khmelenina V, Eshinimaev B, Sorokin D, Fuse H, Lidstrom M, Trotsenko Y (2008) Classification of halo (alkali) philic and halo (alkali) tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int J Syst Evol Microbiol 58:591–596. https://doi.org/10.1099/ijs.0.65317-0
Kalyuzhnaya MG, Puri AW, Lidstrom ME (2015) Metabolic engineering in methanotrophic bacteria. Metab Eng 29:142–152. https://doi.org/10.1016/j.ymben.2015.03.010
Kampman C, Temmink H, Hendrickx TLG, Zeeman G, Buisman CJN (2014) Enrichment of denitrifying methanotrophic bacteria from municipal wastewater sludge in a membrane bioreactor at 20 °C. J Hazard Mater 274:428–435. https://doi.org/10.1016/j.jhazmat.2014.04.031
Karthikeyan OP, Chidambarampadmavathy K, Cirés S, Heimann K (2015) Review of sustainable methane mitigation and biopolymer production. Crit Rev Environ Sci Technol 45:1579–1610. https://doi.org/10.1080/10643389.2014.966422
Karthikeyan OP, Chidambarampadmavathy K, Nadarajan S, Heimann K (2016) Influence of nutrients on oxidation of low level methane by mixed methanotrophic consortia. Environ Sci Pollut Res 23:4346–4357. https://doi.org/10.1007/s11356-016-6174-7
Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs K-J, Stunnenberg HG, Jetten MSM, den Camp HJMO (2011) Autotrophic Methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicumSolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol 193:4438–4446. https://doi.org/10.1128/JB.00407-11
Khadem AF, Wieczorek AS, Pol A, Vuilleumier S, Harhangi HR, Dunfield PF, Kalyuzhnaya MG, Murrell JC, Francoijs K-J, Stunnenberg HG, Stein LY, DiSpirito AA, Semrau JD, Lajus A, Médigue C, Klotz MG, Jetten MSM, den Camp HJMO (2012) Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum Strain SolV. J Bacteriol 194:3729–3730. https://doi.org/10.1128/JB.00501-12
Khalifa A, Lee CG, Ogiso T, Ueno C, Dianou D, Demachi T, Katayama A, Asakawa S (2015) Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int J Syst Evol Microbiol 65:3527–3534. https://doi.org/10.1099/ijsem.0.000451
Khmelenina VN, Rozova ON, But SY, Mustakhimov II, Reshetnikov AS, Beschastnyi AP, Trotsenko YA (2015) Biosynthesis of secondary metabolites in methanotrophs: biochemical and genetic aspects. Appl Biochem Microbiol 51:150–158. https://doi.org/10.1134/S0003683815020088
Khosravi-Darani K, Mokhtari Z-B, Amai T, Tanaka K (2013) Microbial production of poly(hydroxybutyrate) from C1 carbon sources. Appl Microbiol Biotechnol 97:1407–1424. https://doi.org/10.1007/s00253-012-4649-0
Kim HG, Han GH, Kim SW (2010) Optimization of lab scale methanol production by Methylosinus trichosporium OB3b. Biotechnol Bioprocess Eng 15:476–480. https://doi.org/10.1007/s12257-010-0039-6
Kits KD, Campbell DJ, Rosana AR, Stein LY (2015a) Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Terr. Microbiol. https://doi.org/10.3389/fmicb.2015.01072
Kits KD, Klotz MG, Stein LY (2015b) Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol 17:3219–3232. https://doi.org/10.1111/1462-2920.12772
Knief C (2015) Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6:1346. https://doi.org/10.3389/fmicb.2015.01346
Knief C, Dunfield PF (2005) Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol 7:1307–1317. https://doi.org/10.1111/j.1462-2920.2005.00814.x
Lebrero R, Chandran K (2017) Biological conversion and revalorization of waste methane streams. Crit Rev Environ Sci Technol 47:2133–2157. https://doi.org/10.1080/10643389.2017.1415059
Lee SY (1996) Bacterial polyhydroxyalkanoates. Biotechnol Bioeng 49:1–14
Lee H-J, Bae J-H, Cho K-M (2001) Simultaneous nitrification and denitrification in a mixed methanotrophic culture. Biotechnol Lett 23:935–941. https://doi.org/10.1023/A:1010566616907
Lee SG, Goo JH, Kim HG, Oh J-I, Kim YM, Kim SW (2004) Optimization of methanol biosynthesis from methane using Methylosinus trichosporium OB3b. Biotechnol Lett 26:947–950
Lee S-W, Im J, DiSpirito AA, Bodrossy L, Barcelona MJ, Semrau JD (2009) Effect of nutrient and selective inhibitor amendments on methane oxidation, nitrous oxide production, and key gene presence and expression in landfill cover soils: characterization of the role of methanotrophs, nitrifiers, and denitrifiers. Appl Microbiol Biotechnol 85:389–403. https://doi.org/10.1007/s00253-009-2238-7
Li H, Chi Z, Lu W, Wang H (2014) Sensitivity of methanotrophic community structure, abundance, and gene expression to CH4 and O2 in simulated landfill biocover soil. Environ Pollut 184:347–353. https://doi.org/10.1016/j.envpol.2013.09.002
Lidstrom ME (2006) Aerobic methylotrophic prokaryotes. In: The prokaryotes. Springer New York, pp 618–634
Lindner AS, Pacheco A, Aldrich HC, Costello Staniec A, Uz I, Hodson DJ (2007) Methylocystis hirsuta sp. nov., a novel methanotroph isolated from a groundwater aquifer. Int J Syst Evol Microbiol 57:1891–1900. https://doi.org/10.1099/ijs.0.64541-0
López JC, Quijano G, Souza TSO, Estrada JM, Lebrero R, Muñoz R (2013) Biotechnologies for greenhouse gases (CH4, N2O, and CO2) abatement: state of the art and challenges. Appl Microbiol Biotechnol 97:2277–2303. https://doi.org/10.1007/s00253-013-4734-z
López JC, Quijano G, Pérez R, Muñoz R (2014) Assessing the influence of CH4 concentration during culture enrichment on the biodegradation kinetics and population structure. J Environ Manag 146:116–123. https://doi.org/10.1016/j.jenvman.2014.06.026
López JC, Porca E, Collins G, Pérez R, Rodríguez-Alija A, Muñoz R, Quijano G (2017) Biogas-based denitrification in a biotrickling filter: influence of nitrate concentration and hydrogen sulfide. Biotechnol Bioeng 114:665–673. https://doi.org/10.1002/bit.26092
López JC, Arnáiz E, Merchán L, Lebrero R, Muñoz R (2018a) Biogas-based polyhydroxyalkanoates production by Methylocystis hirsuta: a step further in anaerobic digestion biorefineries. Chem Eng J 333:529–536. https://doi.org/10.1016/j.cej.2017.09.185
López JC, Merchán L, Lebrero R, Muñoz R (2018b) Feast-famine biofilter operation for methane mitigation. J Clean Prod 170:108–118. https://doi.org/10.1016/j.jclepro.2017.09.157
Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl DA (2015) Brock biology of microorganisms, 14th edn. Pearson, Boston
Majone M, Massanisso P, Ramadori R (1998) Comparison of carbon storage under aerobic and anoxic conditions. Water Sci Technol 38:77–84. https://doi.org/10.1016/S0273-1223(98)00680-5
Mardina P, Li J, Patel SKS, Kim I-W, Lee J-K, Selvaraj C (2016) Potential of immobilized whole-cell Methylocella tundrae as biocatalyst for methanol production from methane. J Microbiol Biotechnol. https://doi.org/10.4014/jmb.1602.02074
Marín I, Arahal DR (2014) The family Beijerinckiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin, pp 115–133. https://doi.org/10.1007/978-3-642-30197-1_255
Mehta PK, Mishra S, Ghose TK (1987) Methanol accumulation by resting cells of Methylosinus trichosporium (I). J Gen Appl Microbiol 33:221–229. https://doi.org/10.2323/jgam.33.221
Mehta PK, Ghose TK, Mishra S (1991) Methanol biosynthesis by covalently immobilized cells of Methylosinus trichosporium: batch and continuous studies. Biotechnol Bioeng 37:551–556. https://doi.org/10.1002/bit.260370609
Modin O, Fukushi K, Yamamoto K (2007) Denitrification with methane as external carbon source. Water Res 41:2726–2738. https://doi.org/10.1016/j.watres.2007.02.053
Mohanty SR, Bodelier PLE, Floris V, Conrad R (2006) Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl Environ Microbiol 72:1346–1354. https://doi.org/10.1128/AEM.72.2.1346-1354.2006
Murray RJ, Furlonge HI (2009) Market and economic assessment of using methanol for power generation in the Caribbean region. J Assoc Prof Eng Trinidad Tobago 38:88–99
Murrell JC (2010) The aerobic methane oxidizing bacteria (methanotrophs). In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 1953–1966
Myung J, Galega WM, Van Nostrand JD, Yuan T, Zhou J, Criddle CS (2015a) Long-term cultivation of a stable Methylocystis-dominated methanotrophic enrichment enabling tailored production of poly (3-hydroxybutyrate-co-3-hydroxyvalerate). Bioresour Technol 198:811–818
Myung J, Wang Z, Yuan T, Zhang P, Van Nostrand JD, Zhou J, Criddle CS (2015b) Production of nitrous oxide from nitrite in stable type II methanotrophic enrichments. Environ Sci Technol 49:10969–10975. https://doi.org/10.1021/acs.est.5b03385
Myung J, Flanagan JCA, Waymouth RM, Criddle CS (2016a) Methane or methanol-oxidation dependent synthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by obligate type II methanotrophs. Process Biochem 51:561–567. https://doi.org/10.1016/j.procbio.2016.02.005
Myung J, Kim M, Pan M, Criddle CS, Tang SKY (2016b) Low energy emulsion-based fermentation enabling accelerated methane mass transfer and growth of poly(3-hydroxybutyrate)-accumulating methanotrophs. Bioresour Technol 207:302–307. https://doi.org/10.1016/j.biortech.2016.02.029
Myung J, Flanagan JCA, Waymouth RM, Criddle CS (2017) Expanding the range of polyhydroxyalkanoates synthesized by methanotrophic bacteria through the utilization of omega-hydroxyalkanoate co-substrates. AMB Express 7:118. https://doi.org/10.1186/s13568-017-0417-y
Nikiema J, Brzezinski R, Heitz M (2007) Elimination of methane generated from landfills by biofiltration: a review. Rev Environ Sci Biotechnol 6:261–284. https://doi.org/10.1007/s11157-006-9114-z
Nyerges G, Stein LY (2009) Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol Lett 297:131–136. https://doi.org/10.1111/j.1574-6968.2009.01674.x
Ogiso T, Ueno C, Dianou D, Huy TV, Katayama A, Kimura M, Asakawa S (2012) Methylomonas koyamae sp. nov., a type I methane-oxidizing bacterium from floodwater of a rice paddy field. Int J Syst Evol Microbiol 62:1832–1837. https://doi.org/10.1099/ijs.0.035261-0
Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF (2009) Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. https://doi.org/10.1111/j.1758-2229.2009.00022.x
Ordaz A, López JC, Figueroa-González I, Muñoz R, Quijano G (2014) Assessment of methane biodegradation kinetics in two-phase partitioning bioreactors by pulse respirometry. Water Res 67:46–54. https://doi.org/10.1016/j.watres.2014.08.054
Park D, Lee J (2013) Biological conversion of methane to methanol. Korean J Chem Eng 30:977–987. https://doi.org/10.1007/s11814-013-0060-5
Park S, Hanna L, Taylor RT, Droege MW (1991) Batch cultivation of Methylosinus trichosporium OB3b. I: Production of soluble methane monooxygenase. Biotechnol Bioeng 38:423–433. https://doi.org/10.1002/bit.260380412
Park S, Shah NN, Taylor RT, Droege MW (1992) Batch cultivation of Methylosinus trichosporium OB3b: II. Production of particulate methane monooxygenase. Biotechnol Bioeng 40:151–157. https://doi.org/10.1002/bit.260400121
Patel SKS, Mardina P, Kim D, Kim S-Y, Kalia VC, Kim I-W, Lee J-K (2016a) Improvement in methanol production by regulating the composition of synthetic gas mixture and raw biogas. Bioresour Technol 218:202–208. https://doi.org/10.1016/j.biortech.2016.06.065
Patel SKS, Mardina P, Kim S-Y, Lee J-K, Kim I-W (2016b) Biological methanol production by a type II methanotroph Methylocystis bryophila. J Microbiol Biotechnol. https://doi.org/10.4014/jmb.1601.01013
Patel SKS, Selvaraj C, Mardina P, Jeong J-H, Kalia VC, Kang YC, Lee J-K (2016c) Enhancement of methanol production from synthetic gas mixture by Methylosinus sporium through covalent immobilization. Appl Energy 171:383–391. https://doi.org/10.1016/j.apenergy.2016.03.022
Patel SKS, Singh RK, Kumar A, Jeong J-H, Jeong SH, Kalia VC, Kim I-W, Lee J-K (2017) Biological methanol production by immobilized Methylocella tundrae using simulated biohythane as a feed. Bioresour Technol 241:922–927. https://doi.org/10.1016/j.biortech.2017.05.160
Pen N, Soussan L, Belleville M-P, Sanchez J, Charmette C, Paolucci-Jeanjean D (2014) An innovative membrane bioreactor for methane biohydroxylation. Bioresour Technol 174:42–52. https://doi.org/10.1016/j.biortech.2014.10.001
Pfluger AR (2010) A thesis submitted to the Department of Civil and Environmental Engineering and the Committee on Graduate Studies of Stanford University in Partial Fulfillment of Requirements for the Degree of Engineer. Stanford University
Pfluger AR, Wu W-M, Pieja AJ, Wan J, Rostkowski KH, Criddle CS (2011) Selection of type I and type II methanotrophic proteobacteria in a fluidized bed reactor under non-sterile conditions. Bioresour Technol 102:9919–9926. https://doi.org/10.1016/j.biortech.2011.08.054
Pieja AJ, Rostkowski KH, Criddle CS (2011a) Distribution and selection of poly-3-hydroxybutyrate production capacity in methanotrophic proteobacteria. Microb Ecol 62:564–573. https://doi.org/10.1007/s00248-011-9873-0
Pieja AJ, Sundstrom ER, Criddle CS (2011b) Poly-3-hydroxybutyrate metabolism in the type II methanotroph Methylocystis parvus OBBP. Appl Environ Microbiol 77:6012–6019. https://doi.org/10.1128/AEM.00509-11
Pieja AJ, Sundstrom ER, Criddle CS (2012) Cyclic, alternating methane and nitrogen limitation increases PHB production in a methanotrophic community. Bioresour Technol 107:385–392. https://doi.org/10.1016/j.biortech.2011.12.044
Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damsté JSS, Op den Camp HJM, Jetten MSM, Strous M (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. https://doi.org/10.1038/nature04617
Rahalkar M, Bussmann I, Schink B (2007) Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int J Syst Evol Microbiol 57:1073–1080. https://doi.org/10.1099/ijs.0.64574-0
Rahnama F, Vasheghani-Farahani E, Yazdian F, Shojaosadati SA (2012) PHB production by Methylocystis hirsuta from natural gas in a bubble column and a vertical loop bioreactor. Biochem Eng J 65:51–56. https://doi.org/10.1016/j.bej.2012.03.014
Rasigraf O, Kool DM, Jetten MSM, Damsté JSS, Ettwig KF (2014) Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol 80:2451–2460. https://doi.org/10.1128/AEM.04199-13
Reyes M, Borrás L, Seco A, Ferrer J (2015) Identification and quantification of microbial populations in activated sludge and anaerobic digestion processes. Environ Technol 36:45–53. https://doi.org/10.1080/09593330.2014.934745
Romanovskaya VA, Rokitko PV, Shilin SO, Malashenko YR (2006) Emended description of Methylomonas rubra sp. nov. Microbiology 75:689–693. https://doi.org/10.1134/S0026261706060117
Rostkowski KH, Pfluger AR, Criddle CS (2013) Stoichiometry and kinetics of the PHB-producing type II methanotrophs Methylosinus trichosporium OB3b and Methylocystis parvus OBBP. Bioresour Technol 132:71–77. https://doi.org/10.1016/j.biortech.2012.12.129
Scheutz C, Kjeldsen P, Gentil E (2009) Greenhouse gases, radiative forcing, global warming potential and waste management—an introduction. Waste Manag Res 27:716–723. https://doi.org/10.1177/0734242X09345599
Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, Marx A, Vorholt JA (2009) Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol 27:107–115. https://doi.org/10.1016/j.tibtech.2008.10.009
Semrau JD, DiSpirito AA, Yoon S (2010) Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. https://doi.org/10.1111/j.1574-6976.2010.00212.x
Semrau JD, DiSpirito AA, Vuilleumier S (2011) Facultative methanotrophy: false leads, true results, and suggestions for future research: facultative methanotrophy. FEMS Microbiol Lett 323:1–12. https://doi.org/10.1111/j.1574-6968.2011.02315.x
Shah NN, Hanna ML, Taylor RT (1996) Batch cultivation of Methylosinus trichosporium OB3b: V. Characterization of poly-β-hydroxybutyrate production under methane-dependent growth conditions. Biotechnol Bioeng 49:161–171. https://doi.org/10.1002/(SICI)1097-0290(19960120)49:2<161:AID-BIT5>3.0.CO;2-O
Sheets JP, Ge X, Li Y-F, Yu Z, Li Y (2016) Biological conversion of biogas to methanol using methanotrophs isolated from solid-state anaerobic digestate. Bioresour Technol 201:50–57. https://doi.org/10.1016/j.biortech.2015.11.035
Shen L, He Z, Wu H, Gao Z (2015) Nitrite-dependent anaerobic methane-oxidising bacteria: unique microorganisms with special properties. Curr Microbiol 70:562–570. https://doi.org/10.1007/s00284-014-0762-x
Siniscalchi LAB, Vale IC, Dell’Isola J, Chernicharo CA, Calabria Araujo J (2015) Enrichment and activity of methanotrophic microorganisms from municipal wastewater sludge. Environ Technol 36:1563–1575. https://doi.org/10.1080/09593330.2014.997298
Sipkema EM, de Koning W, Ganzeveld KJ, Janssen DB, Beenackers AA (2000) NADH-regulated metabolic model for growth of Methylosinus trichosporium OB3b. Model presentation, parameter estimation, and model validation. Biotechnol Prog 16:176–188. https://doi.org/10.1021/bp990155e
Smith TJ, Trotsenko YA, Murrell JC (2010) Physiology and biochemistry of the aerobic methane oxidizing bacteria. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 765–779
Soliman M, Eldyasti A (2016) Development of partial nitrification as a first step of nitrite shunt process in a sequential batch reactor (SBR) using ammonium oxidizing bacteria (AOB) controlled by mixing regime. Bioresour Technol 221:85–95. https://doi.org/10.1016/j.biortech.2016.09.023
Song H, Zhang Y, Kong W, Xia C (2012) Activities of key enzymes in the biosynthesis of poly-3-hydroxybutyrate by Methylosinus trichosporium IMV3011. Chin J Catal 33:1754–1761. https://doi.org/10.1016/S1872-2067(11)60443-9
Stein LY, Klotz MG (2011) Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem Soc Trans 39:1826–1831. https://doi.org/10.1042/BST20110712
Stone KA, Hilliard MV, He QP, Wang J (2017) A mini review on bioreactor configurations and gas transfer enhancements for biochemical methane conversion. Biochem Eng J 128:83–92. https://doi.org/10.1016/j.bej.2017.09.003
Strong PJ, Xie S, Clarke WP (2015) Methane as a resource: can the methanotrophs add value? Environ Sci Technol 49:4001–4018. https://doi.org/10.1021/es504242n
Strong PJ, Laycock B, Mahamud SNS, Jensen PD, Lant PA, Tyson G, Pratt S (2016) The opportunity for high-performance biomaterials from methane. Microorganisms 4:11. https://doi.org/10.3390/microorganisms4010011
Sun F, Dong W, Shao M, Lv X, Li J, Peng L, Wang H (2013) Aerobic methane oxidation coupled to denitrification in a membrane biofilm reactor: treatment performance and the effect of oxygen ventilation. Bioresour Technol 145:2–9. https://doi.org/10.1016/j.biortech.2013.03.115
Sundstrom ER, Criddle CS (2015) Optimization of methanotrophic growth and production of poly(3-hydroxybutyrate) in a high-throughput microbioreactor system. Appl Environ Microbiol 81:4767–4773. https://doi.org/10.1128/AEM.00025-15
Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, Spang A, Bringel F, Vuilleumier S, Lajus A, Médigue C, Bruce DC, Cheng J-F, Goodwin L, Ivanova N, Han J, Han CS, Hauser L, Held B, Land ML, Lapidus A, Lucas S, Nolan M, Pitluck S, Woyke T (2011) Genome sequence of the arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol 193:6418–6419. https://doi.org/10.1128/JB.05380-11
Taher E, Chandran K (2013) High-rate, high-yield production of methanol by ammonia-oxidizing bacteria. Environ Sci Technol 47:3167–3173. https://doi.org/10.1021/es3042912
Takeguchi M, Okura I (2000) Role of iron and copper in particulate methane monooxygenase of Methylosinus trichosporium OB3b. Catal Surv Jpn 4:51–63. https://doi.org/10.1023/A:1019036105038
Takeguchi M, Furuto T, Sugimori D, Okura I (1997) Optimization of methanol biosynthesis by Methylosinus trichosporium OB3b: an approach to improve methanol accumulation. Appl Biochem Biotechnol 68:143–152
Takeuchi M, Kamagata Y, Oshima K, Hanada S, Tamaki H, Marumo K, Maeda H, Nedachi M, Hattori M, Iwasaki W, Sakata S (2014) Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int J Syst Evol Microbiol 64:3240–3246. https://doi.org/10.1099/ijs.0.063503-0
Tavormina PL, Hatzenpichler R, McGlynn S, Chadwick G, Dawson KS, Connon SA, Orphan VJ (2015) Methyloprofundus sedimenti gen. nov., sp. nov., an obligate methanotroph from ocean sediment belonging to the ‘deep sea-1’ clade of marine methanotrophs. Int J Syst Evol Microbiol 65:251–259. https://doi.org/10.1099/ijs.0.062927-0
Tourova TP, Omel’chenko MV, Fegeding KV, Vasil’eva LV (1999) The phylogenetic position of Methylobacter psychrophilus sp. nov. Microbiology 68:493–495
Trotsenko YA, Murrell JC (2008) Metabolic aspects of aerobic obligate methanotrophy⋆. Adv Appl Microbiol 63:183–229
Trotsenko YA, Medvedkova KA, Khmelenina VN, Eshinimayev BT (2009) Thermophilic and thermotolerant aerobic methanotrophs. Microbiology 78:387–401. https://doi.org/10.1134/S0026261709040018
Tsubota J, Eshinimaev BT, Khmelenina VN, Trotsenko YA (2005) Methylothermus thermalis gen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. Int J Syst Evol Microbiol 55:1877–1884. https://doi.org/10.1099/ijs.0.63691-0
van der Ha D, Hoefman S, Boeckx P, Verstraete W, Boon N (2010) Copper enhances the activity and salt resistance of mixed methane-oxidizing communities. Appl Microbiol Biotechnol 87:2355–2363. https://doi.org/10.1007/s00253-010-2702-4
van der Ha D, Vanwonterghem I, Hoefman S, Vos PD, Boon N (2012a) Selection of associated heterotrophs by methane-oxidizing bacteria at different copper concentrations. Antonie Van Leeuwenhoek 103:527–537. https://doi.org/10.1007/s10482-012-9835-7
van der Ha D, Nachtergaele L, Kerckhof F-M, Rameiyanti D, Bossier P, Verstraete W, Boon N (2012b) Conversion of biogas to bioproducts by algae and methane oxidizing bacteria. Environ Sci Technol 46:13425–13431. https://doi.org/10.1021/es303929s
van Kessel MA, Stultiens K, Slegers MF, Guerrero Cruz S, Jetten MS, Kartal B, Op den Camp HJ (2018) Current perspectives on the application of N-damo and anammox in wastewater treatment. Curr Opin Biotechnol 50:222–227. https://doi.org/10.1016/j.copbio.2018.01.031
van Teeseling MCF, Pol A, Harhangi HR, van der Zwart S, Jetten MSM, den Camp HJMO, van Niftrik L (2014) Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol 80:6782–6791. https://doi.org/10.1128/AEM.01838-14
Vecherskaya M, Dijkema C, Saad HR, Stams AJM (2009) Microaerobic and anaerobic metabolism of a Methylocystis parvus strain isolated from a denitrifying bioreactor. Environ Microbiol Rep 1:442–449. https://doi.org/10.1111/j.1758-2229.2009.00069.x
Visscher AD, Schippers M, Cleemput OV (2001) Short-term kinetic response of enhanced methane oxidation in landfill cover soils to environmental factors. Biol Fertil Soils 33:231–237. https://doi.org/10.1007/s003740000313
Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN (2011) Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol 61:2456–2463. https://doi.org/10.1099/ijs.0.028118-0
Wang J, Xia F-F, Bai Y, Fang C-R, Shen D-S, He R (2011) Methane oxidation in landfill waste biocover soil: kinetics and sensitivity to ambient conditions. Waste Manag 31:864–870. https://doi.org/10.1016/j.wasman.2011.01.026
Wang D, Wang Y, Liu Y, Ngo HH, Lian Y, Zhao J, Chen F, Yang Q, Zeng G, Li X (2017a) Is denitrifying anaerobic methane oxidation-centered technologies a solution for the sustainable operation of wastewater treatment Plants? Bioresour Technol 234:456–465. https://doi.org/10.1016/j.biortech.2017.02.059
Wang Y, Wang D, Yang Q, Zeng G, Li X (2017b) Wastewater opportunities for denitrifying anaerobic methane oxidation. Trends Biotechnol 35:799–802. https://doi.org/10.1016/j.tibtech.2017.02.010
Wartiainen I, Hestnes AG, McDonald IR, Svenning MM (2006a) Methylobacter tundripaludum sp. nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78 N). Int J Syst Evol Microbiol 56:109–113. https://doi.org/10.1099/ijs.0.63728-0
Wartiainen I, Hestnes AG, McDonald IR, Svenning MM (2006b) Methylocystis rosea sp. nov., a novel methanotrophic bacterium from Arctic wetland soil, Svalbard, Norway (78 N). Int J Syst Evol Microbiol 56:541–547. https://doi.org/10.1099/ijs.0.63912-0
Wendlandt K-D, Jechorek M, Helm J, Stottmeister U (2001) Producing poly-3-hydroxybutyrate with a high molecular mass from methane. J Biotechnol 86:127–133. https://doi.org/10.1016/S0168-1656(00)00408-9
Wendlandt K-D, Geyer W, Mirschel G, Hemidi FA-H (2005) Possibilities for controlling a PHB accumulation process using various analytical methods. J Biotechnol 117:119–129. https://doi.org/10.1016/j.jbiotec.2005.01.007
Whittenbury R, Phillips KC, Wilkinson JF (1970) Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–218
Wise MG, McArthur JV, Shimkets LJ (2001) Methylosarcina fibrata gen. nov., sp. nov. and Methylosarcina quisquiliarum sp.nov., novel type 1 methanotrophs. Int J Syst Evol Microbiol 51:611–621. https://doi.org/10.1099/00207713-51-2-611
Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, van Niftrik L (2011) A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’. Biochem Soc Trans 39:243–248. https://doi.org/10.1042/BST0390243
Xin J, Cui J, Niu J, Hua S, Xia C, Li S, Zhu L (2004a) Production of methanol from methane by methanotrophic bacteria. Biocatal Biotransform 22:225–229. https://doi.org/10.1080/10242420412331283305
Xin J, Cui J, Niu J, Hua S, Xia C, Li S, Zhu L (2004b) Biosynthesis of methanol from CO2 and CH4 by methanotrophic bacteria. Biotechnology 3:67–71
Xin J, Zhang Y, Zhang S, Xia C, Li S (2007) Methanol production from CO2 by resting cells of the methanotrophic bacterium Methylosinus trichosporium IMV 3011. J Basic Microbiol 47:426–435. https://doi.org/10.1002/jobm.200710313
Xin J, Zhang Y, Dong J, Song H, Xia C (2013) An experimental study on molecular weight of poly-3-hydroxybutyrate (PHB) accumulated in Methylosinus trichosporium IMV 3011. Afr J Biotechnol 10:7078–7087
Yoo Y-S, Han J-S, Ahn C-M, Kim C-G (2015) Comparative enzyme inhibitive methanol production by Methylosinus sporium from simulated biogas. Environ Technol 36:983–991. https://doi.org/10.1080/09593330.2014.971059
Zahn JA, Bergmann DJ, Boyd JM, Kunz RC, DiSpirito AA (2001) Membrane-associated quinoprotein formaldehyde dehydrogenase from Methylococcus capsulatus Bath. J Bacteriol 183:6832–6840. https://doi.org/10.1128/JB.183.23.6832-6840.2001
Zhang Y, Xin J, Chen L, Song H, Xia C (2008) Biosynthesis of poly-3-hydroxybutyrate with a high molecular weight by methanotroph from methane and methanol. J Nat Gas Chem 17:103–109. https://doi.org/10.1016/S1003-9953(08)60034-1
Zhang Y, Xin J, Chen L, Xia C (2009) The methane monooxygenase intrinsic activity of kinds of methanotrophs. Appl Biochem Biotechnol 157:431–441. https://doi.org/10.1007/s12010-008-8447-1
Zhang X, Kong J-Y, Xia F-F, Su Y, He R (2014) Effects of ammonium on the activity and community of methanotrophs in landfill biocover soils. Syst Appl Microbiol 37:296–304. https://doi.org/10.1016/j.syapm.2014.03.003
Zhang W, Ge X, Li Y-F, Yu Z, Li Y (2016) Isolation of a methanotroph from a hydrogen sulfide-rich anaerobic digester for methanol production from biogas. Process Biochem. https://doi.org/10.1016/j.procbio.2016.04.003
Zhang T, Wang X, Zhou J, Zhang Y (2017a) Enrichments of methanotrophic–heterotrophic cultures with high poly-β-hydroxybutyrate (PHB) accumulation capacities. J Environ Sci. https://doi.org/10.1016/j.jes.2017.03.016
Zhang T, Zhou J, Wang X, Zhang Y (2017b) Coupled effects of methane monooxygenase and nitrogen source on growth and poly-β-hydroxybutyrate (PHB) production of Methylosinus trichosporium OB3b. J Environ Sci 52:49–57. https://doi.org/10.1016/j.jes.2016.03.001
Zhu J, Wang Q, Yuan M, Tan G-YA, Sun F, Wang C, Wu W, Lee P-H (2016) Microbiology and potential applications of aerobic methane oxidation coupled to denitrification (AME-D) process: a review. Water Res 90:203–215. https://doi.org/10.1016/j.watres.2015.12.020
Zúñiga C, Morales M, Le Borgne S, Revah S (2011) Production of poly-β-hydroxybutyrate (PHB) by Methylobacterium organophilum isolated from a methanotrophic consortium in a two-phase partition bioreactor. J Hazard Mater 190:876–882. https://doi.org/10.1016/j.jhazmat.2011.04.011
Acknowledgements
The authors gratefully acknowledge Natural Science and Engineering Research Council of Canada (NSERC) and Seed Fund, York University, and City of Toronto, ON, Canada for their endless support and interest at every stage of this research project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
AlSayed, A., Fergala, A. & Eldyasti, A. Sustainable biogas mitigation and value-added resources recovery using methanotrophs intergrated into wastewater treatment plants. Rev Environ Sci Biotechnol 17, 351–393 (2018). https://doi.org/10.1007/s11157-018-9464-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11157-018-9464-3