Abstract
Effluents of anaerobic digesters are an underestimated source of greenhouse gases, as they are often saturated with methane. A post-treatment with methane-oxidizing bacterial consortia could mitigate diffuse emissions at such sites. Semi-continuously fed stirred reactors were used as model systems to characterize the influence of the key parameters on the activity of these mixed methanotrophic communities. The addition of 140 mg L−1 NH +4 –N had no significant influence on the activity nor did a temperature increase from 28°C to 35°C. On the other hand, addition of 0.64 mg L−1 of copper(II) increased the methane removal rate by a factor of 1.5 to 1.7 since the activity of particulate methane monooxygenase was enhanced. The influence of different concentrations of NaCl was also tested, as effluents of anaerobic digesters often contain salt levels up to 10 g NaCl L−1. At a concentration of 11 g NaCl L−1, almost no methane-oxidizing activity was observed in the reactors without copper addition. Yet, reactors with copper addition exhibited a sustained activity in the presence of NaCl. A colorimetric test based on naphthalene oxidation showed that soluble methane monooxygenase was inhibited by copper, suggesting that the particulate methane monooxygenase was the active enzyme and thus more salt resistant. The results obtained demonstrate that the treatment of methane-saturated effluents, even those with increased ammonium (up to 140 mg L−1 NH +4 –N) and salt levels, can be mitigated by implementation of methane-oxidizing microbial consortia.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Effluents of anaerobic digester installations are often oversaturated with methane (Hartley and Lant 2006). When treating low-strength wastewaters, up to 25% of the produced methane is lost in the effluent (Cakir and Stenstrom 2005; Hartley and Lant 2006). It is important to control these methane (CH4) emissions, as this greenhouse gas is 23 times more effective at trapping heat than carbon dioxide (CO2) (IPCC 2001). Methane-oxidizing consortia could be used to mitigate methane-saturated effluents. Methane-oxidizing bacteria (MOB) or methanotrophs form a unique group of Gram-negative bacteria that are able to use methane as their only source of carbon and energy (Anthony 1982; Hanson and Hanson 1996). The MOB can be subdivided in three major groups. The low-concentration methanotrophs (LCM), also referred to as type I methanotrophs, belong mostly to the γ-proteobacteria and are found under normal atmospheric conditions (Dunfield et al. 1999). The high concentration methanotrophs (HCM), also referred to as type II methanotrophs, are mostly α-proteobacteria, which are abundant at concentrations above 100–1,000 ppmv CH4 (Bender and Conrad 1992). A smaller third group, also referred to as type X methanotrophs, combines the features of the former groups (Hanson and Hanson 1996). Growth factors such as micro- and macronutrients, gas concentrations, and physical parameters have an important influence on the activity and growth rate (Begonja and Hrsak 2001; Bender and Conrad 1995; Scheutz et al. 2009). Generally, MOB have the highest activity in a pH range between 6 and 7 and a temperature range between 25°C and 35°C (Hanson and Hanson 1996; Scheutz et al. 2009; Whittenbury et al. 1970). Although nitrate is commonly used as nitrogen source in growth media for methanotrophs, ammonium can be an alternative. However, it is still unclear which concentrations of ammonium have a stimulating or inhibitory effect on the activity (Bodelier and Laanbroek 2004; Nyerges and Stein 2009).
MOB possess two methane-oxidizing enzyme systems: the particulate methane monooxygenase (pMMO), present in most LCM and HCM, and the soluble methane monooxygenase (sMMO), only present in some HCM (Dalton 2005; Hanson and Hanson 1996). The similarity between those methane-oxidizing enzyme systems is low, although they perform the same task, i.e., oxidizing methane (Dalton 2005). The soluble form, sMMO, has a non-heme diiron active site (Merkx et al. 2001). This cytoplasmic enzyme is mostly found in HCM under copper-limiting growth concentrations up to 0.06 mg Cu(II) L−1 (Jahng and Wood 1996; Koh et al. 1993). The pMMO, which has only been studied to a limited extent, has a copper active site (Balasubramanian and Rosenzweig 2008; Dalton 2005). pMMO is the only active methane monooxygenase above a concentration of 0.52 mg Cu(II) L−1 (Hakemian and Rosenzweig 2007). In between these values, a gradual switch between both enzymes is observed (Hakemian and Rosenzweig 2007; Shah et al. 1992). The differential expression of both enzyme systems is thus controlled by the copper concentration. However, the exact mechanism of this “copper switch” is still not known (Begonja and Hrsak 2001; Hakemian and Rosenzweig 2007).
The responses to elevated salt concentrations remain subject of discussion. Gulledge and Schimel (1998) and Whalen (2000) came to the conclusion that the inhibitory effect of ammonium on methane oxidation was due to Cl−, the counter ion of NH +4 . De Visscher and Van Cleemput (2003) observed a high chloride sensitivity for soil HCM. Other authors found no growth inhibition for 135 methanotrophic strains in a medium with 10 g NaCl L−1 (Bowman et al. 1993).
Most research in the field of MOB focuses on soil processes. Up till now, tests with bioreactors were mostly performed with axenic cultures. Yet, methanotrophs are more active when grown in communities together with other bacteria (Begonja and Hrsak 2001; Helm et al. 2006). The aim of this study was to examine the influence of copper and salt addition on the methane oxidation activity in mixed communities. Moreover, the effect of temperature and nitrogen source was investigated. The outcomes of this study provide a first attempt towards process development for a methane-oxidizing bioreactor.
Materials and methods
Sampling and enrichment
As sources for MOB, samples were taken on sites expected to contain an active and abundant methanotrophic community: an anaerobic sludge effluent tank, an open pig manure container, the top of a composting heap, an effluent vessel of an anaerobic digester, a water sample out of a vessel with rotting plant material, and a soil sample. MOB were enriched in each sample: They were inoculated separately in nitrate mineral salts medium (NMS), without copper addition (Whittenbury et al. 1970). The enrichment cultures were placed on a shaker (120 rpm) during 10 days under standard conditions (28°C, pH 7, 20% (v/v) methane in air). Therefore, airtight bottles, with a volume of 1,150 mL were used (Schott AG, Mainz, Germany), which were closed with a rubber butyl stopper (Ochs GmbH, Bovenden, Germany).
Experimental setup to assess the influence of temperature, nitrogen source, and copper addition (test 1)
The enriched cultures were added in equal volumes (1% in total) to four identical airtight bottles of 1,150 mL. The latter were filled with 400 mL NMS medium and were placed on a shaker at 120 rpm (28°C, pH 7, 20% (v/v) methane in air). However, one parameter was altered for each reactor. Reactor A was kept under the standard conditions as mentioned before. Reactor B was placed at a higher temperature of 35 ± 1°C. In reactor C, the nitrate was replaced by ammonium (140 mg NH +4 –N L−1), which can be used as alternative nitrogen source for methanotrophs. To reactor D, 0.64 mg Cu(II) L−1 was added as CuSO4, as this can be assumed to stimulate the expression of pMMO (Shah et al. 1992).
After each cycle of 70 h, half of the well-mixed liquid phase was replenished with freshly made medium. Hence, the hydraulic retention time (HRT) and sludge retention time (SRT) were both 140 h. The addition of new medium was performed under non-sterile conditions. The reactors were open for at least 30 min, which allowed the gas phase to equilibrate with the outside air. A volume of 150 mL of methane (99.95% pure, Air Liquide, Luik, Belgium) was then added to the closed reactors. During the whole test, the pH of all reactors remained in the range of 6 to 7, which is optimal for most methanotrophs (Hanson and Hanson 1996).
Experimental setup to assess the influence of a higher copper concentration (test 2)
Enrichment and inoculation was identical to test 1. Six reactors were placed under the standard conditions of test 1 (28°C, shaker at 120 rpm, NMS medium without copper addition). However, to three of them, 0.64 mg L−1 Cu(II) was added. Half of the medium was replenished after 46 h (one cycle), unless there was still more than 4% (v/v) oxygen present in the reactor. This gives a HRT and SRT of 92 h. From cycle 7 on, NaCl (Normapur, VWR, Leuven, Belgium) was added to the reactors at different concentrations. As a control, reactors without salt addition were maintained as in the previous cycles. After 16 cycles, half of the medium of each reactor was replaced as mentioned before. Then, they were autoclaved (20 min, 121°C, 1 bar). The airtight bottles were cooled down to room temperature and placed on a shaker, while new methane was added. The methane oxidation rate was followed up during 9 days.
Gas composition analyses
For every cycle, a gas sample was taken right after the addition of methane, after 1 day and at the end of each cycle, by use of a gastight syringe (Hamilton, Sigma Aldrich, Bornem, Belgium). Samples were stored in vacutainers, under atmospheric pressure (Labco limited, Buckinghamshire, UK). Gas phase analyses were performed with a Finnigan Trace GC Ultra (Thermo Fisher Scientific, Zellik, Belgium). CH4 concentrations were measured with a flame ionization detector, while CO2 and O2 were measured with a thermal conductivity detector. The methane oxidation rate (MOR) was expressed as the chemical oxygen demand of methane (CH4-COD) over time. The COD value of methane is 4 mg COD mg−1 CH4.
Anion concentrations and pH
At the end of each cycle, samples were taken for further analysis. Filtered samples (0.45-µm filter, Millipore, Brussels, Belgium) were analyzed for Cl−, NO −3 , NO −2 , SO 2−4 , and PO 3−4 , by means of an 761 Compact Ion Chromatograph, equipped with a conductivity detector (Metrohm, Zofingen, Switzerland). Ammonium concentrations were determined by means of steam distillation, according to Greenberg et al. (1992). The pH was determined with an SP10B pH electrode, connected to a C532 multimeter analyzer (Consort, Turnhout, Belgium).
Biomass assessment
To correlate the methane oxidation and biomass growth, the latter was followed up by measuring the COD content in the water phase at the beginning and end of each cycle, according to Bullock et al. (1996). This analysis was performed with a kit, based on spectrophotometry (Hanna Instruments type HI 93754B-25 MR, Temse, Belgium). In test 2, cell dry weight (CDW) was assessed based on a determination of total solids and volatile solids according to Greenberg et al. (1992). To correlate both methods, simultaneous tests were done on all types of the ongoing reactors.
Naphthalene oxidation assay
A slightly modified version of the naphthalene oxidation assay of Brusseau et al. (1990) was used to quantitatively measure the sMMO activity. A crystal of naphthalene was added to 25-mL cell suspension and incubated at 28°C on a shaker for 2 h. After incubation, 250 μL of freshly prepared tetrazotized-o-dianisidine solution (1.0 g L−1) was added to each test tube, and the formation of a colored diazo-dye was immediately monitored by recording the absorbance at a wavelength of 525 nm via spectrophotometry, as described by Koh et al. (1993). The protein content of the cell suspension was determined by the total protein kit of Sigma Aldrich (Bornem, Belgium) after lysis of the cells. The specific activity of sMMO is expressed as nanomoles of naphthol formed, per milligram of cell protein per hour.
Statistical analyses
Statistical analysis was performed with SPSS for Windows, version 15 (SPSS Inc., Chicago, IL, USA). Linear regression and ANOVA were performed, after tests for homogeneity and normal distribution. A significance level of 0.05 was used (LSD).
DNA extraction, polymerase chain reaction amplification, and DGGE analysis
In test 2, samples were taken from the reactors, respectively after 1, 6 (just before salt addition), and 15 cycles and stored at −20°C. DNA extraction was performed according to Boon et al. (2000). Gel electrophoresis was performed to check if DNA was present. As low-concentration methanotrophs are not assumed to play an important role in the total activity (Henckel et al. 2000), primers specific for the high concentration methanotrophs were chosen (Bodelier et al. 2005). These primers target a large part of the type II MOB diversity and allow detection of Methylosinus and Methylocystis species. Fragments of the 16S rRNA gene fragment were amplified using nested polymerase chain reaction (PCR), according to Bodelier et al. (2005). The total amount of cycles was lowered to 15 and 25 respectively for the first and second round. An INGENY phorU2X2 DGGE-system (Goes, The Netherlands) was used for running 8% (w/v) polyacrylamide DGGE gels with a denaturating gradient ranging from 40% to 60% (Bodelier et al. 2005). The obtained DGGE patterns were subsequently processed, using Bionumerics software version 5.1 (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Effect of temperature, ammonium, and copper on the methane oxidation activity
In test 1, four reactors with the same inoculum were incubated under different conditions: (A) a control reactor at 28°C, (B) a reactor at an elevated temperature (35°C), (C) a reactor with ammonium as nitrogen source (28°C), and (D) a reactor with Cu(II) addition (0.64 mg Cu(II) L−1, 28°C). For reactors A, B, and D, NMS medium was used. In reactor C, NO −3 –N was replaced by the same concentration of NH +4 –N (140 mg L−1).
There was no significant difference in methane oxidation activity observed between the reactors under the different conditions of N source or temperature (reactors A, B, and C). From cycle 4 on, the average MOR of these three reactors was 393 ± 109 mg CH4-COD L−1 day−1 (Table 1). This was observed for 11 consecutive cycles. The average biomass growth, expressed as CDW, also stabilized from the fourth cycle on, with an average value of 162 ± 17 mg CDW-COD L−1 cycle−1. The average growth yield of these reactors was 0.25 ± 0.04 g CDW-COD g−1 CH4-COD. The pH value over the whole test period was 6.4 ± 0.2 for all three reactors. The average nitrogen consumption of the three reactors was 18 ± 4 mg N L−1 per cycle (70 h). In most cases, the nitrite concentration was lower than 2 mg NO −2 –N L−1. In the reactor with NH +4 as N donor, NO −3 –N and NO −2 –N concentrations stayed below the detection limit (1 mg NO −2 L−1).
In reactor D, the methane-oxidizing activity was significantly influenced by a copper(II) addition of 0.64 mg Cu(II) L−1 (p ≤ 0.001). The average oxidation rate of 582 ± 109 mg CH4-COD L−1 day−1 was a factor of 1.5 higher than the average of the other three reactors. Nevertheless, this higher activity had no significant effect on the biomass growth nor growth yield (Table 1). The final gas concentration at the end of each cycle did not differ significantly between the four tested reactors. At the end of each cycle, oxygen levels dropped to values lower than 1% (v/v). The gas phase concentration of methane and carbon dioxide at the end of each cycle was 4.2 ± 1.1% and 5.3 ± 1.2% (v/v), respectively. During a cycle of 70 h, on average, 861 ± 144 mg CH4-COD L−1 was oxidized.
Effect of copper on the methane oxidation activity and the microbial community
To confirm the influence of copper addition on the methane oxidation activity, a new test was performed. In test 2, six reactors were set up as before. In three of them, 0.64 mg L−1 copper was supplemented; the other three reactors served as controls without copper addition. During the first 96 h after inoculation, the biomass concentration increased from less than 22 to 231 ± 45 mg CDW-COD L−1, without significant differences between the reactors (p < 0.05). From the second cycle on, the average biomass growth was stable, with average values of 200 ± 9 and 237 ± 42 mg CDW-COD L−1 cycle−1 for the reactors without and with copper, respectively (Table 2). There was no significant difference in growth yield, nor C/N ratio, between the reactors with and without copper addition (Table 2). The activity was a factor of 1.7 higher in the reactors with copper addition. An average oxidation rate of respectively 452±79 (without copper) and 755 ± 40 (with copper) mg CH4-COD L−1 day−1 was found over the last five cycles (Table 2). The methane concentration after 46 h was generally around 3.5% (v/v) for the reactors with copper addition, while the oxygen level dropped below 0.5% (v/v). The reactors without copper addition did not reach these low levels after 46 h. There was still about 2% (v/v) oxygen and 5% (v/v) of methane present in the gas phase.
No sMMO was detected in the reactors with copper addition. For the reactors without copper addition, a naphthalene oxidation rate of 121 ± 26 nmol mg−1 protein h−1 was observed.
Changes in the HCM composition of the reactors were evaluated by means of DGGE analysis (Fig. 2). After six cycles, different methane-oxidizing species became dominant in the reactors with and without copper respectively, which is reflected in a low Pearson correlation of less than 60%. The communities in the reactors with the same treatment were similar to a high extent, with a Pearson correlation in all cases higher than 80%.
Effect of salt addition on the methane oxidation activity and the microbial community
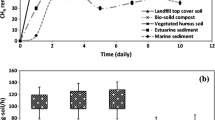
Addition of 7 g NaCl L−1 to the reactors of test 1 did not result in a significant decrease of activity. However, an addition of 8.5 g NaCl L−1 resulted in a decreased MOR in all four reactors. The activity decreased with a factor of 4 in the reactors without copper(II) addition, where it lowered only with a factor of 2 in the reactor with copper(II) addition. This salt concentration was also tested in the reactors of test 2. Addition of 8.5 g NaCl L−1 lowered the MOR to the same extent as in test 1 (Fig. 1). From the eight cycles on, the sodium chloride concentration was increased again, to a final value of 11.2 ± 0.5 g NaCl L−1. This led to a decrease in activity in all the reactors. The highest decrease was observed in the reactors without copper(II) addition. The activity of the latter was a factor of 10 lower, with an average MOR of 39 ± 12 mg CH4-COD L−1 day−1, immediately after the salt addition. The MOR was lower than 60 mg CH4-COD L−1 day−1 during the first 10 days following the salt addition. Moreover, one of the reactors did not show any oxidation during those 10 days. After this period, these reactors showed an increasing performance with a maximum oxidation rate of 439 ± 59 mg CH4-COD L−1 day−1. However, the activity of these reactors showed a large variation.
The influence of salt addition on the methane oxidation rate of reactors without (filled circles) and with (empty circles) copper addition (0.64 mg L−1) during test 2. The average MOR (mg CH4-COD L−1 day−1) and the standard error were calculated for the first day of each cycle. During the seventh cycle, the concentration of NaCl was 8.5 g L−1. From cycle 8 on, the NaCl concentration was 11.2 L−1
The decrease in activity was less in the reactors with copper addition. The MOR decreased 41%, from 715 ± 11 to 422 ± 180 mg CH4-COD L−1 day−1 when 8.5 g NaCl L−1 was added (cycle 8). In the next cycle (cycle 9), the MOR dropped to 241 ± 168 mg CH4-COD L−1 day−1 (Fig. 1). However, in the following cycles, these reactors showed a stable performance between 325 and 475 mg CH4-COD L−1 day−1, with a maximum of 474 ± 61 mg CH4-COD L−1 day−1 in the eighth cycle after the first salt addition (cycle 14) (Fig. 1).
The salt addition had no influence on the nitrogen demand. However, in the reactors with copper addition, nitrite concentrations up to 40 mg NO −2 –N L−1 were detected. The increase of the nitrite concentration corresponded with the decrease in the nitrate level.
There was no significant difference in biomass growth between the reactors with and without salt addition nor between the reactors with and without copper (Table 2). When the cell dry weight was compared with COD measurements, a ratio of 1.49 ± 0.12 mg COD mg−1 CDW was found for samples with, as well as without salt addition. The growth yield was independent of the copper and salt concentration and was on average 0.22 mg CDW-COD mg−1 CH4-COD. The salt addition altered all HCM communities to a large extent. In all reactors, expression of new DGGE bands was observed. The dominant HCM species before salt addition were less abundant after nine cycles of higher salt concentrations, while other HCM species became dominant (Fig. 2).
Changes in DGGE patterns of the HCM community, in the different reactors of test 2, resulting from the addition of copper and/or salts. DGGE clustering was performed, based on the Pearson correlation (given as a percentage). The reactor samples are labeled as follows: inoculum (in triplicate), without Cu(II) (O1 to O3), and with Cu(II) (C1 to C3), after one cycle without salt (cyc1), after six cycles without salt (cyc6), and 15 cycles (nine cycles with salt addition) (cyc15)
From cycle 17 on, the concentration of salt was adjusted to 30 g NaCl L−1. During the test period of 10 days, the MORs were in all reactors below 50 mg CH4-COD L−1 day−1. No methane oxidation could be detected after heat killing the active communities.
Discussion
In this study, efforts are made to identify important parameters that affect methane-oxidizing microbial systems, applicable for the treatment of methane containing effluents. It was found that the MOR was not influenced by either increased temperature, or by the presence of exogenous ammonium supplementation. The addition of copper or NaCl, however, had a significant influence on the MOR.
The influence of temperature and ammonium
A temperature increase from 28° to 35°C did not significantly affect the activity or growth rate of the MOB. It has been reported that most methane-oxidizing bacteria have an optimal temperature of 30°C, which explains why an increase to 35°C was not accompanied by a higher activity (Park et al. 2005; Scheutz and Kjeldsen 2004). Consequently, treatment of effluents in the range between 28°C and 35°C should not give problems.
Changing the nitrogen source from nitrate to ammonium also had no significant influence on the MOR. This result contrasts with those reported by other research groups, who observed inhibition of methane oxidation at ammonium levels of 140 mg N L−1 (Begonja and Hrsak 2001; Hanson and Hanson 1996; King and Schnell 1998; Nyerges and Stein 2009). These conflicting observations suggest that more insight is needed into the specific effects that ammonium has on the microbial methane oxidation process, especially in mixed culture conditions.
The communities in the different reactors behaved similarly under the different environmental conditions. Hence, stable methane-oxidizing communities were present. This was confirmed by DGGE analysis and the presence of active sMMO (Fig. 2). These communities reached a stable MOR after a start-up period of less than 72 h (Table 1). The observed activity, growth yield, and nitrogen consumption were similar to previously described methanotrophic–heterotrophic communities and confirm the presence of high MOB activity in mixed communities (Helm et al. 2006; Hrsak and Begonja 2000; Melse and Van der Werf 2005). The used semi-continuous model system seems a good model for methane-oxidizing bioreactors since the removal rates were similar (Melse and Van der Werf 2005).
It can be assumed that LCM only have a small share in the methane-oxidizing activity, as it is known that they are outcompeted by the HCM under high methane concentrations (> 1% (v/v) CH4) (Amaral and Knowles 1995; Hanson and Hanson 1996). Moreover, fluorescent in situ hybridization revealed that only a small amount of LCM was present in the microbial community (data not shown). The methane concentration at the end of each cycle was, in agreement with the results of all tested reactors, never lower than 3.5% (v/v). It seems that the observed oxygen concentration of 0.5% (v/v) at the end of most cycles was the lowest oxygen level that allowed methane oxidation. This is in agreement with other authors, who found maximum rates of methane oxidation in a range of 0.45% to 20% (v/v) O2 (Henckel et al. 2000; Wilshusen et al. 2004).
The influence of copper
Copper had a significant positive effect on the MOR. The addition of 0.64 mg Cu(II) L−1 increased the average oxidation rate in test 2 by a factor of 1.7 to 755 ± 40 mg CH4-COD L−1 day−1 (Table 2). This is slightly more than reported in studies with axenic methanotrophic cultures, where a factor of 1.2 to 1.5 has been found (Begonja and Hrsak 2001; Joergensen and Degn 1987). Oxygen became a limiting factor for growth before the end of each cycle. Therefore, the higher methane oxidation rate after copper addition did not lead to a significantly higher biomass concentration at the end of each cycle. However, no conclusions can be made about the specific growth yield of the methane-oxidizing bacteria, as only the biomass growth of the whole bacterial community was measured.
The addition of copper had a distinct effect on the HCM community. DGGE analyses of test 2 revealed that the profiles of replicate treatments were very similar throughout the entire test period, although there was no direct cross-inoculation (Fig. 2). In contrast, copper(II) addition had a distinctive selective power, as it shifted the HCM community to an important extent. Moreover, the copper addition had an influence on the MMO enzymes. No sMMO activity was detected in the reactors with copper addition. The latter is in agreement with former observations, where a concentration of 0.64 mg Cu(II) L−1 had an inhibitory effect on sMMO expression while stimulating the pMMO expression (Hakemian and Rosenzweig 2007; Jahng and Wood 1996; Shah et al. 1992).
The influence of salt addition
The addition of NaCl up to a concentration of 7 g NaCl L−1 had no significant influence on the MOR. However, a concentration of 8.5 g NaCl L−1 led to a decrease in methane-oxidizing activity in all reactors. Immediately after the addition of 11.2 g NaCl L−1, the reactors without copper(II) showed almost no activity (< 50 mg CH4-COD L−1 day−1) for 10 days (Fig. 1). These results are in agreement with Schnell and King (1996), who found a 35% lower methane oxidation rate when a salt concentration of 10 g NaCl L−1 was amended to the HCM Methylosinus trichosporium OB3b, although these results contrast with the results of Bowman et al. (1993), who did not observe a high sensitivity for NaCl, when using a rapid titer tray method.
Interestingly, the MOR remained above 250 mg CH4-COD L−1 day−1 in the reactors which received both copper and NaCl (11.2 ± 0.5 g L−1). The different response to increased salt concentrations could be a result of either community and/or enzyme level effects. At the community level, a population shift was seen during the cycles without salt addition. It is possible that a fortuitous selection took place towards more salt tolerant species. This, however, appears unlikely, as the dominant HCM species before and after salt addition were completely different (Fig. 2). Moreover, since no significant activity was seen at a concentration of 30 g NaCl L−1, it is clear that truly halophilic methanotrophs were not present in considerable amounts. However, over time, the fact that methane-oxidizing activity recovered partly in all reactors (Fig. 1) suggests that salt tolerant species were enriched and became more abundant, as shown by DGGE analysis.
The fast decrease in activity after salt addition is probably due to differences at the enzyme level. The immediate effect of the salt addition clearly correlated with the copper concentration (Table 2). As copper is the key element in the expression of both sMMO and pMMO, the differential effect of salt on reactor performance could be related to the type of methane monooxygenases present in the different reactors: The reactors without copper were enriched with methanotrophs that showed sMMO activity, whereas the reactors with added copper addition did not show any sMMO activity. As the only other enzyme in MOB known to oxidize methane with such a high rate is pMMO, the MOB in these reactors appear to have pMMO as the active methane monooxygenase (Begonja and Hrsak 2001; Hakemian and Rosenzweig 2007; Shah et al. 1992). This suggests that sMMO and pMMO possess different sensitivities towards NaCl. To our knowledge, these findings have not previously been reported and need to be confirmed by means of pMMO and sMMO expression assays. However, they are consistent with the work of Trotsenko and Khmelenina (2002), who found that only two of the 30 salt-resistant methanotrophs they analyzed produced sMMO. This difference in sensitivity can also be due to the different locations of the monooxygenase enzymes in the bacterial cells, rather than the intrinsic tolerance of the enzyme itself. pMMOs are embedded in intraplasmatic membranes, which might offer protection against the increased salt concentrations, where sMMOs have less osmotic protection in the cytoplasm (Colby et al. 1977; Dalton 2005). If location is more important than enzyme structure itself, then the difference in sensitivity might also extend to other types of salts or solutes.
After ten cycles with salt addition, the MORs were still a factor of 2 lower than the controls. It has been reported that bacteria use additional energy to regulate their metabolism under hyperosmotic conditions (Csonka 1989). That could lead to a lower bacterial activity. The formation of nitrite could also be a reason for the lower activity. Nitrite concentrations up to 40 mg NO −2 –N L−1 were observed in the reactors where copper(II) and salt were amended. Although the reason for the high nitrite concentration is unknown, similar levels of nitrite have been shown to reduce the methane oxidation activity of at least three axenic MOB strains (Nyerges and Stein 2009).
The findings of this work provide a first attempt for optimizing microbial methane removal systems. The maximum MOR achieved was 280 mg CH4 L−1 reactor day−1. As the maximum solubility of methane in water is 26 mg CH4 L−1 (22°C) (Melse and Van der Werf 2005), a hydraulic retention time of 2.2 h would be required to treat methane-saturated effluents. Because of the low solubility of both methane and oxygen, treatment of the aerated water phase as such would be necessary. Since the MOR is comparable at temperatures of 28°C and 35°C, good methane removal can be achieved at lower temperatures. In situations where increased copper concentrations are acceptable, addition of copper(II) to methanotrophic bioreactors should be considered, as this increased the MORs by more than 50%. Anaerobic effluents can contain salt concentrations of up to 10 g NaCl L−1 (Gourdon et al. 1989; Ismail et al. 2008). As concentrations of up to 7 g NaCl L−1 and 140 mg NH +4 –N L−1 did not seem to have a negative influence on the MOR, treatment of methane-saturated effluents, rich in salts and high in ammonium, should be possible.
References
Amaral JA, Knowles R (1995) Growth of methanotrophs in methane and oxygen counter gradients. FEMS Microbiol Lett 126:215–220
Anthony C (1982) Biochemistry of methylotrophs. Academic, London
Balasubramanian R, Rosenzweig AC (2008) Copper methanobactin: a molecule whose time has come. Curr Opin Chem Biol 12:245–249
Begonja A, Hrsak D (2001) Effect of growth conditions on the expression of soluble methane monooxygenase. Food Technol Biotechnol 39:29–35
Bender M, Conrad R (1992) Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol 101:261–270
Bender M, Conrad R (1995) Effect of CH4 concentrations and soil conditions on the induction of CH4 oxidation activity. Soil Biol Biochem 27:1517–1527
Bodelier PLE, Laanbroek HJ (2004) Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol Ecol 47:265–277
Bodelier PLE, Meima-Franke M, Zwart G, Laanbroek HJ (2005) New DGGE strategies for the analyses of methanotrophic microbial communities using different combinations of existing 16S rRNA-based primers. FEMS Microbiol Ecol 52:163–174
Boon N, Goris J, De Vos P, Verstraete W, Top EM (2000) Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913
Bowman JP, Sly LI, Nichols PD, Hayward AC (1993) Revised taxonomy of the methanotrophs—description of Methylobacter Gen-Nov, emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the Group-I methanotrophs. Int J Syst Bacteriol 43:735–753
Brusseau GA, Tsien HC, Hanson RS, Wackett LP (1990) Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19–29
Bullock CM, Bicho PA, Zhang Y, Saddler JN (1996) A solid chemical oxygen demand (COD) method for determining biomass in waste waters. Water Res 30:1280–1284
Cakir FY, Stenstrom MK (2005) Greenhouse gas production: a comparison between aerobic and anaerobic wastewater treatment technology. Water Res 39:4197–4203
Colby J, Stirling DI, Dalton H (1977) Soluble methane mono-oxygenase of Methylococcus capsulatus (Bath)—ability to oxygenate normal-alkanes, normal-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J 165:395–402
Csonka LN (1989) Physiological and gentic responses of bacteria to osmotic-stress. Microbiol Rev 53:121–147
Dalton H (2005) The Leeuwenhoek Lecture 2000—the natural and unnatural history of methane-oxidizing bacteria. Philos Trans R Soc Lond, Ser B 360:1207–1222
De Visscher A, Van Cleemput O (2003) Induction of enhanced CH4 oxidation in soils: NH +4 inhibition patterns. Soil Biol Biochem 35:907–913
Dunfield PF, Liesack W, Henckel T, Knowles R, Conrad R (1999) High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl Environ Microbiol 65:1009–1014
Gourdon R, Comel C, Vermande P, Veron J (1989) Kinetics of acetate, propionate and butyrate removal in the treatment of a semi-synthetic landfill leachate on anaerobic filter. Biotechnol Bioeng 33:1167–1181
Greenberg AE, Clesceri LS, Eaton AD (eds) (1992) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, American Water Works Association, Water Environment Federation, Washington
Gulledge J, Schimel JP (1998) Low-concentration kinetics of atmospheric CH4 oxidation in soil and mechanism of NH +4 inhibition. Appl Environ Microbiol 64:4291–4298
Hakemian AS, Rosenzweig AC (2007) The biochemistry of methane oxidation. Annu Rev Biochem 76:223–241
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Hartley K, Lant P (2006) Eliminating non-renewable CO2 emissions from sewage treatment: An anaerobic migrating bed reactor pilot plant study. Biotechnol Bioeng 95:384–398
Helm J, Wendlandt KD, Rogge G, Kappelmeyer U (2006) Characterizing a stable methane-utilizing mixed culture used in the synthesis of a high-quality biopolymer in an open system. J Appl Microbiol 101:387–395
Henckel T, Jackel U, Schnell S, Conrad R (2000) Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl Environ Microbiol 66:1801–1808
Hrsak D, Begonja A (2000) Possible interactions within a methanotrophic–heterotrophic groundwater community able to transform linear alkylbenzenesulfonates. Appl Environ Microbiol 66:4433–4439
IPCC (2001) Climate change 2001: the scientific basis. In: Houghton J, Ding Y, Griggs D, van der Linden P (eds) Contribution of working group I to the third assesment report of the intergovernmental panel of climate change. Cambridge University Press, Cambridge
Ismail SB, Gonzalez P, Jeison D, van Lier JB (2008) Effects of high salinity wastewater on methanogenic sludge bed systems. Water Sci Technol 58:1963–1970
Jahng D, Wood TK (1996) Metal ions and chloramphenicol inhibition of soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol 45:744–749
Joergensen L, Degn H (1987) Growth-rate and methane affinity of a turbidostatic and oxystatic continuous culture of Methylococcus capsulatus (Bath). Biotechnol Lett 9:71–76
King GM, Schnell S (1998) Effects of ammonium and non-ammonium salt additions on methane oxidation by Methylosinus trichosporium OB3b and Maine forest soils. Appl Environ Microbiol 64:253–257
Koh SC, Bowman JP, Sayler GS (1993) Soluble methane monooxygenase production and trichloroethylene degradation by a type-I methanotroph, Methylomons methanica-68-1. Appl Environ Microbiol 59:960–967
Melse RW, Van der Werf AW (2005) Biofiltration for mitigation of methane emission from animal husbandry. Environ Sci Technol 39:5460–5468
Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Muller J, Lippard SJ (2001) Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew Chem Int Ed 40:2782–2807
Nyerges G, Stein LY (2009) Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol Lett 297:131–136
Park JR, Moon S, Ahn YM, Kim JY, Nam K (2005) Determination of environmental factors influencing methane oxidation in a sandy landfill cover soil. Environ Technol 26:93–102
Scheutz C, Kjeldsen P (2004) Environmental factors influencing attenuation of methane and hydrochlorofluorocarbons in landfill cover soils. J Environ Qual 33:72–79
Scheutz C, Kjeldsen P, Bogner JE, De Visscher A, Gebert J, Hilger HA, Huber-Humer M, Spokas K (2009) Microbial methane oxidation processes and technologies for mitigation of landfill gas emissions. Waste Manage Res 27:409–455
Schnell S, King GM (1996) Responses of methanotrophic activity in soils and cultures to water stress. Appl Environ Microbiol 62:3203–3209
Shah NN, Park S, Taylor RT, Droege MW (1992) Cultivation of Methylosinus trichosporium Ob3b. 3. Production of particulate methane monooxygenase in continuous culture. Biotechnol Bioeng 40:705–712
Trotsenko YA, Khmelenina VN (2002) Biology of extremophilic and extremotolerant methanotrophs. Arch Microbiol 177:123–131
Whalen SC (2000) Influence of N and non-N salts on atmospheric methane oxidation by upland boreal forest and tundra soils. Biol Fertil Soils 31:279–287
Whittenbury R, Phillips KC, Wilkinson JF (1970) Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–217
Wilshusen JH, Hettiaratchi JPA, De Visscher A, Saint-Fort R (2004) Methane oxidation and formation of EPS in compost: effect of oxygen concentration. Environ Pollut 129:305–314
Acknowledgments
This work was supported by a Ph.D. grant (no. 83259) from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen), a research grant from the Flemish Fund for Scientific Research (FWO-Vlaanderen, 3G070010), and by the Geconcerteerde Onderzoeksactie (GOA) of Ghent University (BOF09/GOA/005). Tim Lacoere’s help with the molecular analyses is greatly acknowledged. Also, thanks to Jan Vermeulen and Samuel Bodé for their support with GC analyses and Vicky D’havé for her assistance. Furthermore, Anthony Hay, Joachim Desloover, Yu Zhang, and Willem De Muynck are gratefully appreciated for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van der Ha, D., Hoefman, S., Boeckx, P. et al. Copper enhances the activity and salt resistance of mixed methane-oxidizing communities. Appl Microbiol Biotechnol 87, 2355–2363 (2010). https://doi.org/10.1007/s00253-010-2702-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2702-4