Abstract
Agricultural soils in oilfields have high risk for polycyclic aromatic hydrocarbon (PAH) pollution. In this study, from the Jianghan Oilfield (Hubei Province, China) with a history of >50 years, 7 soil samples (OS-1 to OS-7) were collected. Subsequently, the bacterial, archaeal, and fungal community structures were investigated by Illumina MiSeq sequencing, and the relationship between microbial community structure and soil PAH content was analyzed. The results indicated that bacterial and archaeal Chao 1 indices showed a significantly negative relationship with soil PAH content, and only the bacterial Shannon index had a significantly negative relationship with soil PAH content. Moreover, the community structure of bacteria (r 2 = 0.9001, p = 0.013) showed a stronger correlation with PAH content than that of fungi (r 2 = 0.7357, p = 0.045), and no significant relationship was found between archaeal community structure (r 2 = 0.4553, p = 0.262) and soil PAH content. In addition, the relative greater abundances of some bacterial genus belonging to Actinobacteria (Mycobacterium and Micromonospora) and Proteobacteria (Pseudomonas, Lysobacter, Idiomarina, Oxalobacteraceae, and Massilia), fungal genus belonging to Ascomycota (Sordariales and Pleosporales), and archaeal phylum (Euryarchaeota) were detected in the soil samples (OS-3 and OS-5) with greater PAH content. In summary, soil PAHs showed an obvious influence and selectivity on the soil microbiota. Furthermore, compared with fungi and archaea, bacteria was more sensitive to soil PAH pollution, and the diversity indices and community structure of bacteria both might be suitable indicators for assessment of soil PAH stress on the soil ecosystem.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Polycyclic aromatic hydrocarbons (PAHs), a group of typically and widespread organic pollutants, could pose a serious threat to the environment and human health due to their intensive carcinogenicity, teratogenicity, and mutagenicity (Couling et al. 2010; Xue and Warshawsky 2005). Moreover, these compounds are difficult to be degraded and could be long-lived in environment due to their stable polycyclic structures and strong hydrophobic property (Eggen and Majcherczyk 1998; Xue and Warshawsky 2005). Soil is an important natural reservoir for PAHs, and PAHs in soil could be transferred through the food chain (Van der Oost et al. 2003). Thus, some key questions about soil PAH pollution, such as their remediation, migration and transformation, and risk assessment, have long concerned people all over the world.
Soil microorganisms are not only the important participants for the natural PAH elimination and transformation, they are also useful indicators for the assessment of soil quality under the pressure of organic or inorganic pollutants (Haritash and Kaushik 2009; Khan et al. 2013; Ruggiero et al. 2011; Ruyters et al. 2010). Therefore, in recent years, to better understand the mechanism of soil PAH transformation and to evaluate the risk of soil PAH pollution, more attentions has been paid to the responses of microbial activities and community structures to soil PAH pollution (Abbasian et al. 2016; Cebron et al. 2015; Chen et al. 2016; Wang et al. 2016; Zhang et al. 2014). Presently, microbial ribosome RNA genes (16S rRNA and 18S rRNA) and fungal ITS (internal transcribed spacer) variable regions contained in ribosomal DNA have been widely used as biomarkers to reflect the responses of soil microbes under PAH pressures, which also could be used as an available tool to identify some novel PAH-degrading microbes (Cebron et al. 2015; Llado et al. 2013; Su et al. 2016; Wang et al. 2013). For example, using bacterial 16S rRNA gene as biomarker, Sawulski et al. (2014) found that bacterial community structure was significantly shifted by PAH amendment, and phenanthrene, fluoranthene, and benzopyrene showed varied selectivity on soil bacterial flora. Some bacterial genera (Chryseobacterium, Pusillimonas, and Sphingobium) and fungal genus (Fusarium) had been identified as important HMW (high-molecular weight)–PAH degraders in PAH-polluted soils, and it had been reported that microbes related to carbon metabolism and nitrogen-transforming were significantly enhanced and restrained by PAH contamination, respectively (Llado et al. 2013, 2015). In addition, some functional genes involved in PAH degradation and some novel techniques based on RNA- or DNA-SIP (stable-isotope probing) have also been widely used to investigate the relationship between soil microorganisms and PAH pollution (Li et al. 2015; Liu et al. 2015; Peng et al. 2014; Song et al. 2015, 2016). It was reported that the abundances of some functional genes had good dose–response relationships with soil PAH contamination under both aerobic and anaerobic conditions (Guo et al. 2011; Han et al. 2014). Therefore, these studies confirmed that the soil microbial community and its functions had a sensitive response to PAH contamination and also identified some unique degraders for different kinds of PAHs in varied soil types, which could provide some essential basic theories and reliable guidance for the remediation and risk assessment of soil PAH pollution.
Agricultural soils near oilfields have a high risk of PAH pollution because they are aged soils with mixed PAH contamination, which might be very representative objects for a study about the interaction between soil microorganisms and PAHs. Very recently, the relationship between soil bacterial community structure and PAH contamination in the regions of several important oilfields in China was reported (Gao et al. 2016; Liao et al. 2015; Liu et al. 2015; Sun et al. 2015a, b). For example, in the soil collected from Dagang Oilfield (North China), Liu et al. (2015) found that the gene copies of AlkB (alkane monooxygenase gene) and Nah (naphthalene dioxygenase gene) were positively and negatively related to the soil PAH contents, respectively. Meanwhile, Sun et al. (2015a) found many known oil-degrading species (Alkanindiges, Arthrobacter, Pseudomonas, Mycobacterium, and Rhodococcus) dominated in nine soil samples collected from the Daqing (Northeast China) and Changqing (Northwest China) oilfields. Therefore, it seemed that PAH contamination had an obvious selection effect on soil microbiota in the area of oilfields and that these studies could provide some essential microbial information for the risk assessment and bioremediation of PAH contamination for local soils. Soil geochemical parameters have been considered as important factors influencing the community structures of soil microorganisms (Ge et al. 2008; Wang et al. 2015b), and indigenous microbial information might be very important for in situ remediation in a given PAH-polluted soil (Sun et al. 2015b). However, the oilfields selected in most present studies were located in the region of Northern China, and no information was available for the oilfield soils in south-central China. Furthermore, soil bacteria, fungi, and archaea all are important components of soil microorganisms, but few studies have taken all of them into consideration simultaneously.
In this study, the Jianghan Oilfield, with a history of >50 years and located in south-central China (Qianjiang City, Hubei Province), was selected, and soil samples were collected from the agricultural field near this oilfield. By means of Illumina MiSeq sequencing, the bacterial, fungal, and archaeal community structures were explored, and the relationship between microbial community structure and soil PAH contents was investigated. The aim was to profile and compare the community structures of different kinds of microorganisms in soil with potential PAH contamination, which might provide some useful microbial information for the risk assessment and in situ bioremediation of the PAH-polluted soil.
Materials and Methods
Site Description and Soil Sampling
Jianghan Oilfield, located in the interior of Jianghan Plain (Qianjiang city, Hubei province, China), one of the most important oilfields in China, has a history of >50 years. This region has a subtropical monsoon moist climate with a mean annual temperature of 16.1 °C and a mean annual precipitation of 1100 mm, and the soil is classified as a Fluvo-aquic soil. In this study, 7 soil samples (OS-1 to OS-7) were collected from the agricultural field (rice–wheat rotation) in the Jianghan Oilfield in July 2014. Detailed geographic information of the sampling sites is shown in Fig. 1. In each site, 10 surface-soil cores (5 cm in diameter; 20 cm in depth) were randomly selected in an area of 100 m2 and then mixed thoroughly to form one representative sample. Three samples were collected from each site as replicates. Soil samples were kept in an ice box and taken back to the laboratory. After removing the fine roots and visible organic debris by passing through a 2-mm sieve, the samples were stored at 4 and −80 °C for chemical analysis and DNA extraction, respectively.
Measurement of Soil Basic Properties and PAH Content
Soil pH was determined with a soil to water ratio of 2:5. Soil organic carbon (SOC) was measured using the K2Cr2O7 oxidation method. Soil-available phosphorus (AP) was determined by the routine method recommended by the Chinese Society of Soil Science (Lu 1999). Soil ammonium and nitrate were extracted with 2 mol L−1 KCl and determined using a continuous flow analyzer (SAN++, Skalar, Holland).
Soil PAH content was determined according to the method of Cao et al. (2009), and the brief procedure was as follows: 10 g Na2SO4 and 2 g copper powder were added to a 250-mL beaker containing 10 g soil (freeze-dried at −80 °C) and mixed thoroughly; the mixture was transferred into an extraction thimble, and 70 mL acetone and hexane (1:1 ratio) was added into the thimble to soak the mixture for 12 h. The thimble was placed into a Suoshi Extraction System for 6 h at 75 °C, and the extract was concentrated to approximately 2 mL by rotary evaporation at 45 °C under −46 kPa. The concentrated extract was passed through a chromatographic column (1 cm silica sand, 12 cm active silica, 6 cm neutral alumina, and 1 cm anhydrous Na2SO4); after being washed with hexane, the column was eluted with 70 mL mixed liquor of dichloromethane and hexane (3:7 ratio), and the collected eluent was concentrated by rotary evaporation and diluted to 1 mL with methanol. Finally, total PAH content was detected by a high-pressure liquid chromatographer (Hitachi High-Technologies Corporation, Japan) with a mixed interior reference containing 16 priority-controlled PAHs (Dr. Ehrenstorfer GmbH, Augsburg, Germany).
Soil DNA Extraction and Illumina MiSeq Sequencing
Soil DNA was extracted from 0.5 g of frozen soil using a Fast DNA SPIN Kit for Soil (Q-BIOgene, Carlsbad, California, USA) according to the manufacturer’s protocol. The extracted DNA was checked using 1% agarose gel electrophoresis and stored at −20 °C before use.
The primer sets of 515F-907R, 817F-1196R, and Arch340F-Arch806R were used to amplify the bacterial 16S rRNA, fungal 18S rRNA, and archaeal 16S rRNA genes, respectively. The sequences of the primer sets and thermal profiles used in the following amplification for each target gene are listed in Table 1. Primer sequences were modified by adding Illumina adaptor A with an 8-nucleotide barcode sequence to the ends of each forward primer and adaptor B to the ends of each reverse primer using TruSeq v1/v2 Kit (Illumina, San Diego, California, USA) according to the manufacturer’s protocol. And then, each of the target genes were amplified on an ABI GeneAmp 9700Cycler (Applied Biosystems, USA). The PCR products were purified using an AxyPrepDNA PCR clean-up system (Axygen Biosciences, Union City, California, USA), and then sequenced on the MiSeq platform (Illumina, San Diego, CA, USA) at Majorbio Bio-Pharm Technology, Company Ltd. (Shanghai, China). Raw fastq files were demultiplexed and quality-filtered using QIIME (version 1.9.1) with the following criteria: The 300-base pair (bp) reads were truncated at any site receiving an average quality score <20 over a 50-bp sliding window, and the truncated reads that were shorter than 50 bp were discarded; exact barcode matching, 2-nucleotide mismatch in primer matching, reads containing ambiguous characters were removed; and only sequences that overlap longer than 10 bp were assembled according to their overlap sequence. Reads that could not be assembled were discarded. Operational taxon units (OTUs) were clustered with 97% similarity cut-off using UPARSE (version 7.1 [http://drive5.com/uparse/]), and chimeric sequences were identified and removed using UCHIME. The taxonomy assignment was performed based on the Silva ribosomal database (release119 [http://www.arb-silva.de]) using a confidence threshold of 70% (Amato et al. 2013). In this study, Chao1 and Shannon indices were calculated using Mothur software (version v.1.30.1 [http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity]) with 97% similarity cut-off (Schloss et al. 2011), and they were used to estimate the potential total number of OTU and evaluate the level of diversity in each sample, respectively. The calculation equations are listed below.
where S obs is the total number of OTU generated from the MiSeq sequencing; n 1 is the number of OTU only containing one sequence and n 2 is the number of OTU containing two sequences; n i is the sequence number in the ith OUT; and N is the total sequence number obtained from the MiSeq sequencing.
Data Analysis
One-way analysis of variance (ANOVA) followed by a Student-Newman-Keuls test was performed using SPSS 11.5 (SPSS, Chicago, USA) to check for quantitative differences between soil samples. p < 0.05 was considered to be difference significant. Linear regression analysis was performed to investigate the relationship between soil PAH contents and microbial diversity indices using SPSS 11.5 software. Correlation analysis between microbial community structures and soil basic properties was calculated with a Mantel test using OTU-based Bray–Curtis distance matrices with 999 permutations in R software (http://www.r-project.org) with Vegan package. Cluster analysis based on the microbial phyla was performed using the Vegan package (Jaccard method) in R software, and the heat maps of microbial community based on the bacterial and fungal genus were also generated from R software with the Pheatmap package.
Results
Soil Properties and PAH Contents
The results of soil basic properties and PAH contents are listed in Table 2. The pH values of all soil samples were >8.0. The SOC contents in OS-4 to OS-7 samples were all greater than those in OS-1 to OS-3 samples, and the greatest SOC content was detected in the OS-5 sample. The highest soil AP content was detected in OS-6, and the AP contents in OS-4, OS-6, and OS-7 were greater than those of OS-5 and OS-1 to OS-3. The average NH4 + and NO3 − contents in these soil samples were in the range of 4.03 (OS-4) to 4.60 (OS-3) and 0.36 (OS-4) to 2.18 (OS-6) mg kg−1, respectively. Relatively greater PAH contents were detected in the OS-3 (0.68 mg kg−1) and OS-5 (0.88 mg kg−1) samples, and no significant difference was found for PAH content in the other soil samples.
Microbial Diversity and Relationship Between Microbial Community and Soil Properties
The results of Chao 1 and Shannon indices generated from the MiSeq sequencing for the soil samples in this oilfield are listed in Table 3. The lower values of bacterial Chao 1 and Shannon were found in the OS-3 (771.27 and 4.38) and OS-5 (806.26 and 4.41) samples, and lowest fungal Chao 1 and Shannon values were also detected in the OS-3 sample (98.2 and 1.20). As for archaea, the lowest Chao 1 values were found in the OS-3, OS-5, and OS-6 samples, but the lowest Shannon value was detected in the OS-1 (3.24). It seemed that the soil samples with greater PAH contents (OS-3 and OS-5) also had lower values of bacterial diversity indices, but this trend was not obvious for the diversity indices of fungi and archaea. Linear correlation analysis between soil PAH contents and microbial diversity indices was performed (Fig. 2), which indicated that the bacterial (p < 0.05) and archaeal (p < 0.05) Chao 1 indices both had a significantly negative relationship with the soil PAH contents, but no obvious correlation was found between soil PAH contents and the fungal Chao 1 index (Fig. 2a). Moreover, a significantly negative relationship (p < 0.01) was only found between the bacterial Shannon index and soil PAH content (Fig. 2b). The result of Mantel correlations analysis between microbial community structure and soil basic properties is listed in Table 4. The contents of SOC (p < 0.01), NH4 + (p < 0.05), and PAHs (p < 0.05) were significantly correlated with soil bacterial community structure, and SOC (p < 0.05) and PAHs (p < 0.05) contents were also significantly correlated with soil fungal community structure. However, only soil SOC contents (p < 0.05) had a significant relationship with soil archaeal community structure.
Microbial Community Structures Based on Phylum
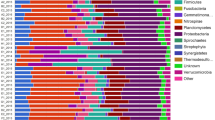
Cluster analysis and the relative abundance of bacterial, fungal, and archaeal community structures based on the phylum are shown in Fig. 3. The result indicated that the bacterial phylum compositions of soil samples with greater PAH contents (OS-3 and OS-5) had a relatively far distance from the other soil samples (Fig. 3a). The dominant bacterial phyla were Proteobacteria and Actinobacteria in OS-3 and OS-5 samples, respectively, but bacterial phylum profiles in the other soil samples were almost evenly occupied by Acidobacteria, Actinobacteria, Chloroflexi, and Proteobacteria (Fig. 3a). The fungal community structures based on the phylum showed a relatively greater ratio of Ascomycota to Basidiomycota in soil samples (OS-3 and OS-5) with greater PAH contents (Fig. 3b), although OS-3 and OS-5 samples could not be separated from the other soil samples by cluster analysis. Based on the archaeal phylum, OS-3, OS-5, and OS-7 samples could be clustered in one group, which had a relatively greater ratio of Euryarchaeota to Thaumarchaeota compared with the other soil samples (Fig. 3c). Although OS-3 and OS-5 soil samples could be clustered in one group, the former had a greater relative abundance of Thaumarchaeota than the latter.
Microbial Community Structures Based on Genus
Based on the genus, heat maps reflecting the bacterial, fungal, and archaeal community structures in these soil samples were created (Figs. 4, 5, 6). Similar to the result of cluster analysis based on phylum, the bacterial community structures in OS-3 and OS-5 samples were different from the other ones (Fig. 4). Compared with other soil samples, an obviously greater abundance of Mycobacterium was found in the OS-3 sample, and the abundances of Pseudomonas, Oxalobacteraceae, Micromonospora, Massilia, Lysobacter, Idiomarina, and Cytophaageaceae were greater in OS-5. The heat map of fungal community structures also indicated that the fungal community structures in the soil samples with greater PAH contents (OS-3 and OS-5) were different from the other soil samples (Fig. 5), and obviously greater abundances of Sordariales and Pleosporales were found in OS-3 and OS-5 samples, respectively. The heat map generated from the archaeal genus is shown in Fig. 6. A relatively far distance is shown between the two soil samples (OS-3 and OS-5) with greater PAH contents, which was quite different from the results of bacteria and fungi. The soil sample OS-3 had a unique archaeal community structure with relatively greater abundances of GOM-Arc-1-norank and Methanobacterium, whereas relatively greater abundances of some archaeal genus (Archaea-unclassified, Euryarchaeota-unclassified, and Methanosarcina) were detected in OS-5.
Discussion
The content of PAHs in the soil near the Jianghan Oilfield was in the range of 0.18–0.88 mg kg−1, which was relatively lower compared with the results of other studies. It has been reported that the 16 priority PAHs in the heavily contaminated soil of the four main oilfields in Northern China were in the range of 480–45,325 mg kg−1 (Wang et al. 2015a), which was in the range of 0.17–1.3 mg kg−1 in the soil of oil-producing zone in Dagang Oilfield (Liu et al. 2015). In this study, the bacterial and archaeal Chao 1 indices showed a significantly negative relationship with soil PAH content, and only the bacterial Shannon index had a negative correlation with soil PAH content. However, no obvious relationship was found between the fungal diversity indices and soil PAH content. This result might indicate that the community structure of bacteria is more sensitive to PAH pressure than those of fungi and archaea. Soil microbial diversity indices were usually used to evaluate the responses of soil microbes to environmental pressures, and the negative effects of PAHs or crude oil on them also has been reported (Abbasian et al. 2016; Elarbaoui et al. 2015; Perez-Leblic et al. 2012). It also has been reported that the bacterial Shannon index decreased with the increasing PAH content in soil and that no obvious relationship was found between fungal or archaeal Shannon index and soil PAH content (Abbasian et al. 2016; Perez-Leblic et al. 2012). Thus, it seems that the community structure of bacteria might be more sensitive to soil PAH contamination than those of fungi and archaea. Moreover, the result of Mantel correlation analysis also showed that bacterial (r 2 = 0.9001, p < 0.05) community structures had a more tightly relationship with soil PAH content than those of fungi (r 2 = 0.7357, p < 0.05), whereas no significant correlation was found between archaeal community structure and soil PAH content. The previous results might indicate that soil bacteria are more sensitive than fungi to PAH pressure and that soil PAH content might have no obvious influence on the soil archaea community structure. Some recent studies also found that soil archaea was not sensitive to some soil properties or PAH addition (Gao et al. 2016; Wu et al. 2016). For example, Wu et al. (2016) investigated the short-term responses of soil thaumarchaeotal community to PAH pollution, and they found that the most important factor affecting the soil thaumarchaeotal community was soil pH but not PAH content. At present, comparative studies of bacterial and archaeal community structure response to soil PAH stress have seldom been reported. However, it has been reported that ammonia-oxidizing bacteria were more susceptible than ammonia-oxidizing archaea to some harsh environmental conditions (low pH, high temperature, high salinity, low oxygen) or to organic inhibitors (allylthiourea and sulfadiazine), possibly because of the different chemical components and structures in their cell walls and membranes (Erguder et al. 2009; Hatzenpichler et al. 2008; Schauss et al. 2009). Such a difference might result in a lower cellular permeability of archaea compared with that of bacteria, which might help archaeal cells maintain a suitable intracellular environment and enable an undisturbed energetic production process under some unfavourable conditions (He et al. 2012; Valentine 2007). Therefore, the distinct responses of bacteria and archaea to the soil PAH stress might be due to the distinctions of cell structure and physiology between bacteria and archaea and the relatively low PAH content of soil. In addition, the result of the Mantel test (Table 4) also indicated that soil SOC was tightly correlated with microbial community structure, and the highest and lowest SOC contents were detected in OS-5 and OS-3 samples, respectively, which might result in the difference of microbial biota in those two samples with relatively greater PAH content.
The result of cluster analysis based on the microbial phylum (Fig. 3) indicated that only bacterial composition in soil samples (OS-3 and OS-5) with greater PAH contents could be completely separated from the other soil samples, which confirmed that bacteria were more sensitive to PAH pressure than fungi and archaea, and relatively greater abundances of Proteobacteria and Actinobacteria were detected in the OS-3 and OS-5 soil samples, respectively. These two bacterial phyla had been reported as dominant bacterial phyla in rhizosphere soil contaminated by PAHs, and Actinobacteria was also found to be dominant in soil amended with crude oil (Abbasian et al. 2016; Peng et al. 2015). Moreover, similar to the result based on the bacterial phyla, the heat map (Fig. 4) based on the bacterial genus also showed that the bacterial community structures in the OS-3 and OS-5 samples were different from those in the other soil samples, and different bacterial genus were enriched in OS-3 (Mycobacterium) and OS-5 (Pseudomonas, Oxalobacteraceae, Micromonospora, Lysobacter, Idiomarina, and Massilia) samples. Among those bacterial genus, well-known PAH-degrading microbes—Mycobacterium (Actinobacteria) and Pseudomonas (Proteobacteria)—have been widely used in bioremediation for soil PAH pollution (Khan et al. 2014; Sun et al. 2014), and also have been frequently detected in soil contaminated by PAHs (Chen et al. 2016; Han et al. 2014; Li et al. 2015; Sun et al. 2015a). For example, using the functional genes concerning PAH degradation, Han et al. (2014) found that in a PAH-polluted soil collected from a coke-factory area, the dominate genotypes of pdo1 (pyrene dioxygenases gene) and nah were Mycobacterium and Pseudomonas, respectively. The other genus belonging to Actinobacteria (Micromonospora), Gammaproteobacteria (Lysobacter and Idiomarina), and Betaproteobacteria (Oxalobacteraceae and Massilia) were also commonly identified in the PAH-contaminated soils (Hou et al. 2015; Luo et al. 2016; Song et al. 2015). As for fungi, the result of cluster analysis based on fungal phylum (Fig. 3b) showed a relatively greater ratio of Ascomycota to Basidiomycota in soil samples (OS-3 and OS-5) with greater PAH content. Furthermore, the heat map (Fig. 5) based on the fungal genus showed that the fungal compositions in OS-3 and OS-5 were also different from those in the other samples, and obviously greater abundances of Sordariales-unclassified and Pleosporales were detected in OS-3 and OS-5 soil samples, respectively, which all belong to Ascomycota. Although some representative PAH-degrading fungi genus—such as Trichoderma, Fusarium, and Pestalotiopsis—all belong to Ascomycota (Andreolli et al. 2016; Cebron et al. 2015; Kristanti and Hadibarata 2015; Llado et al. 2013), the ability of PAH degradation for Sordariales and Pleosporales had not been determined. Although the result of Mantel correlation analysis (Table 4) showed that archaeal community structure had no obvious relationship with soil PAH contents, the result of relative abundance of archaeal phyla (Fig. 3c) indicated that the soil samples (OS-3 and OS-5) with greater PAH contents had greater abundances of Euryarchaeota than most of the other samples (except OS-7). Recently, Euryarchaeota has also been reported as an important archaeal phylum with the potential ability for PAH degradation in the soils nearby some oilfields in Northern China (Sun et al. 2015b). Therefore, it seems that the shift of bacterial community structure, along with some representative bacterial genus (Mycobacterium and Pseudomonas), might be useful indicators to assess the risk of soil PAH contamination. Moreover, fungal (Ascomycota) and archaeal (Euryarchaeota) phyla might also be useful microbial parameters in the evaluation of soil PAH pollution, and further investigation into the relations among soil PAHs, fungal, and archaeal community structures should be performed to provide more reliable evidences in the future.
Conclusions
Taken together, this results of this study further confirmed the intensive selectivity of PAH contamination on the soil microbiota. In this study, relatively low PAH contents were detected in soil samples collected from the Jianghan Oilfield located in south-central China, whereas the community structures of soil microorganisms were obviously different in soil samples with different PAH contents. The correlation analysis between soil properties and microbial diversity indices indicated that the community structures of bacteria, fungi, and archaea showed varied responses to this mild soil PAH pollution, and it seems that the most sensitive and insensitive microorganisms to soil PAH contamination were bacteria and archaea, respectively. Furthermore, relatively greater abundances of some well-known bacterial PAH degraders—such as Mycobacterium and Pseudomonas, belonging to Proteobacteria or Actinobacteria combined with the greater ratios of fungal phylum of Ascomycota and archaeal phylum of Euryarchaeota—were detected in the soil samples with greater PAH contents, which might provide some useful microbial information for the bioremediation and risk assessment of soil PAH pollution in this typical oilfield.
References
Abbasian F, Lockington R, Megharaj M, Naidu R (2016) The biodiversity changes in the microbial population of soils contaminated with crude oil. Curr Microbiol 72:663–670
Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A et al (2013) Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J 7:1344–1353
Andreolli M, Lampis S, Brignoli P, Vallini G (2016) Trichoderma longibrachiatum Evx1 is a fungal biocatalyst suitable for the remediation of soils contaminated with diesel fuel and polycyclic aromatic hydrocarbons. Environ Sci Pollut Res 23:9134–9143
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917
Cao QM, Chen GZ, Liao SY (2009) Distribution correlations of polycyclic aromatic hydrocarbons with organic carbon and black carbon in surface sediments of three mangrove wetlands in the Shantou Wetland Demonstration Site, China. Huanjing Kexue Xuebao 29:861–868
Cebron A, Beguiristain T, Bongoua-Devisme J, Denonfoux J, Faure P, Lorgeoux C et al (2015) Impact of clay mineral, wood sawdust or root organic matter on the bacterial and fungal community structures in two aged PAH-contaminated soils. Environ Sci Pollut Res 22:13724–13738
Chen SC, Peng JJ, Duan GL (2016) Enrichment of functional microbes and genes during pyrene degradation in two different soils. J Soil Sed 16:417–426
Couling NR, Towell MG, Semple KT (2010) Biodegradation of PAHs in soil: influence of chemical structure, concentration and multiple amendment. Environ Pollut 158:3411–3420
Eggen T, Majcherczyk A (1998) Removal of polycyclic aromatic hydrocarbons (PAH) in contaminated soil by white rot fungus Pleurotus ostreatus. Int Biodeterior Biodegrad 41:111–117
Elarbaoui S, Richaed M, Boufahja F, Mahmoudi E, Thomas-Guyon H (2015) Effect of crude oil exposure and dispersant application on meiofauna: an intertidal mesocosm experiment. Environ Sci Process Impacts 17:997–1004
Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33:855–869
Gao PK, Tian HM, Wang YS, Li YS, Li Y, Xie JX et al (2016) Spatial isolation and environmental factors drive distinct bacterial and archaeal communities in different types of petroleum reservoirs in China. Sci Rep UK 6:20174
Ge Y, He JZ, Zhu YG, Zhang JB, Xu ZH, Zhang LM et al (2008) Differences in soil bacterial diversity: driven by contemporary disturbances or historical contingencies? ISME J 2:254–264
Guo GX, Deng H, Qiao M, Mu YJ, Zhu YG (2011) Effect of pyrene on denitrification activity and abundance and composition of denitrifying community in an agricultural soil. Environ Pollut 159:1886–1895
Han XM, Liu YR, Zheng YM, Zhang XX, He JZ (2014) Response of bacterial pdo1, nah, and C12O genes to aged soil PAH pollution in a coke factory area. Environ Sci Pollut Res 21:9754–9763
Haritash AK, Kaushik CP (2009) Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H et al (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105:2134–2139
He JZ, Hu HW, Zhang LM (2012) Current insights into the autotrophic thanumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154
Hou JY, Liu WX, Wang BB, Wang QL, Luo YM, Franks AE (2015) PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 138:592–598
Khan MI, Cheema SA, Tang XJ, Hashmi MZ, Shen CF, Park J et al (2013) A battery of bioassays for the evaluation of phenanthrene biotoxicity in soil. Arch Environ Contam Toxicol 65:47–55
Khan Z, Roman D, Kintz T, Alas MD, Yap R, Doty S (2014) Degradation, phytoprotection and phytoremediation of phenanthrene by endophyte Pseudomonas putida, PD1. Environ Sci Technol 48:12221–12228
Kristanti A, Hadibarata T (2015) Biodegradation and identification of transformation products of fluorene by Ascomycete fungi. Water Air Soil Pollut 226:406
Li XF, Hou LJ, Liu M, Zheng YL, Li Y, Lin XB (2015) Abundance and diversity of polycyclic aromatic hydrocarbon degradation bacteria in urban roadside soils in Shanghai. Appl Microbiol Biotechnol 99:3639–3649
Liao JQ, Wang J, Jiang DL, Wang MC, Huang Y (2015) Long-term oil contamination causes similar changes in microbial communities of two distinct soils. Appl Microbiol Biotechnol 99:10299–10310
Liu QL, Tang JC, Bai ZH, Hecker M, Giesy JP (2015) Distribution of petroleum degrading genes and factor analysis of petroleum contaminated soil from the Dagang Oilfield, China. Sci Rep UK 5:11068
Llado S, Gracia E, Solanas AM, Vinas M (2013) Fungal and bacterial microbial community assessment during bioremediation assays in an aged creosote-polluted soil. Soil Biol Biochem 67:114–123
Llado S, Covino S, Solanas AM, Petruccioli M, Dannibale A, Vinas M (2015) Pyrosequencing reveals the effect of mobilizing agents and lignocellulosic substrate amendment on microbial community composition in a real industrial PAH-polluted soil. J Hazard Mater 283:35–43
Lu RK (1999) Methods of agrochemical soil analysis. China Agricultural Science Press, Beijing
Luo J, Gu HP, Wang HZ, An QL, Xu JM (2016) Complete genome sequence of Massilia sp WG5, an efficient phenanthrene-degrading bacterium from soil. J Biotechnol 218:49–50
Ovreas L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–7373
Peng RH, Fu XY, Tian YS, Zhao W, Zhu B, Xu J et al (2014) Metabolic engineering of Arabidopsis for remediation of different polycyclic aromatic hydrocarbons using a hybrid bacterial dioxygenase complex. Metab Eng 26:100–110
Peng AP, Liu J, Ling WT, Chen ZY, Gao YZ (2015) Diversity and distribution of 16S rRNA and phenol monooxygenase genes in the rhizosphere and endophytic bacteria isolated from PAH-contaminated sites. Sci Rep UK 5:12173
Perez-Leblic MI, Turmero A, Hernandez M, Hernandez AJ, Pastor J, Ball AS et al (2012) Influence of xenobiotic contaminants on landfill soil microbial activity and diversity. J Environ Manag 95:S285–S290
Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG et al (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Ruggiero P, Terzano R, Spagnuolo M, Cavalca L, Colombo M, Andreoni V et al (2011) Hg bioavailability and impact on bacterial communities in a long-term polluted soil. J Environ Monit 13:145–156
Ruyters S, Mertens J, Tseyen I, Springael D, Smolders E (2010) Dynamics of the nitrous oxide reducing community during adaptation to Zn stress in soil. Soil Biol Biochem 42:1581–1587
Sawulski P, Clipson N, Doyle E (2014) Effects of polycyclic aromatic hydrocarbons on microbial community structure and PAH ring hydroxylating dioxygenase gene abundance in soil. Biodegradation 25:835–847
Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S (2009) Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11:446–456
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLOS ONE 6:e27310
Song MK, Luo CL, Jiang LF, Zhang DY, Wang YJ, Zhang G (2015) Identification of benzo[a]pyrene-metabolizing bacteria in forest soils by using DNA-based stable-isotope probing. Appl Environ Microbiol 81:7368–7376
Song MK, Jiang LF, Zhang DY, Luo CL, Wang Y, Yua ZQ et al (2016) Bacteria capable of degrading anthracene, phenanthrene, and fluoranthene as revealed by DNA based stable-isotope probing in a forest soil. J Hazard Mater 308:50–57
Su JQ, Ouyang WY, Hong YW, Liao D, Khan S, Li H (2016) Responses of endophytic and rhizospheric bacterial communities of salt marsh plant (Spartina alterniflora) to polycyclic aromatic hydrocarbons contamination. J Soil Sed 16:707–715
Sun K, Liu J, Gao YZ, Jin L, Gu YJ, Wang WQ (2014) Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp Rh6-gfp. Sci Rep UK 4:5462
Sun WM, Dong YR, Gao P, Fu MY, Ta KW, Li JW (2015a) Microbial communities inhabiting oil-contaminated soils from two major oilfields in Northern China: implications for active petroleum degrading capacity. J Microbiol 53:371–378
Sun WM, Li JW, Jiang L, Sun ZL, Fu MY, Peng XT (2015b) Profiling microbial community structures across six large oilfields in China and the potential role of dominant microorganisms in bioremediation. Appl Microbiol Biotechnol 99:8751–8764
Valentine DL (2007) Adaptations to energy stress dictate the ecology and evolution of the archaea. Nat Rev Microbiol 5:316–323
Van der Oost R, Beyer J, Vermeulen NPE (2003) Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ Toxicol Pharmacol 13:57–149
Wang J, Li FM, Li X, Wang XJ, Li XY, Su ZC et al (2013) Effects of electrokinetic operation mode on removal of polycyclic aromatic hydrocarbons (PAHs), and the indigenous fungal community in PAH-contaminated soil. J Environ Sci Health A 48:1677–1684
Wang J, Cao XF, Liao JQ, Huang Y, Tang XY (2015a) Carcinogenic potential of PAHs in oil-contaminated soils from the main oil fields across China. Environ Sci Pollut Res 22:10902–10909
Wang JT, Cao P, Hu HW, Li J, Han LL, Zhang LM et al (2015b) Altitudinal distribution patterns of soil bacterial and archaeal communities along Mt. Shegyla on the Tibetan Plateau. Microbiol Ecol 69:135–145
Wang LW, Feng L, Zhan Y, Zhu LZ (2016) Shifts in microbial community structure during in situ surfactant-enhanced bioremediation of polycyclic aromatic hydrocarbon-contaminated soil. Environ Sci Pollut Res 23:14451–14461
Wu YC, Zhu QH, Zeng J, Ding QM, Gong Y, Xing P et al (2016) Effects of pH and polycyclic aromatic hydrocarbon pollution on thaumarchaeotal community in agricultural soils. J Soil Sed 16:1960–1969
Xue W, Warshawsky D (2005) Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol 206:73–93
Zhang XZ, Xie JJ, Sun FL (2014) Effects of three polycyclic aromatic hydrocarbons on sediment bacterial community. Curr Microbiol 68:756–762
Zhou JZ, Wu LY, Deng Y, Zhi XY, Jiang YH, Tu QC et al (2011) Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5:1303–1313
Acknowledgements
This work was supported by the Natural Science Foundation of China (Grant No. 41371477) and the Fundamental Research Funds for the Central Universities (Grant No. XDJK2014B047).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, ZF., Wang, MX., Zuo, XH. et al. Comparative Investigation of Bacterial, Fungal, and Archaeal Community Structures in Soils in a Typical Oilfield in Jianghan, China. Arch Environ Contam Toxicol 72, 65–77 (2017). https://doi.org/10.1007/s00244-016-0333-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00244-016-0333-1