Abstract
Crude oil spills resulting from excavation, transportation and downstream processes can cause intensive damage to living organisms and result in changes in the microbial population of that environment. In this study, we used a pyrosequencing analysis to investigate changes in the microbial population of soils contaminated with crude oil. Crude oil contamination in soil resulted in the creation of a more homogenous population of microorganisms dominated by members of the Actinomycetales, Clostridiales and Bacillales (all belonging to Gram-positive bacteria) as well as Flavobacteriales, Pseudomonadales, Burkholderiales, Rhizobiales and Sphingomonadales (all belonging to Gram-negative bacteria). These changes in the biodiversity decreased the ratios of chemoheterotrophic bacteria at higher concentrations of crude oil contamination, with these being replaced by photoheterotrophic bacteria, mainly Rhodospirillales. Several of the dominant microbial orders in the crude oil contaminated soils are able to degrade crude oil hydrocarbons and therefore are potentially useful for remediation of crude oil in contaminated sites.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Crude oil is considered as one of the main sources of energy in the world, and growth in population and industries increases the demand for more crude oil extraction [20]. Leakage of oil into soil around the oil wells, pipelines and pumping stations is a normal phenomenon in the process of extraction of crude oil [20]. Furthermore, crude oil transportation and downstream processes are also high-risk points for crude oil contamination [11, 34]. Although the level of soil contamination with crude oil depends on the site location, this value can rise to around 10 % w/w [18]. Contamination can be spread into other natural habitats by superficial waters or floods especially in the rainy seasons [45]. Crude oils consist of different aliphatic and aromatic hydrocarbons, which are rich in carbon and hydrogen but deficient in other nutritional elements [32, 38, 41]. Since these compounds can be potentially poisonous for different eukaryotic and prokaryotic cells, soil contamination by crude oil can lead to immediate changes in the microbial composition of the given environment [1]. Furthermore, because oil spillages change the chemical composition of soil and thereby the nutrient availability to microorganisms, contamination can lead to prolonged effects on the microbial diversity and their abundances in the contaminated sites [31]. These changes in microbial population depend heavily on the composition of the total microbial community present in the site, the chemical composition of the crude oil and the physicochemical factors governing the particular environment [37].
Since microorganisms are the most effective factors in the natural degradation of crude oil and industrial bioremediation, understanding the changes in the microbial diversity resulting from contamination of soil with crude oil can be useful for the selection of the most effective hydrocarbon degrading microorganisms for crude oil remediation. The most abundant microorganisms in a soil sample contaminated by crude oil are potentially beneficial in facilitating the removal of crude oil contamination [5, 12, 35]. In this study, the changes in microbial diversity and their abundances were analysed using a high throughput pyrosequencing strategy in order to understand these dominant microorganisms in the soils contaminated with crude oil.
Materials and Methods
Preparation of the Column and Oil Spiking
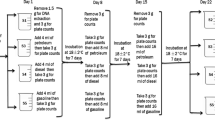
A Gauthier was used to collect the superficial soil samples (6.8 pH at room temperature, 2.8 % moisture and 612 μS conductivity) in order to save the integrity of soil samples. The soil samples were carefully transferred to suitable columns matched to the weight of soil samples (Fig. 1). Among the five columns, one column was used as control without any treatment, and four others were treated with different concentrations (0.5, 2.5, 5 and 10 %) of crude oil. Overall, two sets of these columns were prepared, allowing the reproducibility of the results to be determined; these columns were incubated in room temperature (25 °C) for 1 week.
DNA Extraction and Pyrosequencing Process
Following the incubation period, several samples were obtained from different parts of each column. The samples obtained from each column were mixed thoroughly to make a homogenous sample. The DNA from the soils was extracted using a power soil DNA kit (MO BIO), according to manufacturer’s instructions. First, after a gentle vortex of 0.25 g sediment samples in tubes containing beads and bead buffer, the contents were mixed with 60 μl solution C1. Following a vigorous vortexing for 10 min using a bead beating machine and centrifuging at 10,000×g for 30 S, the supernatants (400–500 μl) were processed according to the manufacturer’s instructions. The levels of DNAs obtained were quantified by a quantifluor dsDNA system (Promega) and were adjusted to a minimum concentration of 100 ng/µl.
The genomic DNAs were sequenced on the GS-FLX platform located at the Australian Genome Research Facility (AGRF). First, the DNAs were amplified using the primers directed for 16SrRNA of bacteria (referred to as 16S: 27F—519R; 5′-AGAGTTTGATCMTGGCTCAG-3′and 5′-GWATTACCGCGGCKGCTG-3′) and archaea (referred to as 16S: 341F-806R; 5′-CCTAYGGGRBGCASCAG-3′and 5′-GGACTACNNGGGTATCTAAT-3′) as well as 18SrRNA of fungi (referred to as ITS: 1F-2R; 5′-CTTGGTCATTTAGAGGAAGTAA-3′and 5′-TCCTCCGCTTATTGATATGC-3′) and algae (referred to as Euk3191F-EukBR; 5′-CTGGTTGATCCTGCCAG-3′ and 5′-ACCAGACTTGCCCTCC-3′). After a DNA quantification of the PCR products by fluorometry and qPCR, the DNA fragments were sequenced using the GS-FLX platform by the Australian Genome Research Facility (AGRF).
Data Analysis
The raw data files obtained from pyrosequencing were uploaded into the MG-RAST (Metagenome Rapid Annotation using Subsystem Technology) [2, 17], and the microbial diversity and their abundances were analysed using the Ribosomal Data Project II (RDP II) annotation source. To minimise the mistakes through analysis on the microbial diversity using 16SrRNA, the data were filtered by minimum percentage identity cutoff (97 %), E-value cutoff (1e-5) and minimum alignment length cutoff (50 bp) [3].
The microbial diversity and their abundances were analysed further using excel software to analyse the Shannon’s index and to investigate the ecological changes as a result of crude oil treatment. The Shannon index (a minus sum of the proportion (n/N) of the number of one particular species in a community divided by the total number of individuals in that community multiplied by the natural logarithm of this proportion) reflects the number of species in a dataset and their distribution in a community [22]. To increase the accuracy of our analysis, the metabolic (chemoheterotrophic, photoautotrophic, photoheterotrophic and chemolithotrophic) activity of all microbial strains identified were extracted from authorised references [6, 19, 39, 44], and the strains with a same metabolic activity and same phylogenetic order were grouped together.
Nucleotide Sequence Accession Numbers
These metagenomic data are publicly available in the MG-RAST system as F44 (control), F45 (treated with 2.5 % crude oil), F47 (treated with 5 % crude oil), F48 (treated with 0.5 % crude oil) and F49 (treated with 10 % crude oil) under project identifiers 4576644.3., 4576645.3., 4576646.3., 4576647.3. and 4576648.3., respectively.
Results
The numbers of reads after quality control, mean length and the numbers of reads fitted in an operational taxonomic unit (OTU) for all samples are listed in Table 1. A full list of the microbial orders found in these columns is shown in Fig. 2. According to the saturated plateaus in the rarefaction curves obtained from the analysis of the control soil sample and the samples treated with different concentrations of crude oil, the abundances of microbial population in these sampling are representative of those present in the columns (data are not shown). Moreover, according to the principal coordinate analysis (PCoA) (data are not shown), these treatments considerably altered the biodiversity present in the treated columns in comparison to the control sample. In total, the abundances of microorganisms in the sediments treated with 0.5 and 2.5 % crude oil were increased in comparison to the control. Although this value reached its peak in the soil sample treated by 0.5 % crude oil, there were gradual decreases in the total abundances of microorganisms in higher concentrations (2.5 and 5 %) to a level slightly more than the control. However, there was a small rise again in the abundance of microorganisms when the soil sample was treated with 10 % crude oil. In addition to total abundances of microorganisms, the treatments decreased the numbers of microbial orders in these samples from 59 in the control (0 %) to 46 in the soil contaminated with 10 % crude oil. Furthermore, there were changes in the ratios of microbial orders in the samples when compared with the total abundances of microorganisms in the corresponding sample. These changes in the abundances of microbial orders were significant after analysis with Shannon index (Table 2).
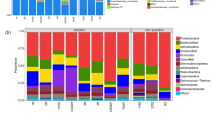
Among these microorganisms, Actinomycetales (20 %) were the dominant microorganism in the control soil, followed by Cytophagales (7.4 %), Solibacterales (3.6 %), Glomerellales (3.5 %), Planctomycetales (3.4 %), Verrucomicrobiales (3.2 %), Clostridiales (3 %), Bacillales (2.5 %) and many other orders with less than 2.5 % abundance (Fig. 3). However, the ratios of microorganism changed in the samples treated with different concentrations of crude oil. In the soil sample treated with 0.5 % crude oil (Fig. 3), there were Actinomycetales (25 %), Flavobacteriales (6.1 %), Glomerellales (2.9 %), Planctomycetales (2.8 %), Verrucomicrobiales (2.7 %), Rhizobiales (2.6 %), Rhodospirillales (2.5 %), Cytophagales (2.5 %), Bacillales (2.1 %) and many other orders with less than 2 % of the population (Fig. 3).
The ratio of Actinomycetales (37.21 %) reached the highest amount in the soil spiked with 2.5 % crude oil, followed by Rhizobiales (3.4 %), Rhodospirillales (3 %), Flavobacteriales (2.9 %), Planctomycetales (2.9 %), Cytophagales (2.5 %), Clostridiales (2.3), Bacillales (2 %) and many other orders that were less than 2 % of the population (Fig. 3). These values changed in the soils contaminated with higher levels of crude oil, as well. Although the ratio of Actinomycetales decreased in the soil sample treated with 5 % crude oil, still it was the dominant microorganism in this soil, followed by Sphingomonadales (5.1 %), Rhizobiales (4.6 %), Burkholderiales (3.4 %), Rhodospirillales (3.0 %), Flavobacteriales (2.9 %), Clostridiales (2.3 %), Rhodocyclales (2.2 %), Bacillales (2.1 %) and many other orders less than 2 % abundance. Furthermore, these changes in the diversity and ratios of microorganisms in the soil sample treated by 10 % crude oil were much severe than other samples. In this sample, Sphingomonadales (16 %) was nominated as the dominant microorganism and Actinomycetales (10.2 %) was placed in the second step, followed by Rhodospirillales (6.3 %), Rhizobiales (2.8 %), Burkholderia (2.4 %), Planctomycetales (2 %) and many other orders with less than 2 % abundances.
Based on the literature and microbial classification handbooks [6, 19, 39, 44], all microbial strains identified in this study were classified into four physiological categories (chemoheterotrophs, photoautotrophs, photoheterotrophs and chemolithotrophs) to analyse the ecological relationships between microorganisms in the soil samples. Thereafter, these strains were re-classified into their orders to facilitate the statistical analysis. Since the members of microbial orders show sometimes very diverse physiology, this procedure of microbial classification from strains to orders reduces the errors in calculations. Based on this study, the majority of microorganisms in the control and all soils treated with different concentrations of crude oil belonged to chemoheterotrophic bacteria (Fig. 4). However, the soil treated with 10 % crude oil showed a significant decrease in the ratio of chemoheterotrophic bacteria (83 %), replaced by photoheterotrophic bacteria (17 %).
Discussion
In this study, a pyrosequencing study was employed to investigate changes in the microbial diversity and the ratios of their populations in soils contaminated with crude oil. Based on the results, crude oil contamination showed severe effects on the total microbial population and their biodiversity in soils, and these effects depended heavily on the concentration of the crude oil. Although the total abundance of microorganisms in the sample treated with 0.5 % crude oil increased in comparison to the control sample, further increases in the levels of crude oil contamination led to decreases in the total abundances of microorganism. Furthermore, these treatments affect negatively the numbers of microbial orders identified in these soil subjects. Therefore, crude oil contamination in soil created a more homogenous environment in terms of microbial diversity, and many of the microorganisms susceptible to constituents of crude oil were lost from these environment. Several microbial orders, such as Fibrobacterales, Thermoanaerobacterales, Nautiliales, Enterobacteriales, Thermotogales, Ustilaginales and Plocamiales, showed a high susceptibility to this treatment and vanished from the soils spiked with low concentrations of crude oil (0.5 and 2.5 %). This list was extended to Aquificales, Thermomicrobiales, Chroococcales (a Cyanobacteria), Deinococcales, Candidatus Brocadiales, Acholeplasmatales and Entomoplasmatales (in the soil samples treated with 5 % crude oil) and to Bacteroidales, Oscillatoriales (an algae), Selenomonadales, Gemmatimonadales, Nitrospirales and Glomerellales (in the soil samples treated with 10 % crude oil).
Furthermore, except for some orders, the ratios of the majority of microorganisms decreased in the soil spiked with crude oil in comparison to the control samples. These changes in microbial diversity led to transformations in the abundant microorganisms in these soils. Members of Actinomycetales, mostly Mycobacterium sp., Nocardia sp. and Pseudonocardia sp., were the dominant microorganisms in the control soil and its ratio even increased in the soils treated with 0.5 and 2.5 % crude oil. Although the ratio of Actinomycetales decreased in higher levels of treatment, the members of this order were still one of the most dominant microorganisms in the soils treated with crude oil. A high level of resistance to crude oil is very common among members of this order, and many genera in this order are commonly used for the degradation of different types of hydrocarbons [23, 25, 46]. Members of Clostridiales (Clostridium sp., Symbiobacterium sp. and Oxobacter sp.) and Bacillales (Bacillus sp. and Brevibacillus sp.) were two other Gram-positive orders with the ability to resist high levels of crude oil contamination, while their ratios decreased significantly in the soils treated with the 10 % treatment. While members of Clostridiales are involved in the degradation of alkanes in anaerobic conditions [16, 30, 40], genera of Bacillales degrade these compounds in aerobic conditions [8, 15, 28].
Several groups of Gram-negative bacteria, including Flavobacteriales, Pseudomonadales, Burkholderiales, Rhizobiales and Sphingomonadales, were identified as dominant microorganisms in these soil samples as well. The highest ratio of Flavobacteriales (mainly Flavobacterium sp.), Pseudomonadales (mainly Pseudomonas sp.), Burkholderiales (mainly Variovorax sp. and Brachymonas petroleovorans) and Rhizobiales (mainly Bradyrhizobium sp.) were observed in the soils treated by 0.5 % crude oil, but higher levels of treatment decreased their values. Among these groups of aerobic bacteria, members of Pseudomonadales [29, 36], Burkholderiales [10] and Flavobacteriales [4, 14, 21, 42] have been commonly used for the bioremediation of crude oil contaminated subjects.
Inversely, increases in the levels of crude oil treatment raised the ratio of Sphingomonadales (mainly Sphingomonas sp.) in the soils, and members of this order became the most dominant microorganisms in the soil treated with 10 % crude oil. This increase in the ratio of Sphingomonas sp. indicates its ability to resist high crude oil pollution and its ability to degrade different types of hydrocarbons as reported in previous studies [7, 9, 24, 26, 43]. Rhodospirillales showed a similar distribution, and members of this order became the second most dominant microorganisms in the soils treated with 10 % crude oil. Rhodospirillales show a photoheterotrophic metabolism in which electrons and hydrogen required for CO2 fixation are originate from some organic molecules [13]. Two orders of Rhodospirillales (Azospirillum sp. and Phaeospirillum sp.) were found in these samples, and their involvement in the degradation of crude oil has been shown in previous studies [27, 33].
These changes in the diversity and ratios of microorganisms affect the total microbial physiology of the soil samples. Although chemoheterotrophic bacteria were dominant in both control soil and soils spiked with crude oil, this value decreased with higher concentrations of crude oil contamination, where they were replaced by photoheterotrophic bacteria, mainly Rhodospirillales. Furthermore, these treatments did not show significant effects on the ratios of both oxygenic and non-oxygenic phototrophic microorganisms. Overall, this study indicated that several types of soil microorganisms are highly susceptible to crude oil contamination. However, there were several other microbial orders, especially Sphingomonadales, which are known as crude oil hydrocarbon degrading organisms that are able to resist these conditions. In addition to adaptations for resistance to the toxic effects of different classes of hydrocarbons and their resistance to heavy metals, which are naturally found in crude oil, these microorganisms are able to produce the enzymes required for the degradation of aliphatic and aromatic hydrocarbons. Such microorganisms can be used as potential candidates for remediation of crude oil in contaminated sites. Furthermore, investigation on the genetic capacity of these microorganisms can provide significant information regarding the genes involved in hydrocarbon bioremediation as well as the production of bio-surfactants.
References
Abbasian F, Lockington R, Mallavarapu M, Naidu R (2015) A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl Biochem Biotech 176:670–699
Abbasian F, Lockington R, Mallavarapu M, Naidu R (2015) The integration of sequencing and bioinformatics in metagenomics. Rev Env Sci Bio/Technol 14:357–383
Abbasian F, Lockington R, Mallavarapu M, Naidu R (2015) A pyrosequencing-based analysis of microbial diversity governed by ecological conditions in the Winogradsky column. World J Microbiol Biotechnol 31:1115–1126
Balba M, Al-Awadhi N, Al-Daher R (1998) Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J Microbiol Methods 32:155–164
Balba M, Al-Daher R, Al-Awadhi N, Chino H, Tsuji H (1998) Bioremediation of oil-contaminated desert soil: the Kuwaiti experience. Environ Int 24:163–173
Boone DR, Brenner DJ, Castenholz RW, De Vos P, Garrity G M, Krieg NR, Staley JT (2011) Bergey’s Manual of Systematic Bacteriology: the Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. ed. Springer
Colombo M, Cavalca L, Bernasconi S, Andreoni V (2011) Bioremediation of polyaromatic hydrocarbon contaminated soils by native microflora and bioaugmentation with 〈i〉 Sphingobium chlorophenolicum 〈/i〉 strain C3R: a feasibility study in solid-and slurry-phase microcosms. Int Biodeterior Biodegradation 65:191–197
Cubitto MA, Moran AC, Commendatore M, Chiarello MN, Baldini MD, Sineriz F (2004) Effects of Bacillus subtilis O9 biosurfactant on the bioremediation of crude oil-polluted soils. Biodegradation 15:281–287
Dai M, Copley SD (2004) Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl Environ Microbiol 70:2391–2397
Dobslaw D, Engesser KH (2014) Degradation of toluene by ortho cleavage enzymes in Burkholderia fungorum FLU100. Microb Biotechnol 8:143–154
Dorn PB, Vipond TE, Salanitro JP, Wisniewski HL (1998) Assessment of the acute toxicity of crude oils in soils using earthworms, microtox and plants. Chemosphere 37:845–860
Gallego JL, Loredo J, Llamas JF, Vázquez F, Sánchez J (2001) Bioremediation of diesel-contaminated soils: evaluation of potential in situ techniques by study of bacterial degradation. Biodegradation 12:325–335
Garrity G, Bell J, Lilburn T (2005) Order I. Rhodospirillales Pfennig and Trüper 1971, 17 AL. Bergey’s Manual of Systematic Bacteriology, 1–95
Gentili AR, Cubitto MA, Ferrero M, Rodriguéz MS (2006) Bioremediation of crude oil polluted seawater by a hydrocarbon-degrading bacterial strain immobilized on chitin and chitosan flakes. Int Biodeterior Biodegradation 57:222–228
Ghazali FM, Rahman RNZA, Salleh AB, Basri M (2004) Biodegradation of hydrocarbons in soil by microbial consortium. Int Biodeterior Biodegradation 54:61–67
Gieg LM, Duncan KE, Suflita JM (2008) Bioenergy production via microbial conversion of residual oil to natural gas. Appl Environ Microbiol 74:3022–3029
Glass EM, Meyer F (2011) The Metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. Handbook of molecular microbial ecology I: metagenomics and complementary approaches, pp. 325–331
Gogoi B, Dutta N, Goswami P, Krishna Mohan T (2003) A case study of bioremediation of petroleum-hydrocarbon contaminated soil at a crude oil spill site. Adv Environ Res 7:767–782
Goodfellow M, Peter K, Busse H-J, Trujillo ME, Ludwig W, Suzuki K-I, Parte A (2012) Bergey’s manual of systematic bacteriology: volume 5: the actinobacteria. ed. Springer, New York
Hussain T, Gondal M (2008) Monitoring and assessment of toxic metals in Gulf War oil spill contaminated soil using laser-induced breakdown spectroscopy. Environ Monit Assess 136:391–399
Kaplan CW, Kitts CL (2004) Bacterial succession in a petroleum land treatment unit. Appl Environ Microbiol 70:1777–1786
Keylock C (2005) Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos 109:203–207
Kim S-J, Kweon O, Sutherland JB, Kim H-L, Jones RC, Burback BL, Graves SW, Psurny E, Cerniglia CE (2015) Dynamic response of Mycobacterium vanbaalenii PYR-1 to BP Deepwater Horizon crude oil. Appl Environ Microbiol 81:4263–4276
Koukkou A-I, Vandera E (2011) Hydrocarbon-degrading soil bacteria: current research. microbial bioremediation of non-metals: current research, vol 93. Caister Academic Press, Norfolk
Kweon O, Kim S-J, Holland RD, Chen H, Kim D-W, Gao Y, Yu L-R, Baek S, Baek D-H, Ahn H (2011) Polycyclic aromatic hydrocarbon-metabolic network in Mycobacterium vanbaalenii PYR-1. J Biotech, JB. 00215-00211
Liang Q, Lloyd-Jones G (2010) Sphingobium scionense sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from contaminated sawmill soil. Int J Syst Evol Microbiol 60:413–416
Liu J-F, Sun X-B, Yang G-C, Mbadinga SM, Gu J-D, Mu B-Z (2015) Analysis of microbial communities in the oil reservoir subjected to CO2-flooding by using functional genes as molecular biomarkers for microbial CO2 sequestration. Front Microbiol 6:236
Margesin R, Schinner F (2001) Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl Microbiol Biotech 56:650–663
Matsui T, Yamamoto T, Shinzato N, Mitsuta T, Nakano K, Namihira T (2014) Degradation of oil tank sludge using long-chain alkane-degrading bacteria. Ann Microbiol 64:391–395
Mbadinga SM, Wang L-Y, Zhou L, Liu J-F, Gu J-D, Mu B-Z (2011) Microbial communities involved in anaerobic degradation of alkanes. Int Biodeterior Biodegradation 65:1–13
Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R (2011) Bioremediation approaches for organic pollutants: a critical perspective. Environ Int 37:1362–1375
Morgan P, Watkinson RJ (1994) Biodegradation of components of petroleum. Biochemistry of Microbial Degradation. Kluwer Academic Publishers, Dordrecht, pp 1–13
Muratova AY, Turkovskaya O, Antonyuk L, Makarov O, Pozdnyakova L, Ignatov V (2005) Oil-oxidizing potential of associative rhizobacteria of the genus Azospirillum. Microbiology 74:210–215
Nicolotti G, Egli S (1998) Soil contamination by crude oil: impact on the mycorrhizosphere and on the revegetation potential of forest trees. Environ Pollut 99:37–43
Odokuma L, Dickson A (2004) Bioremediation of a crude oil polluted tropical rain forest soil. Global J Environ Sci 2:29–40
Pacwa-Płociniczak M, Płaza GA, Poliwoda A, Piotrowska-Seget Z (2014) Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ Sci Pollut Res 21:9385–9395
Rahman P, Rahman T, Lakshmanaperumalsamy P, Banat IM (2002) Occurrence of crude oil degrading bacteria in gasoline and diesel station soils. J Basic Microbiol 42:284–291
Sathishkumar MB, Raj Arthur, Baik S-H, Yun S-E (2008) Biodegradation of crude oil by individual bacterial strains and a mixed bacterial consortium isolated from hydrocarbon contaminated areas. Clean 36:92–96
Schink B, Janssen P, Brune A (2011) Bergey's manual of systematic bacteriology. Springer, New York.
Sherry A, Gray N, Ditchfield A, Aitken C, Jones D, Röling W, Hallmann C, Larter S, Bowler B, Head I (2013) Anaerobic biodegradation of crude oil under sulphate-reducing conditions leads to only modest enrichment of recognized sulphate-reducing taxa. Int Biodeterior Biodegradation 81:105–113
Shi Q, Hou D, Chung KH, Xu C, Zhao S, Zhang Y (2010) Characterization of heteroatom compounds in a crude oil and its saturates, aromatics, resins, and asphaltenes (SARA) and non-basic nitrogen fractions analyzed by negative-ion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Energy Fuels 24:2545–2553
Vinas M, Sabaté J, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018
Wang S, Cui Z, Zheng L, Gao W, Li Q (2012) Characterization of PAHs-degrading bacterium Porphyrobacter sp. D22F and its ecological niche in oil-degrading consortium D22-1. Chin J Appl Environ Biol 18:122–127
Whitman WB, Parte AC (2009) Systematic bacteriology. ed. Springer
Zakaria MP, Takada H, Tsutsumi S, Ohno K, Yamada J, Kouno E, Kumata H (2002) Distribution of polycyclic aromatic hydrocarbons (PAHs) in rivers and estuaries in Malaysia: a widespread input of petrogenic PAHs. Environ Sci Technol 36:1907–1918
Zeinali M, Vossoughi M, Ardestani SK (2008) Naphthalene metabolism in Nocardia otitidiscaviarum strain TSH1, a moderately thermophilic microorganism. Chemosphere 72:905–909
Acknowledgments
The authors would like to appreciate Australian Government for IPRS scholarship and CRC-CARE (Cooperative Research Centre for Contamination Assessment and Remediation of the Environment) for additional funding towards this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Abbasian, F., Lockington, R., Megharaj, M. et al. The Biodiversity Changes in the Microbial Population of Soils Contaminated with Crude Oil. Curr Microbiol 72, 663–670 (2016). https://doi.org/10.1007/s00284-016-1001-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1001-4