Abstract
Purpose
Thaumarchaeota is an ecologically relevant archaeal phylum which may significantly contribute to global nitrogen cycling. Thaumarchaeotal abundance, composition, and activity can be changed by soil pH and pollutants such as toxic metals. This study aims to examine the responses of thaumarchaeotal community to soil pH variation and polycyclic aromatic hydrocarbon (PAH) pollution which may co-occur in agricultural soils.
Materials and methods
Field soil samples were collected from agricultural land impacted by both acidification and PAH contamination. Thaumarchaeotal abundance and composition were assessed using molecular approaches targeting 16S rRNA or amoA genes and were linked to environmental factors by correlation and canonical correspondence analysis (CCA). To evaluate the short-term responses of Thaumarchaeota to PAHs, additional soil microcosms amended with either three selected PAHs were established. Changes in thaumarchaeotal communities during the incubation were monitored.
Results and discussion
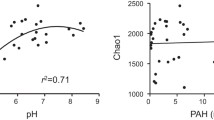
A significant correlation between thaumarchaeotal gene abundance and soil pH was observed within field samples, with the I.1a-associated group enriched when pH <5.0. CCA suggests that the community variation was primarily related to soil pH. In contrast, the effects of PAHs were minimal. In soil microcosms, high concentrations of PAHs persisted after the 4-week incubation. Independent of the PAHs added, thaumarchaeotal amoA abundance slightly increased and the compositions were stable at the end of the incubation. This might be associated with the pollutants bioavailability and potential microbe-PAH interactions in the soil.
Conclusions
Soil pH variation strongly shapes the agricultural soil thaumarchaeotal community, whereas PAH effects appear to be marginal even in the presence of high concentrations of pollutants. The complicated interaction between soil matrix, pollutants, and Thaumarchaeota requires further study.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Thaumarchaeota, previously referred to as the mesophilic Crenarchaeota (Brochier-Armanet et al. 2008), are widespread and are often the most abundant archaea in terrestrial and marine environments. Some thaumarchaeotal clades, including the I.1a, I.1a-associated, and I.1b groups, possess genes encoding putative ammonia monooxygenase and thus may play a crucial role in global nitrogen transformation (Prosser and Nicol 2008). In addition to these potential ammonia-oxidizing Thaumarchaeota (AOA), there are other thaumarchaeotal groups including the groups I.1c, ALOHA, and pSL12 that can be dominant in specific habitats but appear to be ammonia oxidation independent (i.e., non-AOA) (Weber et al. 2015).
Because of their potential contribution to ammonia oxidation, the first and rate-limiting step of nitrification, ammonia-oxidizing Thaumarchaeota in agricultural soils have been extensively studied since their discovery. It is presently known that AOA are influenced by multiple edaphic factors, among which the most significant are substrate supply, organic carbon, and pH (Prosser and Nicol 2012; Shen et al. 2014). The pH was of great concern because of the wide variation of soil acidity across the world (He et al. 2012). Additionally, evidence indicates that there is a niche differentiation of Thaumarchaeota along a pH gradient (Gubry-Rangin et al. 2011). For example, Nitrosotalea devanaterra, the only enrichment of the I.1a-associated group, is capable of optimal growth at pH values between 4.0 and 5.5 (Lehtovirta-Morley et al. 2011), while pure cultures of the groups I.1a and I.1b prefer neutral conditions (Könneke et al. 2005, Tourna et al. 2011). Indeed, the I.1a-associated group appears to be dominant and dictates ammonia oxidation in many acidic soils (Nicol et al. 2008; Yao et al. 2011; Zhang et al. 2012), although growth of the I.1b AOA at low pH is not unusual (Wu and Conrad 2014). Other acidophilic thaumarchaeotal clades include I.1c and I.3, which mainly thrive under strongly acidic conditions (Oton et al. 2016; Weber et al. 2015). In contrast, the I.1b Thaumarchaeota are dominant in neutral soils, as revealed by large-scale meta-analysis (Gubry-Rangin et al. 2011; Oton et al. 2016).
Thaumarchaeota can be sensitive to inorganic and organic xenobiotics. It was demonstrated in soil microcosms that the toxic metals Cu, Zn, Hg, and As considerably reduced the abundance, composition, and activity of AOA (Mertens et al. 2009; Liu et al. 2010; Mertens et al. 2010; Subrahmanyam et al. 2014a). As for organic pollutants, Nitrosopumilus maritimus, a cultivated representative of the group I.1a, was inhibited by the addition of low concentration of organic pollutants to the medium (Urakawa et al. 2012; Sipos and Urakawa 2016). Industrial effluents containing mixed organic compounds also change the soil AOA community (Subrahmanyam et al. 2014b). However, the interactions between organic compounds and Thaumarchaeota are complex. The thaumarchaeotal ammonia monooxygenase belongs to the copper-containing membrane-bound monooxygenase (CuMMO) family, which has a wide substrate range that includes aromatic hydrocarbons (Pester et al. 2011). Furthermore, the growth of Thaumarchaeota could be greatly enhanced by the presence of small organic molecules such as pyruvate (Tourna et al. 2011), which is among the final metabolites of bacterial polycyclic aromatic hydrocarbon (PAH) degradation (Juhasz and Naidu 2000). In view of the vast diversity of organic pollutants, however, the thaumarchaeotal responses to these compounds are poorly understood. For instance, little is known about the effects of PAHs on Thaumarchaeota, although PAHs might be the most widely distributed persistent organic pollutants in the environment (Harvey 1991).
Soil acidification and pollution pose major risks to crop production and can potentially change the characteristics of the soil microbiota. It is well known that the intensive use of mineral nitrogen fertilization may cause considerable pH decline of cropland soil (Guo et al. 2010), which in turn changes the habitat of Thaumarchaeota and the reaction balance of ammonia oxidation (He et al. 2012), the energy source of AOA. On the other hand, PAH pollution of agricultural soils is common in some rapidly industrialized areas due to their continuous emission (Xu et al. 2006; Ping et al. 2007). As a result, the co-occurrence of soil acidification and PAH pollution might not be unusual. In this study, we examined the thaumarchaeotal communities in agricultural soils impacted by both fertilization-induced acidification and PAH pollution, as well as in soil microcosms exposed to a high concentration of selected PAHs. The aims of the study were therefore (1) to reveal the effects of soil acidification and PAH pollution on the in situ thaumarchaeotal community and (2) to assess the effects of PAHs on soil Thaumarchaeota under controlled conditions.
2 Material and methods
2.1 Site description and field sampling
The field under study is located adjacent to Hua-Xi (HX) village in the outskirts of Nanjing, Jiangsu Province, China (31°51′57″N, 118°35′58″E). The soil was classified as an Argosols according to the Chinese Soil Taxonomy. Two sets of experiments, i.e., an in situ survey and an amendment experiment, were performed to reveal the responses of Thaumarchaeota to long- and short-term PAH pollution. For the in situ survey, 20 composite surface soils (0–10 cm, HX-1 to HX-20) were randomly collected on November 13, 2014, from the field of vegetable cultivation. Each sample was obtained by mixing five 50-g subsamples from a 10 × 10 m grid. The soils were immediately transported to the laboratory after sampling, and a 50-g subsample of each soil was stored at −20 °C for molecular analysis. The rest of the soil was air-dried, homogenized, and stored at 4 °C for physicochemical analysis. For the PAH amendment experiment, a bulk soil sample was collected from the field, air-dried, sieved through a 2-mm mesh, and stored at 4 °C before use.
2.2 PAH amendment experiment
The soil used in the PAH amendment experiment had a neutral pH of 7.4. Soil microcosms were established with amendment of either anthracene (three-ring), benz(a)anthracene (four-ring), or benzo(a)pyrene (five-ring) (Sigma-Aldrich). Specifically, PAH solutions in acetone were added to air-dried soils which were then placed in a fume hood overnight to allow the volatilization of the solvent. The PAH-spiked soils were well mixed, and a 5-g fraction was added to serum bottles containing 15-g clean soil, giving the final PAH concentrations of approximately 150 (for anthracene) and 50 mg kg−1 (for benz(a)anthracene and benzo(a)pyrene). After adjusting to 60 % water-holding capacity (soil moisture approximately 19 %), bottles were capped with butyl stoppers and statically incubated in the dark at 25 °C for up to 4 weeks. During the incubation, the microcosms were kept aerobic by flushing the headspaces with air regularly. All treatments were performed in triplicate, and destructive sampling took place at the beginning of the experiment and the first, second, and fourth weeks for the molecular and PAH analyses described below.
2.3 Analysis of soil chemical characteristics
The soil pH was determined with a soil:water solution (1:2.5). Total carbon (TC) and total nitrogen (TN) were measured with a Vario Max element analyzer (Elementar), and the C/N ratio was calculated by directly dividing TC by TN. Total phosphorus (TP) and total potassium (TK) were analyzed by standard methods (Lu 2000). Inorganic N species (NH4 +, NO3 −, and NO2 -) were extracted with 2 mol l−1 KCl and analyzed on a continuous flow analyzer (SA1000, Skalar Analytical).
2.4 PAH analysis
Soxhlet extraction was performed with 10.0 g (field sample) or 1.0 g (microcosm sample, see “2.2”) air-dried and homogenized soils for 24 h. Prior to the ultra-fast liquid chromatography (UFLC) analysis, dichloromethane extracts were concentrated on a rotary evaporator and purified by column chromatography with activated silica gel as described previously (Wu et al. 2008). Fifteen out of the 16 EPA priority PAHs were determined with a Shimadzu UFLC-20 system (Shimadzu, Kyoto, Japan) equipped with a reversed phase C18 column (Shim-pack XR-ODSII, Kyoto, Japan) (Zeng et al. 2013). All PAH concentrations are presented based on the soil dry weight.
2.5 Soil DNA extraction and quantitative PCR
DNA was extracted from 0.5 g of each fresh soil sample with the FastDNA SPIN Kit for soil (MP Biomedicals) following the manufacturer’s instruction. The quality and quantity of DNA were checked by electrophoresis and by a spectrophotometer (NanoDrop 1000). A tenfold dilution of the DNA showed no inhibition of PCR and was thus used in the downstream molecular analysis.
Gene copies were determined by quantitative PCR (qPCR) based on SYBR Green chemistry using a CFX96 Optical Real-Time Detection System (Bio-Rad Laboratories) as previously described (Wu and Conrad 2014). The primer sets 771F/957R (Ochsenreiter et al. 2003) and CamoA-19f/CamoA-616r (Pester et al. 2012) were used to quantify the thaumarchaeotal 16S ribosomal RNA (rRNA) and ammonia monooxygenase (amoA) genes, respectively. The qPCR standards were generated using linearized plasmid DNA from single clones containing the genes. A dilution series of the standard template across seven orders of magnitude for each gene was used in each assay. The blank was run with water as the template instead of DNA extract. Tenfold diluted extracts containing 3.7–10.8 ng DNA were added to the reaction mixture in each assay. The qPCR amplification efficiency for the thaumarchaeotal 16S rRNA gene was 85.4 % with an r 2 value of 0.999; for the thaumarchaeotal amoA gene, the efficiency was 79.0 % and the r 2 was 0.993. Abundance of ammonia-oxidizing bacteria (AOB) was evaluated by qPCR amplification of the bacterial amoA gene with the primer set amoA-1F/amoA-2R (Rotthauwe et al. 1997); this assay had an efficiency of 82.2 % and an r 2 of 0.998. All reactions were performed in triplicate for each assay.
2.6 Terminal restriction fragment length polymorphism
Terminal restriction fragment length polymorphism (T-RFLP) of the thaumarchaeotal 16S rRNA gene was performed following the methods described elsewhere with minor modification (Wu and Conrad 2014). Briefly, an 833-bp fragment was amplified with the primers A109F (Großkopf et al. 1998) and 957R, with Cy5 fluorescent dye attached to the forward primer. After purification, approximately 100 ng of PCR product was digested by two units each of AluI and MseI (New England Biolabs) in a 20-μL volume at 37 °C for 1.5 h. These two enzymes were selected based on an in silico restriction site analysis of available representative sequences from major thaumarchaeotal groups. The reactions were then desalted using the SigmaSpin Post-Reaction Clean-up Columns (Sigma-Aldrich), and 2 μL of the desalted fragment was used in the length polymorphism analysis (CEQ 8000 Genetic Analysis System, Beckman-Coulter). Peaks with relative abundance of >1 % were used to calculate the relative terminal restriction fragment (T-RF) abundance.
For the assignment of major T-RFs, the sequences retrieved in the clone library analysis (see “2.7”) were virtually digested (http://insilico.ehu.es/restriction/main/index.php) and manually assigned to specific T-RFs according to the length of the terminal fragment.
2.7 Cloning, sequencing, and OTU classification
Triplicate PCR amplicons of the 832-bp thaumarchaeotal 16S rRNA gene fragment were pooled, gel purified, and cloned into the pGEM-T Easy Vector (Promega). The recombinant plasmid was transferred into competent E. coli JM109 cells, and ∼30 positive clones were randomly selected and sequenced (MajorBio, Shanghai).
After removing vector and primer sequences, chimeric sequences in each library were identified by Bellerophon (Huber et al. 2004) and excluded from the following analysis. The sequences were aligned against a representative selection of thaumarchaeotal reference sequences and classified into operational taxonomic units (OTUs) at the 95 % identity level using MOTHUR (Schloss et al. 2009).
2.8 Phylogenetic analysis
Representatives for each OTU, their closest hits in the GenBank database obtained by the BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the reference sequences of major thaumarchaeotal clades were used to reconstruct the phylogenetic tree. Neighbor-joining tree and bootstrap support were calculated with MEGA version 4 (Tamura et al. 2007) using the Jukes–Cantor model.
2.9 Canonical correspondence analysis
Canonical correspondence analysis (CCA) was performed with OTUs or T-RFs as species inputs to reveal the regulation of thaumarchaeotal community structure by environmental factors (ter Braak and Šmilauer 2002). CCA was performed using CANOCO 4.53 with default settings (SCIENTIA Software). A binary matrix was constructed by scoring the relative abundance of particular OTUs or T-RFs, and the matrix was used for CCA.
2.10 Statistics
The coefficient of variation (Cv) was calculated by dividing the standard deviation by the mean (SD/mean). Pearson’s correlation analyses were performed to evaluate the relationships among data sets.
2.11 Nucleic acids accession number
Thaumarchaeotal 16S rRNA gene sequences representative of each OTU were deposited with GenBank under the accession numbers of KT449785-KT449799.
3 Results
3.1 Soil characteristics and PAH level
Eight soil characteristics including pH, total carbon, nitrogen, phosphorus, potassium, and inorganic N content were determined (Table 1). The soil pH varied between 4.26 and 8.05 and showed a mosaic spatial distribution. The more variable edaphic factors included ammonium and nitrate, as suggested by high values of Cv, implicating differential fertilization levels during the cultivation of vegetables. In contrast, soil contents of carbon, nitrogen, phosphorus, and potassium were less variable. The sum of 15 PAHs (ΣPAHs) in the soils varied between 0.18 and 20.7 mg kg−1 dry weight (d.w.) (Table 1). There were multiple correlations between the soil characteristics; for example, pH was inversely correlated with the ammonium level in the soils (r = −0.517, P < 0.05, n = 20).
3.2 Abundance of Thaumarchaeota and ammonia-oxidizing bacteria
Soil thaumarchaeotal and AOA abundances were evaluated by qPCR targeting the 16S rRNA and amoA genes, respectively. The copies of these two thaumarchaeotal genes across all soils were directly correlated (r = 0.902, P < 0.01, n = 20), with the 16S rRNA gene varying from 1.58 × 107 to 8.17 × 108 copies g−1 d.w. and the amoA gene varying between 1.98 × 106 and 1.45 × 108 copies g−1 d.w. (Table S1, Electronic Supplementary Material). The abundances of both thaumarchaeotal genes were significantly correlated with pH and C/N (Table 2). Additional correlations were observed between ammonium, PAHs and the thaumarchaeotal amoA gene.
The copy number of the bacterial amoA gene varied from 5.68 × 104 to 2.42 × 107 g−1 d.w., and in most samples were less than that of thaumarchaeotal amoA genes, resulting in thaumarchaeotal:bacterial amoA gene ratios (AOA/AOB) of between 0.9 and 60.5 (Table S1, Electronic Supplementary Material). Bacterial amoA gene abundance was mainly correlated with carbon, nitrogen and, in particular, nitrate (Table 2). The ratios of AOA/AOB were inversely correlated to nitrate levels.
3.3 Community composition of Thaumarchaeota
The composition of the thaumarchaeotal community in the soils was examined by both clone library and T-RFLP analysis of an 832-bp 16S rRNA gene fragment. After removal of chimeras, 575 thaumarchaeotal 16S rRNA gene sequences from 20 libraries were retrieved (Fig. 1a). The sequences were classified into 24 OTUs at 95 % identity, with 9 singleton OTUs. Assignment of representatives from each OTU showed that all sequences were affiliated with the phylum Thaumarchaeota, with 77.7, 19.8, and 1.0 % corresponding to groups I.1b, I.1a-associated, and I.1a, respectively. Additional 1.4 % was affiliated with the group I.1c. A few OTUs that were closely related to Nitrososphaera, Nitrosotalea, soil metagenome fragments 29i4, and 54d9 were abundant (>5 % of the total number of sequences).
With T-RFLP, 10 major terminal restriction fragments (T-RFs) were produced by the combined digestion of AluI and MseI (Fig. 1b). The T-RFs of 102, 103, 280, 310, and 458-bp were affiliated with the group I.1b, as suggested by a virtual enzymatic digestion of library sequences. Among them, T-RF 102 bp was exclusively clustered with the soil fosmid 54d9; T-RF 144 was mostly related to the OTU-5 that corresponds to the I.1a-associated group, while T-RFs 292 and 62 were associated with groups I.1a and I.1c, respectively. The phylogeny of T-RF 459 was not clear because no such T-RF was obtained from retrieved sequences, but the virtual restriction pattern suggested T-RF 459 was very likely related to the I.1a-associated group. It should be noted that the virtual digestion of some sequences from the most acidic samples (pH < 5.0) produced a T-RF of 24 bp, which was too short to be captured with the current T-RFLP method. These sequences comprised OTU-5 and were related to N. devanaterra, a cultivated representative of the I.1a-associated group (Fig. 2).
Neighbor-joining tree of thaumarchaeotal 16S rRNA genes. The representative sequences retrieved in this study for each OTU (in box), key reference sequences (in bold), and closest matched sequences were included in the tree. The corresponding T-RFs are listed after the OTU. Only bootstrap values greater than 50 % are shown near nodes
3.4 Correlation between thaumarchaeotal community composition and environmental factors
To explore the correlation between environmental factors and the thaumarchaeotal community, CCA was performed with OTUs or T-RFs (Fig. 3). The first two CCA axes explained 38.1 and 52.8 % of the variance in the OTU and T-RF compositions, respectively. Among the nine variables tested, pH was the only factor that significantly contributed to the thaumarchaeotal community composition (P < 0.01) regardless of the analysis methods. In contrast, the impact of PAHs on thaumarchaeotal community was minimal.
Moreover, pH was directly correlated with the abundance of specific thaumarchaeotal lineages. For instance, the sum of OTU-3 and 5, both of which are affiliated with the I.1a-associated group, declined along the pH gradient; these OTUs were only highly abundant (>25 %) in samples of pH <5.0. A similar trend was observed for T-RFs 144, 438, and 459. These I.1a-associated lineages were minimal in neutral or alkalic soils (pH >6). In contrast, most group I.1b-related OTUs or T-RFs were enriched in soils with the increased pH (Fig. S1, Electronic Supplementary Material).
3.5 PAH amendment experiment
After a 4-week incubation, no significant pH variation was observed. Anthracene was transformed considerably, although the residual concentration (approximately 100 mg kg−1) was still much higher than the background (Fig. S2, Electronic Supplementary Material). Negligible dissipation of benz(a)anthracene and benzo(a)pyrene was observed at the end of the experiment, suggesting the persistence of these PAHs in the respective soil microcosms. There was no net increase in nitrate in all four treatments during the entire incubation period, but a significant NO3 − accumulation could be observed in anthracene-spiked microcosms from the first to the fourth week (data not shown). The copies of soil thaumarchaeotal 16S rRNA and amoA genes in the control microcosms declined from 6.57 ± 1.10 × 107 and 2.75 ± 0.26 × 106 at the beginning to 4.89 ± 0.27 × 107 and 1.58 ± 0.48 × 106 after the 4-week incubation, respectively. None of the amended PAHs reduced the thaumarchaeotal gene abundances (Fig. 4). After the decrease in the first week, significantly increased thaumarchaeotal amoA gene abundance was observed in the benzo(a)pyrene-supplemented microcosms. Meanwhile, the thaumarchaeotal community compositions were stable after the incubation as revealed by the T-RFLP analysis of 16S rRNA genes (Fig. 5).
Changes of thaumarchaeotal a 16S rRNA and b amoA gene copy numbers in the soil microcosms during the 4-week incubation. The results represent the mean ± SE of three replicate microcosms. CK, ANT, BA, and BaP indicate the control, anthracene-, benz(a)anthracene-, and benzo(a)pyrene-amended microcosms, respectively. Bars with the same lowercase letter on top were not significantly different (P > 0.05)
Thaumarchaeotal communities before and after the incubation as revealed by the T-RFLP of the 16S rRNA gene. The results represent the mean ± SE of three replicate microcosms. CK, ANT, BA, and BaP indicate the control, anthracene-, benz(a)anthracene-, and benzo(a)pyrene-amended microcosms, respectively
4 Discussion
Argosols is typically weakly acidic to neutral; however, significant acidification may occur in cropland Argosols due to the intensive use of mineral N fertilizers (Guo et al. 2010). This is demonstrated by the soil pH variation observed in the field samples (Table 1). Long-term vegetable cultivation may result in the accumulation of inorganic N. As such, the neutral pH may be close to the background of Argosols in this area, while the strong soil acidification was very likely due to the excess use of mineral N fertilizers. This was further supported by the negative correlation between soil inorganic N (NH4 ++NO2 − + NO3 −) and pH (r = −0.48, P < 0.05, n = 20).
pH and substrate supply are major factors controlling the thaumarchaeotal community in terrestrial environments (Gubry-Rangin et al. 2011; Prosser and Nicol 2008). Our findings demonstrate a strong association between thaumarchaeotal communities and soil pH in the investigated soils. Indeed, a decline in the soil pH reduced the abundance of Thaumarchaeota or AOA (Table 2), and the enrichment of Nitrosotalea-related lineages in soils of pH < 5.0 suggests the I.1a-associated group is more adapted to acidic environments. In contrast, most I.1b group taxa favored neutral soils (Fig. S1, Electronic Supplementary Material). N fertilization showed marginal effects on Thaumarchaeota despite the wide variation in inorganic N concentrations (Table 1), whereas the abundance of AOB could be strongly correlated with nitrate (Table 2). Overall, these and previous observations (Wu et al. 2011) suggest that pH rather than inorganic N is a determinant of the thaumarchaeotal communities in these agricultural soils affected by fertilization-induced acidification.
In addition to pH, the effects of PAH pollution on soil Thaumarchaeota were explored in this study. Two orders of magnitude gradient of PAHs were observed across the soils, with the ΣPAHs in some samples higher than normally found in contaminated agricultural soils from this region (Yangtze River Delta, east China) (Ping et al. 2007), suggesting serious soil pollution. Nevertheless, no stress of the natural occurrence of PAH contamination on soil Thaumarchaeota was found (Table 2 and Fig. 3), although the inhibition of soil nitrification activity by PAH contamination was widely recognized. To avoid the confounding influence of other soil factors, short-term incubation of soil microcosms spiked with individual PAH was carried out. Limited transformation of PAHs, indicative of persistent PAH stress across the incubation period (Fig. S2, Electronic Supplementary Material), was observed. The decreased abundance of thaumarchaeotal genes (Fig. 4) in the control microcosms might be due to the degradation of free DNA molecules or dead microbial cells. Four-week exposure to 50–150 mg kg−1 PAHs, which is rarely observed in real situations, did not show any adverse effect on soil Thaumarchaeota.
The absence of response may be mainly associated with the bioavailability of PAHs in soil, because the saturated concentration of PAHs in pore water should be extremely low due to their hydrophobicity and the aging process in soil (Alexander 2000). Most PAH molecules may be tightly attached to soil particles, making them inaccessible and thus less toxic to microbes. This is supported by the high effect concentration of PAHs on soil nitrification activity. For example, significant nitrification inhibition was observed at approximately 1000 mg kg−1 of benzo(a)pyrene, one the most toxic PAHs (Sverdrup et al. 2007). Nevertheless, cultivated Thaumarchaeota could be quite sensitive to organic pollutants presented in the medium. N. maritimus, a cultivated I.1a Thaumarchaeota, was inhibited by low concentration of crude oil and methylene blue (Sipos and Urakawa 2016; Urakawa et al. 2012). We also observed a growth inhibition of one I.1b thaumarchaeotal enrichment by benzo(a)pyrene (unpublished data).
The enhanced growth of soil ammonia-oxidizing Thaumarchaeota in the presence of benzo(a)pyrene is interesting (Fig. 4). In a previous study (de Menezes et al. 2012), phenanthrene amendment increased the relative abundance of thaumarchaeotal transcripts in soil. These observations implicate complicated interactions between Thaumarchaeota and PAHs in soil, and many possibilities exist. First, PAH degradation by soil microbes produces small organic molecules (Juhasz and Naidu 2000), which may improve thaumarchaeotal growth even at a low concentration (Tourna et al. 2011). Second, the changes in the soil microbial community caused by PAH stress may in turn affect Thaumarchaeota (Zhalnina et al. 2013). Third, amoA belongs to a family of copper-containing membrane-bound monooxygenases (CuMMO), which has a wide substrate range including aromatic hydrocarbons (Pester et al. 2011). PAH degradation has been demonstrated with AOB (Chang et al. 2002), but whether Thaumarchaeota have a similar function merits further study.
The gene amoA has often been used as a marker to reveal the community characteristics of thaumarchaeotal ammonia oxidizers due to its taxonomic coincidence with the 16S rRNA gene (Pester et al. 2012). However, amoA-based analysis cannot cover the entire phylum of Thaumarchaeota because some clades appear to lack the function of ammonia oxidation (Weber et al. 2015). In this study, the presence of the I.1c group in some of the most acidic samples (pH <5.0), as revealed by clone library and T-RFLP analysis of the 16S rRNA gene (Fig. 1), confirms its preference for acidic conditions (Oton et al. 2016). Moreover, limited by the resolution of these two community analysis methods, no other thaumarchaeotal clades than AOA and I.1c were retrieved; however, these groups could be rare in soil (Oton et al. 2016). Application of next generation sequencing technologies may be helpful to reveal the vast diversity of soil Thaumarchaeota.
5 Conclusions
Taken together, we examined the thaumarchaeotal community in agricultural soils impacted by both acidification and PAH pollution. The results demonstrate a major influence of pH on both thaumarchaeotal abundance and composition. PAHs, even at a high concentration, did not inhibit soil Thaumarchaeota in these aged contaminated soils or short-term exposed microcosms. The influence of PAH on growth demonstrates the complexity of the interactions between Thaumarchaeota and organic pollutants, which should be addressed in future studies.
References
Alexander M (2000) Aging, bioavailability, and overestimation of risk from environmental pollutants. Environ Sci Technol 34:4259–4265
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252
Chang S, Hyman M, Williamson K (2002) Cooxidation of naphthalene and other polycyclic aromatic hydrocarbons by the nitrifying bacterium, Nitrosomonas europaea. Biodegradation 13:373–381
de Menezes A, Clipson N, Doyle E (2012) Comparative metatranscriptomics reveals widespread community responses during phenanthrene degradation in soil. Environ Microbiol 14:2577–2588
Großkopf R, Janssen PH, Liesack W (1998) Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64:960–969
Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW (2011) Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci 108:21206–21211
Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KWT, Vitousek PM, Zhang FS (2010) Significant acidification in major Chinese croplands. Science 327:1008–1010
Harvey RG (1991) Polycyclic aromatic hydrocarbons: chemistry and carcinogenicity. Cambridge University Press, Cambridge
He J-Z, Hu H-W, Zhang L-M (2012) Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Juhasz AL, Naidu R (2000) Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: a review of the microbial degradation of benzo[a]pyrene. Int Biodeter Biodegrad 45:57–88
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci 108:15892–15897
Liu Y-R, Zheng Y-M, Shen J-P, Zhang L-M, He J-Z (2010) Effects of mercury on the activity and community composition of soil ammonia oxidizers. Environ Sci Pollut Res 17:1237–1244
Lu R (2000) Analytical methods for soil and agrochemistry. China Agricultural Science and Technology Press, Beijing
Mertens J, Broos K, Wakelin SA, Kowalchuk GA, Springael D, Smolders E (2009) Bacteria, not archaea, restore nitrification in a zinc-contaminated soil. ISME J 3:916–923
Mertens J, Wakelin SA, Broos K, McLaughlin MJ, Smolders E (2010) Extent of copper tolerance and consequences for functional stability of the ammonia-oxidizing community in long-term copper-contaminated soils. Environ Toxicol Chem 29:27–37
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C (2003) Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797
Oton EV, Quince C, Nicol GW, Prosser JI, Gubry-Rangin C (2016) Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. ISME J 10:85–96
Pester M, Schleper C, Wagner M (2011) The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol 14:300–306
Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M (2012) amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539
Ping LF, Luo YM, Zhang HB, Li QB, Wu LH (2007) Distribution of polycyclic aromatic hydrocarbons in thirty typical soil profiles in the Yangtze River Delta region, east China. Environ Pollut 147:358–365
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531
Rotthauwe J, Witzel K, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shen J-P, Xu Z, He J-Z (2014) Frontiers in the microbial processes of ammonia oxidation in soils and sediments. J Soils Sediments 14:1023–1029
Sipos AJ, Urakawa H (2016) Differential responses of nitrifying archaea and bacteria to methylene blue toxicity. Lett Appl Microbiol 62:199–206
Subrahmanyam G, Hu H-W, Zheng Y-M, Gattupalli A, He J-Z, Liu Y-R (2014a) Response of ammonia oxidizing microbes to the stresses of arsenic and copper in two acidic alfisols. Appl Soil Ecol 77:59–67
Subrahmanyam G, Shen J-P, Liu Y-R, Archana G, He J-Z (2014b) Response of ammonia-oxidizing archaea and bacteria to long-term industrial effluent-polluted soils, Gujarat, Western India. Environ Monit Assess 186:4037–4050
Sverdrup LE, Hagen SB, Krogh PH, van Gestel CAM (2007) Benzo(a)pyrene shows low toxicity to three species of terrestrial plants, two soil invertebrates, and soil-nitrifying bacteria. Ecotoxicol Environ Saf 66:362–368
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
ter Braak CJF, Šmilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide. Wageningen, Biometris
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci 108:8420–8425
Urakawa H, Garcia JC, Barreto PD, Molina GA, Barreto JC (2012) A sensitive crude oil bioassay indicates that oil spills potentially induce a change of major nitrifying prokaryotes from the Archaea to the bacteria. Environ Pollut 164:42–45
Weber EB, Lehtovirta-Morley LE, Prosser JI, Gubry-Rangin C (2015) Ammonia oxidation is not required for growth of group 1.1c soil Thaumarchaeota. FEMS Microbiol Ecol. doi:10.1093/femsec/fiv001
Wu Y, Conrad R (2014) Ammonia oxidation-dependent growth of group I.1b Thaumarchaeota in acidic red soil microcosms. FEMS Microbiol Ecol 89:127–134
Wu Y, Luo Y, Zou D, Ni J, Liu W, Teng Y, Li Z (2008) Bioremediation of polycyclic aromatic hydrocarbons contaminated soil with Monilinia sp.: degradation and microbial community analysis. Biodegradation 19:247–257
Wu Y, Lu L, Wang B, Lin X, Zhu J, Cai Z, Yan X, Jia Z (2011) Long-term field fertilization significantly alters community structure of ammonia-oxidizing bacteria rather than archaea in a paddy soil. Soil Sci Soc Am J 75:1431–1439
Xu SS, Liu WX, Tao S (2006) Emission of polycyclic aromatic hydrocarbons in China. Environ Sci Technol 40:702–708
Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L, Han W, Singh BK (2011) Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625
Zeng J, Lin X, Zhang J, Zhu H, Chen H, Wong M (2013) Successive transformation of benzo[a]pyrene by laccase of Trametes versicolor and pyrene-degrading Mycobacterium strains. Appl Microbiol Biotechnol 97:3183–3194
Zhalnina K, de Quadros PD, Gano KA, Davis-Richardson A, Fagen JR, Brown CT, Giongo A, Drew JC, Sayavedra-Soto LA, Arp DJ, Camargo FAO, Daroub SH, Clark IM, McGrath SP, Hirsch PR, Triplett EW (2013) Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front Microbiol 4:104
Zhang L-M, Hu H-W, Shen J-P, He J-Z (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045
Acknowledgments
We thank Dr. Junli Hu for his helpful suggestions to the manuscript. This study was supported by the 973 Program of Ministry of Science and Technology of China (2014CB441106), the National Natural Science Foundation of China (41371310, 41201301), and the Natural Science Foundation of Jiangsu Province (BK20131462). Y. Wu acknowledges the fellowships from the Youth Innovation Promotion Association, Chinese Academy of Sciences and the State Key Laboratory of Soil and Sustainable Agriculture (Y212000014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Jizheng He
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM. 1
(DOCX 136 kb)
Rights and permissions
About this article
Cite this article
Wu, Y., Zhu, Q., Zeng, J. et al. Effects of pH and polycyclic aromatic hydrocarbon pollution on thaumarchaeotal community in agricultural soils. J Soils Sediments 16, 1960–1969 (2016). https://doi.org/10.1007/s11368-016-1390-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-016-1390-9