Abstract
Potato is a globally important food crop. In addition to various other factors, potato suffers from many biotic stresses. The important diseases are late blight, viruses, bacterial wilt, bacterial soft rot, dry rot, charcoal rot, common scab, black scurf and wart; and insect-pests are like aphid, whitefly, mite, potato tuber moth, potato cyst nematode, potato leaf hopper and white grub. Of which, late blight is the most devastating disease, whereas aphids and whiteflies are more important pests. These biotic factors limit crop growth and reduce tuber yields. The genus Solanum is one of richest source of genetic diversity and provides great opportunities for genetic enhancement of potato applying classical genetics, traditional breeding and modern genomics tools. With the available knowledge on potato genetic resources, genetic diversity, molecular markers, mapping, gene tagging, marker-assisted selection and high-resolution maps, there had been a considerable advancement in potato. The availability of the potato genome sequence and recently sequenced some more wild species, next-generation breeding tools like genome editing, high-throughput genotyping using single nucleotide polymorphism array and genotyping by sequencing, phenomics, genome wide association mapping, genomic selection and other omics resources further provide tremendous opportunities for next-generation breeding of potato. This chapter highlights on genomic designing for biotic stress resistance in potato.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Introduction
Potato (Solanum tuberosum L.) is the fourth most important crop of the world after rice, wheat and maize and is a major food crop consumed by over a billion people globally (Chakrabarti et al. 2017). It is also one of the most efficient food crops. It is a good source of high-quality protein and rich in minerals, vitamins and phytochemicals. Potato can produce over twice as much dry matter and calories per unit area and time compared to wheat, rice, and maize. Besides being an efficient food producer, it has a broader flexibility in planting and harvesting time and, as such, can fit into many prevalent cropping sequences, thereby giving potato growers a wider choice of crops (Singh et al. 2020). These virtues make potato a good candidate crop for providing food and nutritional security to the developing world. Keeping this in view, FAO declared it as the “food for future” and the year 2008 was announced as the International Year of Potato by the United Nations.
Potato is affected by various diseases and pests, which causes severe yield reduction. Among them, late blight, viruses, bacterial wilt and storage rots are the major diseases, whereas aphids, whiteflies, thrips, mites and potato cyst nematodes are economically important insect-pests. Late blight is the most devastating, which can cause complete loss of crop within a week time if severe infestation is found, whereas viruses are serious from the seed potato quality point of view. Viruses transmit over the generations through clonally propagated materials like tubers and degenerate seed quality, ultimately cause yield reduction. Bacterial wilt is becoming more important in the high temperature growing regions; unfortunately, very limited resistant sources are available against this disease. Soil and tuber-borne diseases impacting tuber storage are dry rot, charcoal rot, bacterial soft rot and common scab (Singh et al. 2020).
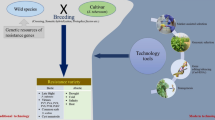
With the rising global temperature under climate change scenario, management of these biotic stresses is inevitable for sustainable potato production to meet the world population food requirement in the future. To achieve this, traditional breeding methods have impacted a lot to improve potato varieties. However, breeding efforts suffer from many problems; among them the polyploid nature of cultivated S. tuberosum, hybridization barriers, poor selection efficiency, and length of breeding programs (it takes over 10 years to release a superior variety), and cumbersome trait phenotyping methods in field conditions. Hence, there is a need to strengthen potato research by applying genomics resources and modern genomics tools. After the potato genome sequencing in 2011 (Xu et al. 2011), the application of structural and functional genomics is still limited in potato. Therefore, discovering novel genes and the deployment of genomics resources is mandatory to enhance tuber production and quality. This chapter aims to update knowledge on modern breeding strategies to obtain improved varieties for sustainable crop production. Figure 2.1 depicts technologies to be used for genomic designing for biotic stress tolerance in potato.
2.2 Biotic Stresses in Potato
2.2.1 Late Blight
Late blight, caused by the oomycete Phytophthora infestans (Mont.) de Bary, is the most devastating disease of potato worldwide (Tiwari et al. 2013). In mid nineteenth century, late blight caused a complete wipeout of crop in the European countries, particularly in Ireland and popularly known as ‘Irish Famine’ (1845). Since then, it spread worldwide, causing now crop loss of up to 10–12 billion US dollars per annum world over. It affects all plant parts (leaves, stems and tubers), causing water-soaked black necrotic spots on leaves. Sporangia of the pathogen can be seen on the lower surface of leaves in the form of white cottony growth around the necrotic lesions. The favorable conditions for disease development are mild temperature (18 ± 2 °C) and high relative humidity (>90%). Due to these specific environmental needs, disease forecasting systems have been developed as complementary tools to manage this disease.
Late blight is controlled mainly through fungicides application and the use of resistant varieties. Besides, cultural practices like sanitation, crop rotation, fertilizers and crop geometry can be successfully applied. Spray of fungicides like metalaxyl and cymoxanil is the most common way to control late blight. Several resistance (R) genes from wild or cultivated potatoes have been exploited through either conventional breeding approaches or biotechnological tools. Among donor wild species, worth to mention are S. demissum, S. bulbocastanum, S. microdontum, S. pinnatisectum, S. cardiophyllum and S. verrucosum (Tiwari et al. 2013). Biopesticides and biocontrol agents like Trichoderma viride and Pseudomonas aeruginosa have also been attempted as safer options to manage this disease. Nevertheless, there is a need for genomic designing for durable resistance to late blight.
2.2.2 Viruses
Potato is infected by more than 30 viruses, which cause yield reduction depending upon disease severity. The major potato viruses are Potato virus X (PVX), Potato virus Y (PVY), Potato virus S (PVS), Potato virus M (PVM), Potato leaf roll virus (PLRV), Tomato leaf curl New Delhi virus-potato (ToLCNDV) in India, and Potato spindle tuber viroid (PSTVd). In general, crop losses are higher in the case of mixed infections. PVY and PLRV are the most devastating, causing up to 80% yield losses, whereas PVX, PVS and PVM are mild, causing up to 30% yield loss (Tiwari et al. 2012). Groundnut bud necrosis virus (GBNV) causes stem necrosis disease in high-temperature regions of central and western India in early planted crop. The newly emerged white fly-transmitted ToLCNDV causing apical leaf curl disease is also serious in northern India. Most viruses are transmitted through contact, mechanical, infected seeds and/or vectors like aphids, whiteflies and thrips. Aphid transmit viruses in two ways (i) persistent and circulative such as PLRV, (ii) non-persistent like PVY, PVA, PVS, and PVM. These viruses cause mosaic or leaf curl and various other mixed symptoms on plants. Circulative virus like PLRV can be managed through the control of aphids.
In general, virus management methods include: prevention of viral transmission, eradication of infected sources, control and avoidance of vectors, and use of virus-free healthy seeds, resistant varieties and biotechnological tools. Conventional and molecular breeding approaches have been applied to breed resistant varieties. The Ryadg gene from S. tuberosum Gp. Andigena has been found effective against several strains of PVY. Besides, there are many genes like Rysto from S. stoloniferum conferring PVY resistance and Rlretb from S. etuberosum providing resistance to PLRV (Tiwari et al. 2012). Resistance to virus vectors would be important to control potato viruses; particularly interesting in this context is wild S. berthaultii, which confers resistance to aphids. Moreover, transgenic approaches have been developed either through pathogen-derived coat protein genes or through disruption of host translation elongation factor.
2.2.3 Soil and Tuber Borne Diseases
Potato is affected by several soil and tuber-borne diseases, causing heavy losses, particularly during storage (Sagar and Sanjeev 2020). Dry rot, charcoal rot and bacterial soft rot cause losses during storage, whereas black scurf and common scab impact tuber appearance and reduce marketable value. Bacterial wilt is another serious potato disease. Dry rot, caused by a fungus F. oxysporum (F. sulphureum/F. sambucinum), is an important post-harvest disease-causing losses during transport and storage. The favorable temperature range for fungus growth is between 15 and 28 °C. Dry rot is managed by sanitation, use of disinfected and healthy quality seeds and boric acid (3%) treatment. Charcoal rot disease, caused by the fungus Macrophomina phaseolina, is more prevalent at high soil moisture combined with high temperature (over 28–30 °C). This disease is managed by crop rotation and rescheduling in planting dates. Bacterial soft rot, caused by Pectobacterium atrosepticum and P. carotovorum (syn. Erwinia carotovora subsp. atroseptica and Erwinia carotovora subsp. carotovora, respectively), is another devastating disease of the potato during harvest, transport and storage. The infected tubers develop wounds, holes and rots. This disease is managed by cultural practices, correct harvesting (e.g., avoid harvesting when temperature rises above 28 °C), curing, boric acid (3%), treatment of harvested tubers and cold storage. Black scurf is caused by Rhizoctonia solani and affects tuber quality and causes moderate yield losses. The optimum temperature for the development of stem lesions is 18 °C. This disease is managed using healthy seeds, boric acid treatment (3%), soil solarization and crop rotation. Common scab is caused by Streptomyces scabies and causes lesions on tuber skin. Congenial environments for disease development is pH (5.2 to >8.0), temperature (20–30 °C) and low soil moisture. The disease is managed by using healthy seeds, boric acid treatment (3%), cultural practices, and crop rotation. Bacterial wilt or brown rot is one of the most damaging diseases of potato caused by Ralstonia solanacearum. The favorable soil temperature for disease development is between 15 and 35 °C. Since there is no resistant source of resistance available, this disease is mainly controlled by the use of healthy seeds and crop rotation (avoidance of other Solanaceous crops). Potato wart, caused by Synchytrium endobioticum, is a problem of hilly regions, like Darjeeling hills in India. An average temperature less than 18 °C and rainfall nearly 700 mm favor disease development. It is managed by sanitation and crop rotation. Very few resistant varieties are available for most of these soil and tuber borne diseases (Sagar and Sanjeev 2020), and genomics assisted breeding is critical to solve this issue.
2.2.4 Insect-Pests
Potato is infested by many insect-pests such as aphids, whiteflies, thrips, white grubs, cutworms, leaf hopper, potato tuber moth and mites (Shah et al. 2020). Aphids (mainly Myzus persicae) are vectors of potato viruses (PVY, PVA, PLRV, PVS and PVM); they transmit viruses from infected to healthy plants causing mosaic and leaf curl symptoms. Whitefly (Bemisia tabaci) is a severe emerging problem in potato under climate change scenario; it transmits ToLCNDV-potato virus and causes mosaic, chlorosis and curling of apical leaves symptoms. Potato leaf hopper (Amrasca biguttula biguttula) also damages the potato crop, causing hopper burn symptoms. Thrips (Thrips palmi) are vectors of groundnut bud necrosis virus, causing stem necrosis. They are particularly common in high-temperature regions. White grub (Brahmina coriacea) is destructive in hilly regions, damaging potato by large shallow and circular holes in the tubers. Cutworm (Agrotis segetum) is another destructive pest of potato in both hills and plains. Mature larvae damage the stem at the ground and make irregular holes in the tubers. Potato tuber moth (Phthorimaea operculella) is a serious pest causing damage in storage and fields. Mite (Polyphagotarsonemus latus) damages early planted crop when temperatures are high. Mostly, these insect-pests are managed by cultural practices and insecticides (Shah et al. 2020). Additionally, potato cyst nematodes (PCN) (Globodera rostochiensis and G. pallida) are the major problems in the world particularly temperate or hilly regions such as Nilgiri and Kufri hills in India, and European countries (Fig. 2.2).
2.3 Genetic Resources
The cultivated potato, S. tuberosum L. belongs to the genus Solanum in the family Solanaceae. According to Hawkes’ classification, the genus is very large and contains over 2000 species, of which nearly 235 are tuber bearing (Hawkes 1990). Potato species are rich sources of genes for resistance to various biotic stresses. They form a polyploid series ranging from diploid (2n = 2x = 24) to hexaploid (2n = 6x = 72), with the basic chromosome number of 12. About 73% of the tuber-bearing Solanum species are diploids, 4% triploids, 15% tetraploids and 8% pentaploids/hexaploids (Hawkes 1990). The potato cultivated worldwide, S. tuberosum, is a tetraploid (2n = 4x = 48). The recent classification by Spooner et al. (2007) distribute the cultivated potato species as following: (i) Solanum tuberosum Andigenum Group of upland Andean genotypes containing diploids (2x), triploids (3x) and tetraploids (4x); and Solanum tuberosum Chilotanum Group of tetraploids (4x) of lowland Chilean landraces, (ii) S. ajanhuiri (2x), (iii) S. juzepczukii (3x), and (iv) S. curtilobum (5x) (Table 2.1). S. tuberosum is generally divided into two subspecies, namely subsp. tuberosum, the universally cultivated potato, and subsp. andigena, a primitive taxon cultivated to a limited extent in the Andes region. An ”effective ploidy” of potato species is represented by the endosperm balance number (EBN). The EBN is a number, ranging from 1 to 4, assigned to each potato species following intra/interploidy crosses (Johnston et al. 1980). It is a powerful tool to predict the success of interploidy/interspecific hybridization, in that normal endosperm development occurs only when there is a 2:1 maternal to paternal EBN ratio in the hybrid endosperm.
The primary genepool includes the cultivated potato (S. tuberosum spp. tuberosum) (4 EBN); no sexual barriers occur within genotypes of the primary genepool. The secondary genepool refers to 4 EBN wild species such as S. demissum, which can be crossed with cultivated S. tuberosum, and nearly 180 diploid 2 EBN wild species. The tertiary genepool includes wild species that are not crossable with cultivated/wild species due to differences in ploidy number and EBN; among them S. bulbocastanum and S. commersonii, both 2x (1 EBN). Many useful genes from wild sources cannot be transferred into the cultivated genepool through conventional techniques because of sexual incompatibilities, primarily due to differences in ploidy and EBN. A few strategies have been proposed to overcome this problem. For example, to introgress resistance to R. solanacearum and P. carotovorum possessed by 1EBN species S. commersonii, Carputo et al. (1997) proposed a breeding scheme based on doubling the chromosome number of S. commersonii, and on the production of triploid and pentabloid bridges. Additional methods to circumvent sexual barriers are manipulation of ploidy and EBN, mentor pollination and embryo rescue, hormone treatment, and reciprocal crosses (Jansky 2006). Notably, also somatic hybridization has been extensively used in potato to overcome sexual barriers and produce genetic variability at both nuclear and cytoplasmic DNA. Interspecific somatic hybridization is a multi-step process involving protoplast isolation and fusion, culture and regeneration of fusion products and, finally, the identification of somatic hybrids among regenerants. Since several important traits exhibit wide variation in the somatic hybrids produced, further breeding efforts are necessary before a genotype combining several useful characteristics is identified. A recent review by Tiwari et al. (2018b) provides progress in somatic hybridization research in potato over 40 years. For example, somatic hybrid between S. tuberosum dihaploid ‘C-13’ (2n = 2x = 24) and diploid (2n = 2x = 24) wild species S. cardiophyllum (Chandel et al. 2015), and S. pinnatisectum (Sarkar et al. 2011) for late blight resistance with wider genetic base.
Potato genetic resources are collected and preserved worldwide. The International Potato Centre (CIP) (https://cipotato.org/), Lima, Peru is a CGIAR research centre for global potato research and development. CIP is amongst the largest international potato gene bank in the world, which provides potato germplasm throughout the world. Other potato gene banks are the US Potato Genebank (NRS) USA, the CGN Potato Collection at the Centre for Genetic Resources, the Netherlands (CGN), and the Commonwealth Potato Collection (CPC) of the Vavilov Institute (VIR) etc. The European Cultivated Potato Database (ECPD) (https://www.europotato.org/) is an online database of the European cultivated potato varieties; an online potato pedigree database resource is also available (http://www.plantbreeding.wur.nl/PotatoPedigree/). UK has also developed its own potato variety database (http://varieties.ahdb.org.uk/). Worth to mention is also the PotatoPro (https://www.potatopro.com/) database, which describes potato statistics at the world level.
2.4 Classical Genetics and Breeding
The cultivated potato is tetraploid and highly heterozygous suffering from acute inbreeding depression. The clonal propagation of tubers preserves the heterozygosity in commercial cultivars. Conventional breeding is a cumbersome task because most traits show tetrasomic inheritance and also chromatid segregation. Due to the small size of potato chromosomes, it is very difficult work at cytological level. In the past, chromosome identification and mapping were carried out using a set of restriction fragment length polymorphism (RFLP) marker-anchored bacterial artificial chromosomes (BAC) as fluorescence in situ hybridization (FISH) probes (Dong et al. 2000). This led to assign genetic linkage groups to specific chromosomes and was also used for chromosome numbering. Potato breeding objectives mostly focus on high tuber yield, quality and resistance to diseases, insect pests and abiotic stresses. More than fifty desirable traits are to be combined while developing a new potato variety. These traits include morphological features, yielding ability, tuber characters, ability to withstand various stresses, wider adaptability, quality parameters, consumer and industrial acceptability. It is perhaps a herculean task to combine all traits in a single variety because of the complex heterozygous potato nature. The genetic base of the existing and newly released potato varieties is relatively narrow compared to that available in the genepools. Only a fraction of useful genes from wild species have been successfully introgressed into potato varieties. The history of conventional potato breeding reveals that many varieties took nearly 30 or more years from hybridization and clonal selection before being released. Conventional potato breeding is often carried out at the tetraploid level, involving selection of specific tetraploid S. tuberosum parents, hybridization and phenotypic recurrent selection in seedling and clonal generations at targeted locations for a wide range of desirable characters. Alternatively, it is performed at the diploid level, involving hybridization between diploid species and S. tuberosum dihaploids (2n = 2x = 24). Haploid-species hybrids are selected for traits of interests and for a propensity to produce 2n gametes to be employed in sexual polyploidization crossing schemes. At both ploidy levels, however, conventional breeding strategies usually take many years and seek huge investments. In addition, they involve human resources and, compared to molecular breeding, they delay the accessibility of the targeted variety to stakeholders.
2.5 Molecular Markers to Assess Genetic Diversity
To study the diversity of plant species, various type of markers can be employed. Classical tools for such investigations are morphological markers. They are easily identifiable simply on the phenotype of an organism. Later, biochemical markers (or isozyme) were discovered; they are based on the relative mobility of enzyme isoforms. However, both markers are influenced by the environment and plant growth stage. A quantum jump towards genome mapping was made possible after introducing DNA markers, which are based on the DNA sequence variation and are least affected by the environment and growth stages. A wide range of molecular markers have been developed over the past four decades. Different types of molecular markers have been developed. The first molecular marker technique was RFLP, followed by PCR-based marker systems such as RAPD, SSR, AFLP, SSCP and CAPS. Sequencing technologies allowed the detection of single nucleotide polymorphisms (SNPs) markers. The progress of next-generation sequencing (NGS) technologies and the decreasing prices for sequence runs have led to a number of novel techniques for the detection of polymorphic markers. Some recent examples are genotyping by sequencing (GBS) and SNP array chip.
Genetic diversity analysis, marker-assisted selection and DNA fingerprinting (genotyping) are the important applications of molecular markers. Unlike morphological descriptors, profiles created by using molecular data are independent of environmental effect. Therefore, the International Union for the Protection of (New) Plant Varieties (UPOV) has constituted a working group to critically examine the feasibility of using biochemical and molecular techniques (BMT) for variety identification. DNA fingerprints can be used to establish distinctness and check the uniformity and stability of a particular variety. A wide range of genetic diversity studies have been carried out in potato world over. Consequently, genetic relationships have been established in cultivated and wild potato species. Various molecular markers such as RAPD, ISSR, SSR, AFLP, DArT and SNP have been extensively used for diversity analysis in potato germplasm and varieties. Isozyme, RAPD and AFLP markers were used for diversity analysis and to test the genetic integrity of potato after micropropagation and long-term conservation (Aversano et al. 2011). SSR has been used to analyze genetic diversity within wild species showing noteworthy resistances to R. solanacearum and aphids. A new set of 24 highly informative SSR markers (two from each linkage group) named as the Potato Genome Identification (PGI) kit has been used worldwide in potato varieties (Tiwari et al. 2018a) and wild species (Tiwari et al. 2019). The role of cytoplasmic markers (T/β, W/α, W/γ and A/ε) has also been studied in potato using plastome- and chondriome-specific markers (Tiwari et al. 2014). A wide range of SNP arrays have recently been developed and applied in potato to characterize germplasm and gene discovery.
2.6 Association Mapping
Linkage mapping is the genetic association of traits with segregating alleles of molecular markers in a defined mapping population. It detects genomic regions that explain phenotypic variation in a trait of interest and subsequently identifies genes/QTLs in that region. QTL mapping in potato is mainly carried out at the diploid level due to the potato highly heterozygous nature. Many QTLs for resistance to biotic stresses like P. infestans and root cyst nematodes are known. On the contrary, association mapping is a method to identify genes or QTLs associated with phenotypic variation in natural populations based on historical recombination events related by descent. The method takes advantage of historical meiotic recombinations and linkage disequilibrium (Flint-Garcia et al. 2003). For association mapping, a population consisting of diverse germplasm including cultivars, breeding clones and landraces is assembled and phenotyped for the complex traits of interest. Molecular markers are then analyzed in the population and marker-trait associations between phenotypic and genetic variation are detected. Marker-trait association approaches are known to identify markers linked with the genes/QTLs. In potato, tetraploid or diploid potatoes have been utilized for association mapping for desirable agronomic traits and biotic stresses. Gebhardt et al. (2004), for the first time, used association mapping in tetraploid potato germplasm to identify markers for late blight resistance and maturity traits in 600 potato cultivars. Further, association mapping was applied based on candidate genes for resistance against Verticillium dahliae (Simko et al. 2004) and Phytophthora infestans (Pajerowska-Mukhtar et al. 2009). Genome wide association mapping (GWAS) has been deployed to identify genes/QTLs at whole genome level in potato by D’hoop et al. (2008). GBS is effectively used for SNP discovery and trait association mapping for multiple traits including biotic stresses (Uitdewilligen et al. 2013; Sharma et al. 2018).
2.7 Molecular Mapping of Resistance Genes and QTLs
Gene mapping is foremost important for molecular breeding. RFLPs were first used to create linkage maps in potato, which showed conserved markers with tomato. Later, PCR-based molecular markers such as SSRs and AFLPs were applied for mapping of genes and QTLs. More than 10,000 AFLP markers were used to generate dense maps exploited by the “Potato Genome Sequencing Consortium”. Many genes for biotic stress resistances have been mapped in potato. Particular focus has been given to simply inherited genes, such as those conferring resistance to late blight, viruses and nematodes. Disease scoring provides clear patterns of qualitative evaluation of genotypes. A number of techniques like bulked segregant analysis have been extensively used to identify markers. A recent study describes mapping of H2 resistance effective against Globodera pallida pathotype Pa1 in tetraploid potato (Strachan et al. 2019). A summary of some work on mapping and molecular markers is presented in Tables 2.2, 2.3 and 2.4.
2.8 Marker-Assisted Breeding
Germplasm characterization is the foremost important part of molecular breeding. In addition to novelty, important characteristics for the release of new varieties are distinctness, uniformity and stability. Therefore, integration of marker-assisted selection (MAS) in conventional breeding is inevitable for the rapid release of new varieties. MAS method has been applied mostly in simply inherited traits like late blight, viruses, and potato cyst nematodes resistance, but limited in case of complex inherited traits like yield contributing traits. Moreover, gene pyramiding through MAS has been executed to accelerate the use of genetic resources in potato breeding. PCR-based markers are the best for MAS due to their ease in application, especially in a resource-poor developing country. Advances in molecular marker technology, large-scale whole genome sequencing and an expanding genetic map of potato chromosomes have progressed significantly. In the future, it would essentially improve the prospects of identification of resistance gene clusters with common sequence motifs for mapping and cloning of more R genes. Thus, it may lead to the development new diagnostic markers for MAS for biotic stress traits in potato.
MAS is particularly useful in the case of introgression breeding from the donor (e.g., wild) to the recipient (e.g., cultivated) genotype. In usual practice, this is achieved by recurrent backcrossing and selection cycles. Markers tightly linked to the gene of interest are used to identify progenies at the seedling stage, thereby decreasing the number of breeding cycles. Tightly linked markers for many qualitative and quantitative traits have been published and made available for MAS such viruses and late blight resistance (Tiwari et al. 2012, 2013) (Tables 2.5 and 2.6). For example, markers linked to extreme resistance to PVY and late blight resistance were validated in triplex parental lines and their progeny. Breeding of multiple disease resistance is a major priority in most potato-growing counties. Recently, MAS has been deployed to develop a new variety Kufri Karan at ICAR-CPRI, Shimla. This variety is highly resistant to late blight, viruses and moderately resistant to potato cyst nematode. MAS can also be performed in programs based upon interspecific hybridization when markers are not associated with the trait under selection. In these cases, MAS can be employed to estimate the wild genome content of the recurrent parent at each backcross, and can help to identify hybrids combining useful traits with the lowest percentage of wild genome content. This type of MAS, termed negative assisted selection, has been successfully applied at various steps of breeding programs aimed at transferring resistance traits from S. commersonii into S. tuberosum (Carputo et al. 2002).
2.9 Genomics-Aided Breeding
2.9.1 The Potato Genome
Unlike diploid crops, tetraploid potato varieties have four copies of each of the 12 chromosomes. This makes it very difficult to follow inheritance patterns, especially concerning the many complex traits with which breeders are compelled to work with. Moreover, the S. tuberosum genome high heterozygosity makes it recalcitrant to current sequencing technologies and bioinformatics programs. Therefore, the genes affecting many important agronomic traits remain still undiscovered and their locations on the 12 chromosomes are often imprecise. In 2011, the “Potato Genome Sequencing Consortium” (PGSC)—formed by 26 international institutes belonging to 14 countries—successfully solved these problems by sequencing a homozygous doubled monoploid (DM 1–3 516 R44, referred as to DM) of S. tuberosum Group Phureja (2n = 2x = 24), in which there were only two copies of each chromosome and, more importantly, each copy was identical. The PGSC deciphered 840 Mb of the potato genome in 2011. This represented the first genome of a plant belonging to the Asterid clade of eudicot, representing 25% of flowering plant species. A total of 39,031 protein-coding genes were predicted in the 840 Mb genome size of the potato. Numerous publications witness the usefulness of DM genome, but undoubtedly its quality and potential are limited by the technology exploited a decade ago. Now the advanced sequencing technologies and the improved software are enabling the generation of a high-quality potato genome assembly that will facilitate research aimed at improving potato agronomic traits and understanding genome evolution (Zhou et al. 2020). At present, a chromosome-scale long-read reference genome assembly of the potato genome has been constructed (Pham et al. 2020). In recent years, a few more wild or cultivated potato genotypes have been sequenced. They highlight evolutionary relationship, adaptation mechanism and novel resistance/tolerance genes in wild species such as Solanum commersonii (Aversano et al. 2015), tuber-bearing Solanum species (Hardigan et al. 2017), S. chacoense ‘M6’ (Leisner et al. 2018), cultivated potato taxa (Kyriakidou et al. 2020), and S. pinnatisectum derived somatic hybrid (Tiwari et al. 2021). Further genome analyses have identified markers for agronomically important traits (Li et al. 2018).
2.9.2 Functional and Comparative Genomics
The potato genome sequence has opened up new vistas in potato research. Functional genomics allows the mining of novel genes in potato germplasm/varieties for traits of economical and industrial importance through transcriptome analysis. Further, genome sequence and reference map allow association of genes to target traits in the genome. Such regions can then be used to define further markers for fine-scale mapping; alternatively, candidate genes can be sought directly from the genome sequence and associated annotation data. This step-change, facilitating sequence-based genomics and aiding molecular breeding in potato, would accelerate trait-gene discovery and gene isolation. This would further shorten the time to breed new varieties and also significantly improve parental genotypic assessment. Genome tagged molecular marker studies would be more meaningful and enable more accurate estimates of population genetic and linkage disequilibrium parameters. The shift towards sequence-based polymorphism rather than fragment-based will virtually replace centiMorgan position by sequence co-ordinates and greatly increase the information output and accuracy of mapping procedures. The integrated potato genetic and physical reference map forms an important resource for genetic mapping efforts and will alleviate many of the complicating aspects of potato genetics. With the release of the genome of the other economically important Solanaceous crop, i.e., the tomato, comparative genomics and sequence-based synteny analysis among Solanaceae have been made feasible. Given the biological and economic importance of many Solanaceous species and the diversity of their phenotypes/products (agriculturally useful parts tubers, berries, etc., growth habits, wide geographical growing range, clonal propagation, regeneration), comparative genomics provides a fundamental framework for tackling both applied and basic questions.
With the completion of genome sequencing of more and more organisms, research focus has now been shifted from sequencing to delineating the biological functions of genes. Methodologies of biological research are evolving from the “one gene in one experiment” to the “multiple genes in one experiment” paradigm. It is not possible to perform analysis on a large number of genes using traditional methods. Earlier DNA microarray and now RNA sequencing are the technologies that enable researchers to investigate and address issues that first were thought to be non-traceable. Functional genomics involves the use of high-throughput methods for the study of large numbers of gene set in parallel. Indirect information on cellular or developmental function can be obtained from spatial and temporal expression patterns. For example, the presence of mRNA and/or protein in different cell types, during development, during pathogen infection, or in different environments. The subcellular localization and post-translational modifications of proteins can be informative as well. The potato genome sequence can be used for functional validation of gene function. The techniques used for functional genomics in potato include RNA-Seq and microarray at the whole genome level and reverse genetic approaches, like gene knockout by RNAi (RNA interference) and VIGS (virus-induced gene silencing), at the gene level. They allow to analyze the expression of many genes in a single reaction quickly and efficiently. Besides, allele mining application has shown discovery in novel alleles of the genes for late blight resistance in wild potato species (Tiwari et al. 2015).
2.9.3 Next-Generation Potato Breeding
Potato improvement through the application of next-generation breeding techniques is essential to shorten the usually long period (over 10 years) required for developing a new variety. Successful completion of potato genome sequencing enables discovering a large number of genes regulating multiple traits like biotic stresses and yield attributes etc. Besides, bi-parental linkage mapping, population genetics by GWAS, genome editing (GE) and genomic selection (GS) coupled with GBS and SNP array chip platform with integrated high-throughput genotyping (HTG) and high-throughput phenotyping (HTP) facilities have emerged as powerful techniques for completion of breeding cycles in shorter time. There is immense potential to apply these new breeding techniques for rapid potato improvement.
2.9.3.1 Genomic Selection (GS)
Potato breeders have to deal with more than 50 characters (biotic stress, abiotic stress, quality traits, yield attributing and tuber traits) to develop a new variety. Although MAS has been a powerful tool in plant breeding and potato has been applied to improve resistance traits, it has limitations for complex inheritance traits, like yield. With the availability of the potato genome, there is immense opportunity to work at the whole genome level. Hence, GS or genome-wide selection, or genomic-assisted breeding can enable the integration of phenotyping and high-throughput genotyping data of pedigree/segregating generations to enhance the selection of superior genotypes and accelerate breeding cycles. GS works on the principle of linkage disequilibrium (LD) with a minimum of one marker per locus in the breeding population without gene mapping. GS accelerates breeding cycles with an increase in genetic gain per unit time and reduces costs as well. It combines molecular and phenotypic data in a training population (TP) to acquire the genomic estimated breeding value (GEBV) of individuals in a TP that have been genotyped but not phenotyped. GS determines genetic association and diversity in different landraces/cultivars/varieties/breeding lines/wild species with variation in topography and ecology. With the identification of genome rearrangements and SNP discovery at whole-genome level, GS can be efficiently applied in the near future. GS has been successfully applied in animals and reported to some extent in plants like maize, wheat, sugar beet. In potato its application has been very limited so far, a few like resistance to late blight and common scab (Enciso-Rodriguez et al. 2018). This might be due to the unavailability of SNP markers distributed throughout the genome, trait association, SNP calling rate and software uses. However, the rapid advancement in genotyping techniques (SNP and haplotypes), high-throughput phenotyping and trait association would lead to reality potato GS in the near future.
2.9.3.2 Genotyping by Sequencing (GBS)
GBS is one of the high-throughput techniques currently being used to generate genotyping data for several crop species including potato (Bastien et al. 2018). With the reducing NGS costs, a considerable amount of high-throughput data has been developed. GBS has been designed for several studies, including genetic analysis, population studies, molecular characterization of germplasm, SNP discovery. To breed varieties, knowledge about genes and environment and their interaction is essential for using GBS to select advanced breeding lines with desirable traits (Schönhals et al. 2017). Besides GBS, SNP chip-based markers are an additional high-throughput genotyping platform available in potato for genotyping. Various platforms such as 20 K SNPs Affymetrix Axiom (SolSTW array) (Vos et al. 2015), Infinium 12 K V2 Potato Array (Illumina platform) (Ellis et al. 2018), and 8 K SNPs (Illumina Infinium BeadChip) (Obidiegwu et al. 2015; Schönhals et al. 2017) are currently available.
2.9.3.3 Genome-Wide Association Studies (GWAS)
GWAS (or linkage disequilibrium mapping) has been applied in potato and many other crops to examine simple and complex traits taking advantages of linkage disequilibrium. It is a family-based linkage mapping approach to identify the link between genotyped markers and phenotypes of interest scored in a large number of individuals with broad genetic and phenotypic diversity (landraces, wild and cultivated species, varieties, core collection). Usually, the rationale behind GWAS is to assess SNPs that influence phenotypes. Currently, the use of Next Generation Sequencing (NGS) allows to genotype large populations with a higher density of markers. This offers the unique opportunity to increase mapping resolution. In line with this, specialized mapping populations have also been developed that significantly enhance the power and efficiency of these association studies. GWAS requires a detailed understanding of population structure to minimize false-positive and false-negative associations; for this purpose, various statistical methods have been developed over the years. Softwares like STRUCTURE and EIGENSTRAT are very popular within the scientific community working with GWAS. Recently, additional software specifically tailored for the tetraploid potato (i.e., GWASpoly) has been made available (Rosyara et al. 2016). In potato, GWAS has been conducted for various traits like common scab resistance (Yuan et al. 2020), Verticillium resistance and quality traits (Khlestkin et al. 2019).
2.9.3.4 Genome Editing (GE)
Genome editing is a targeted alteration in a genome that creates new allelic variation. Sequence-specific nucleases (SSNs) have been applied for genome editing and genetic manipulations. The SSNs technology is rapidly becoming important in plants, which uses three major nuclease systems like Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR- associated proteins (CRISPR/Cas9). Among these, CRISPR/Cas9 is the most widely used today for genome editing and is based upon an RNA-guided method to target DNA sequence. This is being used widely due to its simplicity, multiplexing capability, cost-effectiveness, and high efficiency. An important issue with this technology is the off-target mutations due to mismatch base pairing between gRNA and DNA. Genome editing is an essential tool to create new variants with desirable gene combinations. Unlike genetic transformation methods (see in later section), which creates stable integration of cisgenes or transgenes, genome editing provides crop improvement opportunities where no foreign gene is introduced. In potato, TALENs and CRISPR/Cas9 have been used for site-directed mutagenesis and gene silencing (Nadakuduti et al. 2018). The main targets were traits like cold-induced sweetening and glycoalkaloid (solanine and chaconine) content, acetochalactate synthase, granule bound starch synthase (review by Dangol et al. 2019).
2.9.3.5 High Throughput Phenotyping (HTP)
High-throughput precision phenotyping is essential to utilize ther ultimate potential of a genotype. Present methods of phenotyping are often slow, time consuming, laborious and inaccurate, often destructive or with limited phenotyping ability. High-throughput phenotyping platforms are essential for precision phenotyping and modern breeding applications. They are usually based on automation, sensors, high resolution imaging capability, robotics etc. In potato, a few technologies have been applied to roots and shoot traits. For example, Phenofab and Keytrack System (KeyGene, The Netherlands) have been developed for measuring plant growth and other traits using multiple imaging systems and thermal sensors with automated handling under controlled environments. However, high correlations between pots and field-grown plants are essential.
2.10 Genetic Transformation
Genetic transformation has many advantages for plant breeding, and these advantages are even more striking in crops with complex polyploid inheritance, such as the potato. While conventional breeding manipulates genomes in a mostly uncontrolled fashion, requiring generations of selection to assemble and fix the maximum number of desirable traits, genetic transformation offers a direct approach, allowing introgression of a single, distinct gene without linkage drag. Thus, it enables rapid and often powerful improvement of crop plants, and is not limited by compatibility barriers. In cases where genetic diversity among sexually compatible relatives of crop species is insufficient for a particular trait, genetic transformation may represent the only possibility for trait improvement. It offers a highly effective means of adding a single gene to existing elite potato clones with no or very minimal disturbances. Potato, being highly amenable to genetic transformation, has been subjected to genetic transformation to confer resistance to a wide range of diseases (late blight, viruses, bacterial wilt and soil and tuber borne diseases) and pests (aphids, white fly, potato tuber moth). It should be pointed out that genetic transformation is not simply a faster alternative to conventional breeding. Rather, it is a complementary way to exploit plant genetic diversity that may require time to create and evaluate the most desirable expression of the transgene.
The availability of a suitable regeneration protocol is a pre-requisite for undertaking genetic transformation. A rapid and efficient Agrobacterium tumefaciens mediated transformation protocol based on direct organogenesis from inter-nodal stem explants of in vitro potato plants is available in potato. On the other hand, biological balistics (i.e. gene gun) plant transformation methods have also been applied. In these cases, either tungsten or gold particles coated with DNA are accelerated to a high speed to bombard target tissues. Although potato is highly amenable to Agrobacterium mediated transformation, the use of gene gun is necessary for plastid transformation and enhances transformation efficiency. The technique has been successfully used to transfer the plastid specific cassette for tuber-specific expression of cry1Ab and a fused cry1Ab + cry1B genes to develop transgenic potato resistance to potato tuber moth.
Many efforts have been devoted to the characterization, mapping, and cloning genes or resistance breeding in potato. For example, the RB gene (Bradeen et al. 2009) and an osmotin-like gene were cloned and sequenced from wild S. bulbocastanum and S. chacoense and used for developing transgenics with late blight resistance. The RB gene has been found the most effective to confer durable resistance against late blight in the F1 progenies (Sundaresha et al. 2018). Similarly, virus and potato cyst nematode-resistant genes were cloned, the sequence was characterized and utilized for transgenics development. Post-transcriptional gene silencing (PTGS) or RNA interference (RNAi) are mechanisms of gene regulation in eukaryotes. In plants, PTGS acts as surveillance system against the invading molecular parasites like viruses, transposons and transgenes. PTGS is being utilized for transgenics development against pathogens and for functional genomics aimed at elucidation of gene functions. PTGS has been targeted for the avr3a gene for late blight resistance, and phosphatidic acid phosphatase 2 (PAP2) gene for bacterial wilt resistance. Viral gene sequences were also cloned and characterized. The coat protein gene from an Indian isolate of potato leaf roll virus (PLRV) was targeted through PTGS. Similarly, the coat protein gene of PVY, PLRV and potato apical leaf curl virus (PALCV), replicase associated protein gene of PALCV, and movement protein gene of potato stem necrosis virus were cloned, sequenced and used for the development of virus-resistant transgenics by RNAi technology. Technologies are available for easy and efficient transformation and protocols are applicable to carry out the southern, northern and western blotting for characterizing transgenic events.
2.11 Bioinformatics
Bioinformatics is now an inevitable tool in plant science with increasing advancements in genomics technologies. Bioinformatics plays significant role in generating new tools and databases; it increases the efficiency and precision of data analysis of huge genomics information. Genomics and post-genomics research requires modern bioinformatics tools to integrate genomics, transcriptomics, proteomics, metabolomics and phenomics data. The development of modern and customized bioinformatics tools and advanced databases has become mandatory to handle the increasingly enormous amount of large datasets in crop species. This helps us to systematically store, organize, and analyze large amounts of biological information computationally. With the advent of bioinformatics tools, an enormous amount of DNA, RNA and protein sequences are currently stored in gene data banks. Major public gene banks of the DNA and protein sequences are GenBank NCBI (National Centre of Biological Information) in USA (http://www.ncbi.nlm.nih.gov), EMBL (European Molecular Biology Laboratory) in Europe (http://www.ebi.ac.uk/embl/), and DDBJ (DNA Data Bank) in Japan (http://www.ddbj.nig.ac.jp).
The potato genome sequence was deciphered initially in 2011 by the PGSC and now the database is maintained by the SpudDB, Potato Genomics Resources, Michigan State University, USA (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml). It represents one of the most important tools for potato biotechnologists and breeders worldwide. This database has been recently updated with the genome sequence of wild tuber-bearing S. chacoense. Additional bioinformatics resources are available. Genome sequences of a few more wild potato species are available at the NCBI. Solanaceae Genomics Network (http://solgenomics.net/) is a collection of maps, genomes, and tools for Solanaceae species. SolRgene database (http://www.plantbreeding.wur.nl/SolRgenes) provides a comprehensive dataset to explore disease resistance genes in Solanum species. PoMaMo (Potato Maps and More) (https://gabi.rzpd.de/PoMaMo.html) contains molecular maps of all potato chromosomes with about 1000 mapped elements, sequence data, gene functions, BLAST search, SNP and InDel information etc. The PlantGDB (http://www.plantgdb.org/StGDB/) database describes genomes, gene models, alignments, gene structure annotations, annotated protein alignments etc.
2.12 Social, Political and Regulatory Issues
Conventional breeding is commonly practiced without any concern to develop new potato varieties. Transgenics development has raised serious biosafety issues in public and therefore, transgenics products are fully regulated worldwide with some exceptions. Recently, genome editing technology has emerged as a safer strategy to produce new, improved genotypes. All the technologies result into varieties having desirable agronomics traits. Hence, right of the inventors needs to be protected through legal means. Intellectual property rights (IPRs) have been created to protect these rights. Usually, IPRs are protected under categories such as patents, copyrights, trademarks, trade secrets, geographical indicators, design and layout design of integrated circuits. Patents are the most important form of protection for research and developmental activities. Another important example of intellectual property is given by Plant Breeders’ Rights (PBRs), which refer to the legal protection offered to a breeder or developer or owner over a newly developed variety. Thus, it prevents any third party from the commercial exploitation of the new variety without a developer’s authorization. For example, India has enacted the “Protection of Plant Varieties and Farmers’ Rights” (PPV&FR) Act, 2001 as sui generis system of plant variety protection, which was based on the UPOV Act 1991. The PPV&FR Act 2001 protects plant varieties such as newly bred varieties, extant varieties (released but not completed 15 years on the date of application), farmers varieties (traditionally cultivated or landraces or wild/native) and essentially derived varieties (derived from an initial variety but little difference). For a registration of a new variety, the criteria of novelty (N), distinctness (D), uniformity (U), and stability (S) must be met. Plant variety protection is allowed for 25 years for trees and vines and 20 years for others (while 20 years for tree and vines and 15 years for others by PPV&FRA in India). Moreover, researchers’ rights have been provided to use the protected varieties for research purpose as parents in breeding to develop new varieties. Overall, social, political and regulatory issues need to be addressed before acceptance of any crop variety.
2.13 Future Perspectives
Biotic stresses are one of the major limiting factors of yield reduction in potato. Moreover, in the climate change scenario, management of these biotic stresses would be more challenging where emergence of new pathotypes/strainal variations is common in pathogens and pests. The increasingly interest in genomics research combined with available knowledge on genetics, breeding (conventional and marker assisted selection) have enabled better scope for potato improvement. With the knowledge on a huge genetic diversity in the genus Solanum, phylogenetics relationship, molecular markers, gene mapping and cloning have also paved pathways for genetic enhancement of this crop at molecular level. The potato genome sequence alongwith a few more potato genomes sequences, and functional genomics provide immense opportunities to discover new genes and markers for breeding and biotechnological applications. With the next-generation breeding tools like high-throughput genotyping coupled with high-throughput phenotyping (phenomics), SNP array, GBS, GWAS, GS and genome editing provide powerful technologies fast development of climate resilient potato varieties resistance to biotic stress. Taken together, there is a need of genome wide characterization of whole germplasm collection at global level using robust SNP arrays or other technologies, robust phenotyping under controlled as well as natural environments on multiple locations, marker-trait association analysis, develop trait-specific driven molecular markers or haplotype-based next-generation potato breeding. Simultaneously, discovery of novel genes using transcriptomics approaches and other modern omics/biotechnological tools like proteomics, metabolomics etc. would strengthen the management of biotic stresses in potato.
References
Aversano R, Dato FD, Matteo AD, Frusciante L, Carputo D (2011) AFLP analysis to assess genomic stability in Solanum regenerants derived from wild and cultivated species. Plant Biotech Rep 5:265–271
Aversano R, Contaldi F, Ercolano MR, Grosso V, Iorizzo M et al (2015) The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell 27:954–968
Bastien M, Boudhrioua C, Fortin G, Belzile F (2018) Exploring the potential and limitations of genotyping-by-sequencing for SNP discovery and genotyping in tetraploid potato. Genome 61:449–456
Bonierbale MW, Plaisted RL, Tanksley SD (1988) RFLP Maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120:1095–1103
Bradeen JM, Iorizzo M, Mollov DS, Raasch J, Kramer LC et al (2009) Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance level. Mol Plant-Microbe Interact 22:437–446
Bradshaw JE, Bryan GJ, Lees AK, McLean K, Solomon-Blackburn RM (2006) Mapping the R10 and R11 genes for resistance to late blight (Phytophthora infestans) present in the potato (Solanum tuberosum) R-gene differentials of Black. Theor Appl Genet 112:744–751
Carputo D, Barone A, Cardi T, Sebastiano A, Frusciante L, Peloquin SJ (1997) Endosperm balance number manipulation for direct in vivo germplasm introgression to potato from a sexually isolated relative (Solanum commersonii Dun.). Proc Natl Acad Sci USA 94:12013–12017
Carputo D, Frusciante L, Monti L, Parisi M, Barone A (2002) Tuber quality and soft rot resistance of hybrids between Solanum tuberosum and the incongruent wild relative S. commersonii. Amer J Potato Res 79:345–352
Celebi-Toprak F, Slack SA, Jahn MM (2002) A new gene, Nytbr, for hypersensitivity to potato virus Y from Solanum tuberosum maps to chromosome IV. Theor Appl Genet 104:669–674
Chakrabarti SK, Conghua X, Tiwari JK (2017) The potato genome. Springer Nature, Switzerland, 326 pp
Champouret N (2010) Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. PhD thesis. Wageningen University, Wageningen
Chandel P, Tiwari JK, Ali N, Devi S, Sharma S et al (2015) Interspecific potato somatic hybrids between Solanum tuberosum and S. cardiophyllum, potential sources of late blight resistance breeding. Plant Cell Tiss Org Cult 123:579–589
D’Hoop BB, Paulo MJ, Mank RA, van Eck HJ, van Eeuwijk FA (2008) Association mapping of quality traits in potato (Solanum tuberosum L.). Euphytica 161:47–60
Dangol SD, Barakate A, Stephens J, Çaliskan ME, Bakhsh A (2019) Genome editing of potato using CRISPR technologies: current development and future prospective. Plant Cell Tiss Org Cult 139:403–416
De Jong W, Forsyth A, Leister D, Gebhardt C, Baulcombe DC (1997) A potato hypersensitive resistance gene against potato virus X maps to a resistance gene cluster on chromosome V. Theor Appl Genet 95:246–252
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Ellis D, Chavez O, Coombs J, Soto J, Gomez R et al (2018) Genetic identity in genebanks: application of the SolCAP 12K SNP array in fingerprinting and diversity analysis in the global in trust potato collection. Genome 61:523–537
Enciso-Rodriguez F, Douches D, Lopez-Cruz M, Coombs J, de Los Campos G (2018) Genomic selection for late blight and common scab resistance in tetraploid potato (Solanum tuberosum). G3 (Bethesda) 8:2471–2481
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Foster SJ, Park T-H, Pel M, Brigneti G, Śliwka J et al (2009) Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact 22:589–600
Gebhardt C, Ritter E, Debener T, Schachtschabel U, Walkemeier B et al (1989) RFLP analysis and linkage mapping in Solanum tuberosum. Theor Appl Genet 78:65–75
Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schüler K (2004) Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed 13:93–102
Ghislain M, Spooner DM, Rodriguez F, Villamon F, Nunez J et al (2004) Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theor Appl Genet 108:881–890
Hämäläinen JH, Sorri VA, Watanabe KN, Gebhardt C, Valkonen JPT (1998) Molecular examination of a chromosome region that controls resistance to potato Y and A potyviruses in potato. Theor Appl Genet 96:1036–1043
Hardigan MA, Laimbeer FPE, Newton L, Crisovan E, Hamilton JP et al (2017) Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc Natl Acad Sci USA 114:E9999–E10008
Hawkes JG (1990) The potato, evolution, biodiversity and genetic resources. Belhaven Press, London, 259 pp
Hein I, Birch PRJ, Danan S, Lefebvre V, Odeny DA et al (2009) Progress in mapping and cloning qualitative and quantitative resistance against Phytophthora infestans in potato and its wild relatives. Potato Res 52:215–227
Heldák J, Bežo M, Štefúnová V, Galliková A (2007) Selection of DNA markers for detection of extreme resistance to potato virus Y in tetraploid potato (Solanum tuberosum L.) F1 progenies. Czech J Genet Plant Breed 43:125–134
Hosaka K, Hosaka Y, Mori M, Maida T, Matsunaga H (2001) Detection of a simplex RAPD marker linked to resistance to potato virus Y in a tetraploid potato. Amer J Pot Res 78:191–196
Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N et al (2005) Comparative genomics enabled the cloning of the R3a late blight resistance gene in potato. Plant J 42:251–261
Jacobs MMJ, Vosman B, Vleeshouwers VGAA, Visser RGF, Henken B, van den Berg RG (2010) A novel approach to locate Phytophthora infestans resistance genes on the potato genetic map. Theor Appl Genet 120:785–796
Jansky S (2006) Overcoming hybridization barriers in potato. Plant Breed 125:1–12
Johnston SA, den Nijs TP, Peloquin SJ, Hanneman RE Jr (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor Appl Genet 57:5–9
Kasai K, Morikawa Y, Sorri VA, Valkonen JPT, Gebhardt C, Wantabe KN (2000) Development of SCAR markers to the PVY resistance gene Ryadg based on a common feature of plant disease resistance genes. Genome 43:1–8
Kelley KB, Whitworth JL, Novy RG (2009) Mapping of the potato leafroll virus resistance gene, Rlretb, from Solanum etuberosum identifies interchromosomal translocations among its E-genome chromosomes 4 and 9 relative to the A-genome of Solanum L. sect Petota. Mol Breed 23:489–500
Khlestkin VK, Rozanova IV, Efimov VM, Khlestkina EK (2019) Starch phosphorylation associated SNPs found by genome-wide association studies in the potato (Solanum tuberosum L.). BMC Genetic 20(Suppl 1):29
Kuhl JC, Hanneman RE, Havey MJ (2001) Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genom 265:977–985
Kyriakidou M, Achakkagari SR, Gálvez López JH, Zhu X, Tang CY et al (2020) Structural genome analysis in cultivated potato taxa. Theor Appl Genet 133:951–966
Leisner CP, Hamilton JP, Crisovan E, Manrique-Carpintero NC, Marand AP et al (2018) Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J 94:562–570
Li Y, Colleoni C, Zhang J, Liang Q, Hu Y et al (2018) Genomic analyses yield markers for identifying agronomically important genes in potato. Mol Plant 11:473–484
Marczewski W, Flis B, Syller J, Schäfer-Preg R, Gebhardt C (2001) A major quantitative trait locus for resistance to Potato leafroll virus is located in a resistance hotspot on potato chromosome XI and is tightly linked to N-Gene-like markers. Mol Plant Microbe Interact 14:1420–1425
Marczewski W, Hennig J, Gebhardt C (2002) The Potato virus S resistance gene Ns maps to potato chromosome VIII. Theor Appl Genet 105:564–567
Marczewski W, Flis B, Syller J, Strzelczyk-Żyta D, Hennig J, Gebhardt C (2004) Two allelic or tightly linked genetic factors at the PLRV.4 locus on potato chromosome XI control resistance to potato leafroll virus accumulation. Theor Appl Genet 109:1604–1609
Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R (1998) Isolation, characterization and mapping of simple sequence repeat loci in potato. Mol Gen Genet 259:233–245
Nadakuduti SS, Buell CR, Voytas DF, Starker CG, Douches DS (2018) Genome editing for crop improvement—applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front Plant Sci 9:1607
Obidiegwu JE, Sanetomo R, Flath K, Tacke E, Hofferbert HR et al (2015) Genomic architecture of potato resistance to Synchytrium endobioticum disentangled using SSR markers and the 8.3k SolCAP SNP genotyping array. BMC Genet 16:38
Pajerowska-Mukhtar K, Stich B, Achenbach U, Ballvora A, Lubeck J et al (2009) Single nucleotide polymorphisms in the Allene Oxide Synthase 2 gene are associated with field resistance to late blight in populations of tetraploid potato cultivars. Genetics 181:1115–1127
Pankin A, Sokolova E, Rogozina E, Kuznetsova M, Deahl K et al (2011) Allele mining in the gene pool of wild Solanum species for homologues of late blight resistance gene RB/Rpi-blb1. Plant Genet Res Characteriz Utiliz 9:305–308
Park T-H, Foster S, Brigneti G, Jones JDG (2009) Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica 165:269–278
Pel MA, Foster SJ, Park T-H, Rietman H, van Arkel G et al (2009) Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol Plant Microbe Interact 22:601–615
Pham GM, Hamilton JP, Wood JC, Burke JT, Zhao H et al. (2020) Construction of a chromosome-scale long-read reference genome assembly for potato. Gigascience 9(9):giaa100
Rietman H (2011) Putting the Phytophthora infestans genome sequence at work; identification of many new R and Avr genes in Solanum. PhD thesis, Wageningen University
Ritter E, de Galarreta JIR, van Eck HJ, Sanchez I (2008) Construction of a potato transcriptome map based on the cDNA-AFLP technique. Theor Appl Genet 116:1003–1013
Ritter E, Debener T, Barone A, Salamini F, Gebhardt C (1991) RFLP mapping on potato chromosomes of two genes controlling extreme resistance to potato virus X (PVX). Mol Gen Genet 227:81–85
Rosyara UR, De Jong WS, Douches DS, Endelman JB (2016) Software for genome-wide association studies in autopolyploids and its application to potato. Plant Genome 9(2):1–10
Sagar V, Sanjeev S (2020) Soil and tuber borne diseases of potato and their management. In: Singh AK, Chakrabarti SK, Singh B, Sharma J, Dua VK (2020) Potato science and technology for sub-tropics. New India Publishing Agency, New Delhi, pp 247–266
Sarkar D, Tiwari JK, Sharma SH, Poonam SSA et al (2011) Production and characterization of somatic hybrids between Solanum tuberosum L. and S. pinnatisectum Dun. Plant Cell Tiss Org Cult 107:427–440
Schönhals EM, Ding J, Ritter E, Paulo MJ, Cara N et al (2017) Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L.) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array. BMC Genom 18:642
Shah MA, Bairwa A, Naga KC, Subhash S, Raghavendra KV et al. (2020) Important potato pests and their management. In: Singh AK, Chakrabarti SK, Singh B, Sharma J, Dua VK (2020) Potato science and technology for sub-tropics. New India Publishing Agency, New Delhi, pp 295–326
Sharma SK, MacKenzie K, McLean K, Dale F, Daniels S, Bryan GJ (2018) Linkage disequilibrium and evaluation of genome-wide association mapping models in tetraploid potato. G3 (Bethesda) 8:3185–3202
Simko I, Costanzo S, Haynes KG, Christ BJ, Jones RW (2004) Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor Appl Genet 108:217–224
Singh AK, Chakrabarti SK, Singh B, Sharma J, Dua VK (2020) Potato science and technology for sub-tropics. New India Publishing Agency, New Delhi, p 382
Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E (2006) The novel, major locus Rpi-phu1 for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Genet 113:685–695
Smilde WD, Brigneti G, Jagger L, Perkins S, Jones JD (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor Appl Genet 110:252–258
Sokolova E, Pankin A, Beketova M, Kuznetsova M, Spiglazova S et al (2011) SCAR markers of the R-genes and germplasm of wild Solanum species for breeding late blight-resistant potato cultivars. Plant Genet Res Characteriz Utiliz 9:309–312
Song Ye-Su, Hepting L, Schweizer G, Hartl L, Wenzel G, Schwarzfischer A (2005) Mapping of extreme resistance to PVY (Rysto) on chromosome XII using anther-culture-derived primary dihaploid potato lines. Theor Appl Genet 111:879–887
Sorri VA, Watanabe KN, Valkonen JPT (1999) Predicted kinase-3a motif of a resistance gene analogue as a unique marker for virus resistance. Theor Appl Genet 99:164–170
Spooner DM, Núñez J, Trujillo G, Herrera RM, Guzmán F, Ghislain M (2007) Extensive simple sequence repeat genotyping of potato landraces supports a major re-evaluation of their gene pool structure and classification. Proc Nat Acad Sci USA 104:19398–19403
Strachan SM, Armstrong MR, Kaur A, Wright KM, Lim TY et al (2019) Mapping the H2 resistance effective against Globodera pallida pathotype Pa1 in tetraploid potato. Theor Appl Genet 132:1283–1294
Sundaresha S, Sharma S, Shandil RK, Sharma S, Thakur V et al (2018) An insight into the down-stream analysis of RB gene in F1 RB transgenic potato lines imparting field resistance to late blight. Funct Plant Biol 45:1026–1037
Szajko K, Chrzanowska M, Witek K, Strzelczyk-Zyta D, Zagórska H et al (2008) The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor Appl Genet 116:297–303
Tan MYA (2008) Genetic mapping and pyramiding of resistance genes in potato. PhD Thesis. Wageningen University, Wageningen
Tiwari JK, Gopal J, Singh BP (2012) Marker-assisted selection for virus resistance in potato: options and challenges. Potato J 39:101–117
Tiwari JK, Sundaresha S, Singh BP, Kaushik SK, Chakrabarti SK et al (2013) Molecular markers for late blight resistance breeding of potato: an update. Plant Breed 132:237–245
Tiwari JK, Chandel P, Singh BP, Bhardwaj V (2014) Analysis of plastome and chondriome genome types in potato somatic hybrids from Solanum tuberosum x Solanum etuberosum. Genome 57:29–35
Tiwari JK, Devi S, Sharma S, Chandel P, Rawat S, Singh BP (2015) Allele mining in Solanum germplasm: cloning and characterization of RB-homologous gene fragments from late blight resist1ant wild potato species. Plant Mol Biol Rep 33:1584–1598
Tiwari JK, Ali N, Devi S, Kumar V, Zinta R, Chakrabarti SK (2018a) Development of microsatellite markers set for identification of Indian potato varieties. Sci Hort 231:22–30
Tiwari JK, Devi S, Ali N, Luthra SK, Kumar V et al (2018b) Progress in somatic hybridization research in potato during the past 40 years. Plant Cell Tiss Org Cult 132:225–238
Tiwari JK, Ali S, Devi S, Zinta R, Kumar V, Chakrabarti SK (2019) Analysis of allelic variation in wild potato (Solanum) species by simple sequence repeat (SSR) markers. 3 Biotechnol 9:262
Tiwari JK, Rawat S, Luthra SK, Zinta R, Sahu S et al (2021) Genome sequence analysis provides insights on genomic variation and late blight resistance genes in potato somatic hybrid (parents and progeny). Mol Biol Rep 48(1):623–635
Uitdewilligen JG, Wolters AM, D’hoop BB, Borm TJ, Visser RG, van Eck HJ (2013) A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8:e62355
van der Vossen EAG, Sikkema A, Hekkert BL, Gros J, Stevens P et al (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J 36:867–882
van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D et al (2005) The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad spectrum late blight resistance in potato. Plant J 44:208–222
van Os H, Andrzejewski S, Bakker E, Barrena I, Bryan G et al (2006) Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 73:1075–1087
Velásquez AC, Mihovilovich E, Bonierbale M (2007) Genetic characterization and mapping of major gene resistance to potato leafroll virus in Solanum tuberosum ssp. andigena. Theor Appl Genet 114:1051–1058
Villamon FG, Spooner DM, Orrillo M, Mihovilovich E, Pérez W, Bonierbale M (2005) Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana). Theor Appl Genet 111:1201–1214
Vos PG, Uitdewilligen JG, Voorrips RE, Visser RG, van Eck HJ (2015) Development and analysis of a 20K SNP array for potato (Solanum tuberosum): an insight into the breeding history. Theor Appl Genet 128:2387–2401
Wang M, Allefs S, van den Berg RG, Vleeshouwers VGAA, van der Vossen EAG, Vosman B (2008) Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor Appl Genet 116:933–943
Xu X, Pan S, Cheng S, Zhang B, Mu DJ et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
Yuan J, Bizimungu B, Koeyer DD, Rosyara U, Wen Z, Lagüe M (2020) Genome-wide association study of resistance to potato common scab. Potato Res 63:253–266
Zhou Q, Tang D, Huang W, Yang Z, Zhang Y et al (2020) Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat Genet 52:1018–1023
Acknowledgements
The authors thank Competent Authority, ICAR-CPRI, Shimla for necessary support under the biotechnology program and CABin scheme.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Tiwari, J.K. et al. (2022). Genomic Designing for Biotic Stress Resistance in Potato. In: Kole, C. (eds) Genomic Designing for Biotic Stress Resistant Vegetable Crops. Springer, Cham. https://doi.org/10.1007/978-3-030-97785-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-97785-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-97784-9
Online ISBN: 978-3-030-97785-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)