Abstract
For breeding potato varieties resistant to late blight, identification of resistance genes to Phytophthora infestans (Rpi genes) is essential. Introduction of Rpi genes from wild Solanum species into cultivated potato is likely to be a good method to achieve durable resistance to P. infestans. In this study, we identified two Rpi genes (Rpi-ber1 and Rpi-ber2) derived from two different accessions of Solanum berthaultii. These two genes are closely linked on the long arm of chromosome 10. There are similarities between the predicted genetic locations of the previously identified Rpi-ber and Rpi-ber1, which given the common origin of these genes, may indicate that they are the same. However, the genetic positions of Rpi-ber1 and Rpi-ber2 are different. Rpi-ber1 is positioned between CT214 and TG63, whereas Rpi-ber2 is located below both of these two markers. In addition, the sequences of four linked markers to both R genes showed different polymorphisms indicating the two Rpi genes could be transmitted from different haplotypes (chromosomes).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum L.) is the fourth most important crop and the most important non-cereal food crop in the world. In potato cultivation, the major natural factor which limits yield is late blight caused by the oomycete pathogen Phytophthora infestans (Mont.) de Bary. This devastating disease can result in complete loss of crop yield (Świeżyński and Zimnoch-Guzowska 2001). Fungicide treatment is currently the most common method to control late blight. However, the high cost of fungicide application is problematic, especially in developing countries. Moreover, because fungicide application can impact on health and environmental safety, the use of chemicals is becoming restricted. In addition, the pathogen quickly evolves and some of the new variants are insensitive to commonly used fungicides (Day and Shattock 1997; Goodwin et al. 1996). Therefore, the introduction of genetic resistance into cultivated potato is a potentially valuable method to achieve durable resistance to late blight.
Two main types of resistance to late blight have been described in potato (Umaerus and Umaerus 1994). First, general resistance is often based on a major quantitative trait locus (QTL) and a few minor QTLs and results in partial resistance. Second, specific resistance is based on major dominant resistance (R) genes. In early breeding programs during the first half of last century, 11 R genes (R1–R11) derived from S. demissum were identified. Nine R genes, R3 (now separated as R3a and R3b) and R5–R11 are localized on chromosome 11 (Bradshaw et al. 2006; El-Kharbotly et al. 1994, 1996; Huang et al. 2004; Huang 2005). Other R genes originating from S. demissum were mapped to different locations including R1 on chromosome 5 (El-Kharbotly et al. 1994; Leonards-Schippers et al. 1992) and R2 on chromosome 4 (Li et al. 1998). All R genes introgressed from S. demissum to cultivated potatoes have been overcome by the pathogen as new strains rapidly evolve that are virulent on the previously resistant hosts (Umaerus and Umaerus 1994). Consequently, partial resistance conferred by QTLs was thought to be more durable than resistance conferred by single R genes (Turkensteen 1993). However, partial resistance is strongly correlated with maturity type and it makes resistance breeding more difficult (Wastie 1991). Also the genetic positions of QTLs often correspond to the region of R gene clusters (Gebhardt and Valkonen 2001; Grube et al. 2000). Hence, recent efforts to identify late blight resistance have focused on major R genes conferring broad-spectrum resistance derived from diverse wild Solanum species. Beside S. demissum, other wild Solanum species such as S. acaule, S. chacoense, S. berthaultii, S. brevidens, S. bulbocastanum, S. microdontum, S. sparsipilum, S. spegazzinii, S. stoloniferum, S. sucrense, S. toralapanum, S. vernei, and S. verrucosum have been reported as new sources for resistance to late blight (reviewed by Jansky 2000; Hawkes 1990). To date, three R genes, RB/Rpi-blb1, Rpi-blb2, and Rpi-blb3 from S. bulbocastanum have been mapped on chromosome 8, 6, and 4, respectively (Naess et al. 2000; Park et al. 2005a; van der Vossen et al. 2003, 2005). Another R gene, Rpi-abpt, probably from S. bulbocastanum, has been localized on chromosome 4 (Park et al. 2005b). Rpi1 from S. pinnatisectum on chromosome 7 (Kuhl et al. 2001), Rpi-mcq1 from S. mochiquense (Smilde et al. 2005) and Rpi-phu1 from S. phureja on chromosome 9 (Śliwka et al. 2006) have also been identified.
S. berthaultii originating from Bolivia, South America has also been identified as a source of resistance to P. infestans (reviewed by Jansky 2000; Hawkes 1990). Previously, Rber derived from S. berthaultii was mapped to chromosome 10 (Ewing et al. 2000). Subsequently a finer and higher-resolution genetic map of Rpi-ber (same as Rber) was constructed (Rauscher et al. 2006). In our study, we screened accessions of wild Solanum species S. berthaultii to identify new resistances to P. infestans and mapped two R genes originating from S. berthaultii on chromosome 10. We used several molecular markers from those references and compared genetic positions and allelic differences of these S. berthaultii-derived R genes.
Materials and methods
Plant materials
In total, 22 accessions of S. berthaultii were obtained from three different gene banks: two with BGRC gene bank number from Braunschweig Genetic Resources Center, Braunschweig, Germany, 17 with CGN gene bank number from Centre for Genetic Resources, Wageningen, The Netherlands and three with PI gene bank number from the US Potato Genebank (NRSP-6), Sturgeon Bay, Wisconsin, USA (Table 1). Various numbers of seeds of each accession were sown and tested for resistance to P. infestans. Depending on the phenotypic assignment, identified resistant plants were crossed with susceptible plants to generate mapping populations. We generated 40 different intraspecific or interspecific mapping populations.
In the present study, we selected two core populations to further characterize and identify R genes to P. infestans derived from S. berthaultii depending on the number of seeds produced and the results of segregation. One population is named Stenber219 and was derived from a cross between a resistant individual from accession PI473331 of S. berthaultii and a susceptible individual from accession CGN19035 of S. stenotomum. The initial mapping population for Stenber219 consisted of 44 plants. The second population is named Sphuber237 and was derived from a cross between a resistant plant from accession PI265858 of S. berthaultii and a susceptible line, 96H14-10 used as a Sli gene donor (Phumichai et al. 2006). The initial mapping population for Sphuber237 consisted of 50 plants.
Resistance assay
To identify resistance we used two different P. infestans isolates (98.170.3 and Met+) kindly provided by Dr. David Shaw at Bangor University, UK. Race-specificity of these two isolates was determined on the standard Black differentials (Malcolmson and Black 1966). Isolate 98.170.3 is race 1.3.4.10.11 and Met+ is race 1.3.4.7.8.10.11.
A detached leaf assay was performed using a method modified from Vleeshouwers et al. (1999). Fresh sporangia were propagated on detached leaves of a suitable sensitive plant from a maintenance plate of cleared pea broth agar (1%) supplied with 100 μg/ml carbenicillin at 18°C with 18 h light and 6 h dark condition for a week. The fresh sporangia were harvested by rinsing leaves with sterilized water and incubated at 10°C for 1–4 h to induce zoospore release. Zoospores were re-suspended to a concentration of 5 × 105 zoospore/ml. Fully expanded young leaf material was taken from greenhouse plants and placed on a 9 cm petri-dish with a piece of wet filter paper. Inoculation was performed on the abaxial leaf surface with a 10 μl droplet of the zoospore inoculum. Petri-dishes were wrapped with plastic film and incubated at 18°C with 18 h light and 6 h dark condition for 7–12 days. Experiments were repeated independently using two leaflets per plant at least three times to confirm phenotypes, classified as susceptible, moderately susceptible, moderately resistant, and resistant.
Marker development and map construction
Genomic DNA was extracted from young leaf tissue according to Bendahmane et al. (1997). To identify the genetic position of the R loci, both AFLP markers (Vos et al. 1995) and PCR-based markers were employed.
AFLP analysis was performed as described by Vos et al. (1995). Primary templates were prepared using a PstI/MseI restriction enzyme combination, followed by ligation to appropriate adaptors. Templates were diluted 10 times prior to selective pre-amplification. The pre-amplified products (secondary templates) were checked on a 1% agarose gel and diluted 10 times again. Bulked Segregant Analysis (BSA; Michelmore et al. 1991) was carried out to perform AFLP analysis on resistant and susceptible bulks comprising the secondary templates of eight resistant and eight susceptible genotypes selected based on the results of phenotypes. The bulks were screened with 24 primer combinations that decrease marker density. For the selective amplification, radioactively labeled (33P) P+2 primers were used in combination with M+3 primers. The 33P-labeled PCR products were loaded on the gel after 30 min of pre-run. The amplified DNA fragments were separated on a 6% polyacrylamide gel in 1X TBE buffer for two and half hours. The gels were dried on Whatman papers for 2 h using a vacuum dryer and exposed to X-ray films for 4–6 days. AFLP markers were named according to the enzyme, primer combination, and the mobility of the fragment as described in reference autoradiograms by Keygene NV, Wageningen, The Netherlands. PCR-based markers were also developed to construct genetic linkage maps of the R genes using primers derived from published RFLP markers, tomato BAC sequences and tomato unigene sequences available in a public database (http://www.sgn.cornell.edu/cview/index.pl) and primers published by Rauscher et al. (2006). Genetic linkage maps were constructed according to recombination frequencies between marker loci or between marker loci and the R loci. The PCR-based markers used to construct the genetic linkage maps of the R genes are presented in Table 2.
Results
Identification of resistance
To identify resistance in wild species, we collected 22 different wild accessions of S. berthaultii from three different gene banks. From the seed packages of these 22 accessions, a total of 195 individual plants were tested for resistance to P. infestans in two replications using two different isolates, 98.170.3 and Met+. When the inoculated leaf showed sporulating lesions on the inoculated spot, it was scored as susceptible and when the leaf showed no symptoms or necrosis without sporulation, it was scored as resistant. When the two leaves did not show the same reaction or sporulation was not clear, it was classified as either moderately susceptible or moderately resistant. In the database of accessions at the CGN gene bank, the 22 accessions of S. berthaultii have already been evaluated for resistance to late blight and most of these accessions were assigned as resistant except four accessions. However, the results of the resistance assay from our study were very different from those in the CGN database. A lot of variation was also observed between accessions and within accessions as shown in Table 1. Based on the results of screening S. berthaultii accessions, resistant plants were selected to generate mapping populations. These resistant plants were crossed with susceptible plants of S. berthaultii accessions, other susceptible wild species or cultivated diploid lines. Seeds were successfully obtained from 40 crosses. However, the number of seeds and the frequency of seed germination were variable. Eleven of these 40 populations were selected to be tested for resistance. Approximately 50 seeds from each population were sown in vitro and the in vitro plants were transplanted in the greenhouse. In the Stenber219 population derived from a cross between a resistant S. berthaultii (PI473331) and a susceptible S. stenotomum (CGN19035), 20 resistant and 24 susceptible plants were observed. In the Sphuber237 population derived from a cross between a resistant S. berthaultii (PI265858) and a susceptible line (96H14–10), 25 resistant and 25 susceptible plants were observed. In other mapping populations derived from different accessions, the phenotypes were not clear, segregation was skewed to resistance, or all plants were resistant (data not shown). Therefore, two mapping populations (Stenber219 and Sphuber237) were chosen to construct genetic linkage maps.
Localization of the R loci
The two R loci identified in the Stenber219 and Sphuber237 populations were named Rpi-ber1 and Rpi-ber2 (R gene to P . i nfestans derived from S. berthaultii), respectively. To localize these two R loci on the genetic linkage map of potato, we selected eight resistant and eight susceptible plants of each population to compose resistant and susceptible bulks. These bulks and both parents were tested with chromosome 10 marker TG63 as previously one R gene derived from S. berthaultii had been localized on chromosome 10 (Ewing et al. 2000; Rauscher et al. 2006). TG63 was found to be linked genetically to the resistant phenotypes in both Stenber219 and Sphuber237 populations. On subsequent analysis of individual plants, it was 10.2 and 5.0 cM distant from Rpi-ber1 and Rpi-ber2, respectively. In addition, 24 Pst+2/Mse+3 primer combinations were tested on the bulks and parents, four and five of which produced candidate AFLP markers putatively linked to the R loci, Rpi-ber1, and Rpi-ber2, respectively. To confirm linkage between the candidate AFLP markers and the R loci, the 16 individuals that comprised the two bulks were also screened for these markers. Subsequently, one and three AFLP markers were identified to be linked to Rpi-ber1 and Rpi-ber2, respectively. However, all of these AFLP markers were distant from the R loci or linked as repulsion phase markers. PAGMAGG_324[R] was mapped 43.1 cM from Rpi-ber1 towards the telomere. PAGMGG_227[R] and PAGMGG_228 were mapped 17 cM and PAGMGG_167[R] was mapped 11 cM from Rpi-ber2 towards the centromere.
To obtain more markers linked to the R loci, we used PCR-based markers. Published RFLP markers, tomato BAC sequences, tomato unigene sequences in a public database (http://www.sgn.cornell.edu/cview/index.pl) and markers linked to Rpi-ber on chromosome 10 (Rauscher et al. 2006) were tested on both populations. In the Stenber219 population for Rpi-ber1, six PCR-based markers were identified to be linked to the R locus. CT214 mapped 16 cM towards the centromere and TG63, U221455[R], TG206[R], and TG422 mapped 10.2 cM towards telomere from the R locus. In the Sphuber237 population for Rpi-ber2, ten PCR-based markers were identified to be linked to the R locus. CT238, TG63[R], U221455, TG422[R], and T0724 mapped 5 cM towards centromere and TG403 mapped 12 cM towards telomere from the R locus. All PCR-based markers localized on both maps are described in Table 2 and the genetic linkage maps are shown in Fig. 1.
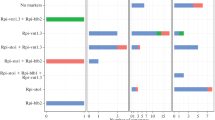
Integrated genetic linkage map of the different late blight resistance genes (a) Rpi-ber, (b) Rpi-ber1, and (c) Rpi-ber2 on chromosome 10. The map of (a) Rpi-ber is modified from Rauscher et al. (2006). The map distances (cM) are indicated on the right of each map
Comparative genetics
Two genetic linkage maps for Rpi-ber1 and Rpi-ber2 were compared to each other and to the map for Rpi-ber published by Rauscher et al. (2006) as shown in Fig. 1. The polymorphic pattern of PCR-based markers bridged between the Rpi-ber1 and Rpi-ber2 maps was also compared as shown in Table 2 and Fig. 2.
Several PCR-based markers were linked to each other on three different maps. CT214 and TG63 were present on all three maps. CT11, CT238, and TG403 were bridged between the Rpi-ber map and the Rpi-ber2 map. U221455, TG422, and BA81l15t3 were bridged between the Rpi-ber1 map and the Rpi-ber2 map. The order of all bridge markers was the same. However, the genetic position of the three R loci was not. Rpi-ber and Rpi-ber1 were located on north of TG63, but genetic distances are 0.6 cM from Rpi-ber and 10.2 cM from Rpi-ber1. Although Rpi-ber and Rpi-ber1 were located between CT214 and TG63, Rpi-ber2 was located south of these two markers.
As the types and the polymorphic patterns of PCR-based markers are variable in Stenber219 and Sphuber237, we compared 11 PCR-based markers linked to Rpi-ber1, Rpi-ber2, or both (Table 2 and Fig. 2). There were more polymorphic markers in Stenber219 (10 markers) than in Sphuber237 (6 markers). One and five of these markers were allele specific in Stenber219 and Sphuber237, respectively. Five markers were bridged between genetic linkage maps of two populations as shown in Fig. 1. However, only one marker (BA81l15t3) was shown to be identical in the type of marker and the polymorphic pattern. The rest of the bridge markers were all different in both populations. For instance, CT214 was a CAPS marker generated by HinfI digestion in Stenber219, but was an allele specific marker in Sphuber237. TG63 and TG422 were CAPS markers generated by Sau96I digestion in both populations. However, these markers were linked to Rpi-ber1 in coupling phase, whereas they were linked to Rpi-ber2 in repulsion phase. Also the restriction patterns of these two markers were different. Four bridge markers showing different marker types and polymorphic patterns in between two populations are shown in Fig. 2.
Discussion
In this study, we identified two R genes derived from the wild Solanum species, S. berthaultii. We screened 195 individual plants and 65 and 66 plants were assigned as susceptible and resistant, respectively. The rest of the plants were assigned as moderately susceptible or moderately resistant due to unclear phenotypes. Based on these results, we generated 40 mapping populations, 11 of which were screened for segregation with about 50 offspring of each population. Two populations, Stenber219 and Sphuber237, showed clear segregation. In other populations, resistance could be associated with QTLs or the resistance allele in the resistant parents could be homozygous.
The R genes designated as Rpi-ber1 and Rpi-ber2 were localized on the long arm of chromosome 10. One R gene (Rber/Rpi-ber) originating from S. berthaultii was previously mapped to a similar genetic region (Ewing et al. 2000; Rauscher et al. 2006). As shown in Fig. 1, all molecular markers linked to each other in these three molecular maps are colinear and there were two molecular markers, CT214 and TG63 which could bridge these three maps. The genetic positions of these two markers demonstrate that Rpi-ber2 could be distinct from the others as Rpi-ber and Rpi-ber1 are positioned between the two markers, whereas Rpi-ber2 is located below the two markers. The accession from which Rpi-ber2 is derived also differs from that of Rpi-ber and Rpi-ber1 although all of these have been identified in the same wild Solanum species. However, identity of Rpi-ber and Rpi-ber1 could not be excluded because these two genes originate from the same accession of S. berthaultii. In addition, although the genetic distances between the R loci and CT214 or TG63 are quite different, it is not conclusive whether Rpi-ber and Rpi-ber1 are the same or not because genetic distance highly relies on the size of population. CT214 and TG63 are 8.2 and 4.8 cM away from Rpi-ber defined using 158 individuals in the preliminary genetic map of Rpi-ber (named as Rber) reported by Ewing et al. (2000), but 5.1 and 0.6 cM away from Rpi-ber defined using 665 individuals in their higher-resolution map (Rauscher et al. 2006). Similarly, two flanking markers to Rpi-abpt and R2-like were 4.7 and 3.8 cM distant in the primary maps constructed from analysing 233 and 78 individuals, but 7.1 and 6.4 cM distant in the high-resolution maps which comprised 1,383 and 1,582 individuals, respectively (Park et al. 2005b, c).
Many characterized R genes are clustered in the same genomic region and members of a cluster can confer resistance to not only one pathogen, but also different pathogens (reviewed by Gebhardt and Valkonen 2001; Hulbert et al. 2001). Two different R genes, Gpa2 conferring resistance to potato cyst nematode and Rx conferring resistance to potato virus X derived from the same ancestor showed physically tight linkage (van der Rouppe Voort et al. 1999) and even one gene, Mi-1 conferred resistance to a nematode, potato aphid and white fly (Milligan et al. 1998; Rossi et al. 1998; Vos et al. 1998; Nombela et al. 2003). In some cases, the homology shared between R genes has facilitated the cloning of R genes such as R3a (Huang et al. 2005) and Rpi-blb2 (van der Vossen et al. 2005). Clustering of R genes with different specificities has also been demonstrated for the Cf-2 and Cf-5 genes for resistance to two distinct isolates of the fungus Cladosporium fulvum (Dixon et al. 1996, 1998), for R3a, R3b, and R5-R11 to P. infestans (Huang et al. 2004; Huang 2005) and R2, R2-like, Rpi-abpt, and Rpi-blb3 to P. infestans (Li et al. 1998; Park et al. 2005a, b, c). This led van der Vossen et al. (2005) to conclude that intragenic recombination or unequal crossover has expanded the Rpi-blb2 locus in S. tuberosum compared with that in S. lycopersicon, in agreement with Hulbert et al. (2001). They suggested that R genes showing distinct resistance specificities within the same R gene cluster could be generated by intragenic or intergenic recombination and unequal crossover at R loci. This theory could explain that Rpi-ber1 and Rpi-ber2 identified in this study differ from each other although they are positioned on the genetically similar region of chromosome 10. Another evidence is that, as shown in Table 2 and Fig. 2, alleles linked to both R genes also showed different polymorphisms, indicating that the two R genes are present on distinct origin of haplotypes (chromosomes). However, no evidence has been found to indicate whether Rpi-ber1 identified in this study is same as Rpi-ber or not (Ewing et al. 2000; Rauscher et al. 2006). Although the two genes originate from the same accession of S. berthaultii and are genetically located on the same genomic region, this does not rule out the possibility that they are distinct genes. We also detected several R genes showing different race-specificities in certain populations in our contemporary researches for potato late blight resistance, even though all the R genes belong to the same R gene cluster and some originate from the same accession of certain wild species (unpublished data).
As a part of our continuing efforts to identify sources of late blight resistance, two R genes derived from S. berthaultii have been identified in this study. Based on the results of the mapping positions of the two R genes and different polymorphic patterns of linked markers to the two R genes, we expect that they could be different each other. However, we cannot conclude it because they are also possibly allelic or/and same genes. Ultimately, therefore, we plan to clone these two genes. Additionally we have identified more targets to clone R genes derived from other wild Solanum species. In combination with other Rpi genes, such as Rpi-mcq1 from S. mochiquense (Smilde et al. 2005), we envisage that a polyculture approach (Dangl and Jones 2001) combined with cisgenic GM technology (Schouten et al. 2006a, b) could be applied to achieve durable resistance for potato late blight.
References
Bendahmane A, Kanyuke K, Baulcombe DC (1997) High-resolution genetical and physical mapping of the Rx gene for extreme resistance to potato virus X in tetraploid potato. Theor Appl Genet 95:153–162
Bradshaw JE, Bryan GJ, Lees AK, McLean K, Solomon-Blackburn RM (2006) Mapping the R10 and R11 genes for resistance to late blight (Phytophthora infestans) present in the potato (Solanum tuberosum) R-gene differentials of Black. Theor Appl Genet 112:744–751
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Day JP, Shattock RC (1997) Aggressiveness and other factors relating to displacement of populations of Phytophthora infestans in England and Wales. Eur J Plant Pathol 103:379–391
Dixon M, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84:451–459
Dixon M, Hatzixanthis K, Jones DA, Harrison K, Johns JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10:1915–1925
El-Kharbotly A, Leonards-Schippers C, Huigen DJ, Jacobsen E, Pereira A, Stiekema WJ, Salamini F, Gebhardt C (1994) Segregation analysis and RFLP mapping of the R1 and R3 alleles conferring race-specific resistance to Phytophthora infestans in progeny of dihaploid potato parents. Mol Gen Genet 242:749–754
El-Kharbotly A, Palomino-Sanchez C, Salamini F, Jacobsen E, Gebhardt C (1996) R6 and R7 alleles of potato conferring race-specific resistance to Phytophthora infestans (Mont.) de Bary identified genetic loci clustering with the R3 locus on chromosome XI. Theor Appl Genet 92:880–884
Ewing EE, Simko I, Smart CD, Bonierbale MW, Mizubuti ESG, May GD, Fry WE (2000) Genetic mapping from field of qualitative and quantitative resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii. Mol Breed 6:25–36
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39:79–102
Goodwin SB, Sujkowski LS, Fry WE (1996) Widespread distribution and probable origin of resistance to metalaxyl in clonal genotypes of Phytophthora infestans in the United States and Western Canada. Phytopathology 86:793–800
Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Hawkes JG (1990) The potato: evolution, biodiversity and genetic resources. Belhaven Press, London
Huang S (2005) The discovery and characterization of the major late blight resistance complex in potato: genomic structure, functional diversity and implications. PhD thesis, Wageningen University, Wageningen
Huang S, Vleeshouwers VGAA, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E (2004) The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant Microbe Interact 17:428–435
Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J 42:251–261
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Jansky S (2000) Breeding for disease resistance in potato. Plant Breed Rev 19:69–155
Kuhl JC, Hanneman RE Jr, Havey MJ (2001) Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genomics 265:977–985
Leonards-Schippers C, Gieffers W, Salamini F, Gebhardt C (1992) The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Mol Gen Genet 233:278–283
Li X, van Eck HJ, van der RouppeVoort JNAM, Huigen DJ, Stam P, Jacobsen E (1998) Autotetraploids and genetic mapping using common AFLP markers: the R2 allele conferring resistance to Phytophthora infestans mapped on potato chromosome 4. Theor Appl Genet 96:1121–1128
Malcolmson JF, Black W (1966) New R genes in Solanum demissum Lindl. and their complementary races of Phytophthora infestans (Mont.) de Bary. Euphytica 15:199–203
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root-knot nematode resistance gene Mi from tomato is a member of the leucien zipper nucleotide binding leucine-rich repeat family of plant genes. Plant Cell 10:1307–1319
Naess SK, Bradeen JM, Wielgus SM, Haberlach GT, McGrath JM, Helgeson JP (2000) Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor Appl Genet 101:697–704
Nombela G, Williamson VM, Muniz M (2003) The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant Microbe Interact 16:645–649
Park T-H, Gros A, Sikkema A, Vleeshouwers VGAA, Muskens M, Allefs S, Jacobsen E, Visser RGF, van der Vossen EAG (2005a) The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Mol Plant Microbe Interact 18:722–729
Park T-H, Vleeshouwers VGAA, Hutten RCB, van Eck HJ, van der Vossen E, Jacobsen E, Visser RGF (2005b) High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol Breed 16:33–43
Park T-H, Vleeshouwers VGAA, Huigen DJ, van der Vossen EAG, van Eck HJ, Visser RGF (2005c) Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet 111:591–597
Phumichai C, Ikeguchi-Samitsu Y, Fujimatsu M, Kitanishi S, Kobayashi A, Mori M, Hosaka K (2006) Expression of S-locus inhibitor gene (Sli) in various diploid potatoes. Euphytica 148:227–234
Rauscher GM, Smart CD, Simko I, Bonierbale M, Mayton H, Greenland A, Fry WE (2006) Characterization and mapping of Rpi-ber, a novel potato late blight resistance gene from Solanum berthaultii. Theor Appl Genet 112:674–687
Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95:9750–9754
Rouppe Voort J, Kanyuka K, van der Vossen E, Bendahmane A, Mooijman P, Klein-Lankhorst R, Stiekema W, Baulcombe D, Bakker J (1999) Tight physical linkage of the nematode resistance gene Gpa2 and the virus resistance gene Rx on a single segment introgressed from the wild species Solanum tuberosum subsp. andigena CPC 1673 into cultivated potato. Mol Plant Microbe Interact 12:197–206
Schouten HJ, Krens FA, Jacobsen E (2006a) Cisgenic plants are similar to traditionally bred plants: international regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep 7:750–753
Schouten HJ, Krens FA, Jacobsen E (2006b) Do cisgenic plants warrant less stringent oversight? Nature Biotechnol 24:753
Śliwka J, Jakuczum H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E (2006) The novel, major locus Rpi-phu1 for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Genet 113:685–695
Smilde WD, Brigneti G, Jagger L, Perkins S, Jones JD (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor Appl Genet 110:252–258
Świeżyński KM, Zimnoch-Guzowska E (2001) Breeding potato cultivars with tubers resistant to Phytophthora infestans. Potato Res 44:97–117
Turkensteen LJ (1993) Durable resistance of potatoes against Phytophthora infestans. In: Jacobs Th, Parlevliet JE (eds) Durability of disease resistance. Kluwer Academic Publisher, Dordrecht, pp 115–124
Umaerus V, Umaerus M (1994) Inheritance of resistance to late blight. In: Bradshaw JE, Mackay GR (eds) Potato genetics. CAB International, Oxon, pp 365–401
van der Vossen E, Sikkema A, Hekkert BL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J 36:867–882
van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S (2005) The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J 44:208–222
Vleeshouwers VGAA, van Dooijeweert W, Paul Keizer LC, Sijpkes L, Govers F, Colon LT (1999) A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur J Plant Pathol 105:241–250
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Onstenk J, de Both M, Peleman J, Liharska T, Hontelez J, Zabeau M (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nature Biotechnol 16:1365–1369
Wastie RL (1991) Breeding for resistance. In: Ingram DS, Williams PH (eds) Advances in plant pathology 7: Phytophthora infestans the cause of late blight of potato. Academic Press, New York, pp 193–223
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, TH., Foster, S., Brigneti, G. et al. Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica 165, 269–278 (2009). https://doi.org/10.1007/s10681-008-9784-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-008-9784-4