Abstract
Foxtail millet (Setaria italica), a drought-tolerant plant, is grown in drylands all over the world. However, the molecular basis of drought tolerance in S. italica is not yet understood. Previously, we comprehensively characterised the SiWRKY genes and discovered that SiWRKY89, a homologue of AtWRKY57, had a noticeably higher expression level during dry conditions. In this study, a transgenic experiment was carried out in Arabidopsis to investigate the function of SiWRKY89 in conferring drought tolerance. Phenotypic analysis showed that the root length of seedlings and the survival rates of mature transgenic Arabidopsis were greater than those of the control plants under drought conditions. Additionally, compared to the control plants, the transgenic plants had higher proline content and antioxidant activity. Furthermore, qRT-PCR investigation for abiotic stress-responsive genes revealed that SiWRKY89-overexpressing plants had higher expression levels than their control counterparts. Additionally, the yeast one-hybrid experiment demonstrated that SiWRKY89 could bind to the W-box elements of AtNCED3. By upregulating the downstream gene AtNCED3 and activating the reactive oxygen species scavenging mechanisms, SiWRKY89 overexpression improved Arabidopsis drought tolerance. Thus, we provide a molecular and biochemical basis for drought tolerance and a candidate gene for crop breeding for drought tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to Cramer et al. (2011), drought stress affects crop output in two-thirds of the world’s land and is predicted to get worse with rising temperatures brought on by climate change (Zandalinas et al. 2018). Additionally, the IPCC (2014) estimates that by 2050, the world’s population would have reached over 9 billion, necessitating a 70–100% increase in crop production to meet demand for food (Godfray et al. 2010; Gupta et al. 2020). The challenges in crop breeding in this environment are to boost food production and enhance tolerance to drought stress. Foxtail millet (Setaria italica (L.) P. Beauv.) is mainly cultivated in dry areas, but drought stress during later growth stages (jointing and grain filling stages) can cause grain yield loss (Zhang et al. 2010; Tang et al. 2017). Some millet germplasms are drought tolerant in the seedling development stage but not in the booting stage (Zhang et al. 2010). Using conventional methods to breed drought-tolerant cultivars with higher yields is complex and time-consuming. Therefore, the key is to understand the basis of drought tolerance and identify key genes that can considerably increase plant drought tolerance throughout the growing period.
In order to identify key genes that regulate the drought response pathway in S. italica, several genetic and transcriptomic studies have been performed (Qie et al. 2014; Qin et al. 2020; Wang et al. 2021; Ceasar 2022). Using high-throughput transcriptomic sequencing, whole genome expression profiling studies have revealed numerous drought response transcription factors (TFs), including WRKY, NAC, AP2/ERF, bHLH, and bZIP, which could regulate the expression of several downstream genes (Rabara et al. 2014; Qin et al. 2020).
The WRKY family is an important TF family for plant growth and development. The WRKY domain is characterised by the highly conserved WRKYGQK domain at the N-terminus and the less conserved zinc-finger domain at the C-terminus (Eulgem et al. 2000; Ulker and Somssich 2004; Rushton et al. 2010). The WRKY TFs can regulate the expression of their target genes by binding to the W-box in the target gene promoter (Ulker and Somssich 2004). The WRKY family members have been characterised in various species (Xie et al. 2005; Bencke-Malato et al. 2014; Okay et al. 2014; Zhang et al. 2017); they play important roles in plant growth, development, and biotic and abiotic stress tolerance (Chen et al. 2012; Jiang et al. 2012, 2016; Guo et al. 2022).
The functions of the WRKY genes in response to drought stress have been extensively established. For instance, Jiang et al. (2012) demonstrated that overexpression of AtWRKY57 enhances Arabidopsis’ ability to withstand drought. It was discovered that AtWRKY57 can bind to the promoters of RD29A and NCED3, increasing the expression of those genes. Additionally, it was shown that AtWRKY63 can bind to the RD29A promoter and regulate how plants respond to ABA and drought stress (Ren et al. 2010). In rice, research has shown that drought and heat tolerance can be enhanced by the overexpression of OsWRKY11 driven by an HSP101 promoter (Wu et al. 2009) while overexpression of OsWRKY45 in Arabidopsis enhances the tolerance to drought stress (Tao et al. 2011). Also, by altering the root architecture’s length, SbWRKY30 has been reported to make rice more tolerant to drought stress (Yang et al. 2020). Additionally, it has been suggested that the WRKY genes from a few other non-model crops, including soybean, sorghum, and wheat, are crucial for drought tolerance. For instance, GsWRKY20 overexpression in soybean enhances Arabidopsis’ ability to withstand drought by regulating the wax biosynthesis process (Luo et al. 2013), while constitutive expression of wheat TaWRKY2 confers salt and drought tolerance in transgenic Arabidopsis by directly binding and activating the drought-responsive gene RD29B (Niu et al. 2012).
Setaria italica has high water use efficiency, and it is considered an excellent model for investigating the WRKY regulation mechanism of drought tolerance (Lata et al. 2010). We previously performed genome-wide identification and expression profiling of SiWRKY genes against drought stress (Zhang et al. 2017). SiWRKY89, a homologous gene of Arabidopsis gene AtWRKY57, was found to be upregulated under drought stress. In this study, the role of SiWRKY89 in drought tolerance was examined by exploring the effects of overexpressing the gene in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Seeds of Yugu1 (a sequenced S. italica cultivar) were grown in a greenhouse as described in our previous study (Zhang et al. 2017). The seeds of Nicotiana benthamiana and Arabidopsis thaliana (Columbia-0) were obtained from the Tobacco Research Institute of the Chinese Academy of Agricultural Sciences, Qingdao, China. Nicotiana benthamiana and Arabidopsis seeds were sown in pots containing soil and vermiculite, and grown in a greenhouse at 22 °C under a 16 h photoperiod, with the relative humidity set to 70%.
Bioinformatic analysis of SiWRKY89
The full protein sequence of WRKY TFs from Arabidopsis and S. italica were obtained and used in a multiple sequence alignment following the method described in our previous study (Zhang et al. 2017). A phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis (MEGA) 5.0 program. Bootstrap values were calculated with 1000 iterations. The amino acid sequences of SiWRKY89 and AtWRKY57 were aligned with BioEdit software (Tom et al. 2011). To predict the regulatory mechanisms in which SiWRKY89 is involved, potential regulatory elements within the 2 kb promoter region upstream of the SiWRKY89 start codon were analysed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Subcellular localisation of SiWRKY89
The coding sequence of SiWRKY89 (without stop codon) with restriction sites KpnI and BamHI was amplified from the cDNA of Yugu1 with the specific primers SiWRKY89 CDS-F and SiWRKY89 CDS-R (Table S1). The amplified fragment was then digested with KpnI and BamHI and cloned into the corresponding sites of the pEGFP vector to generate the recombinant construct SiWRKY89-GFP driven by CaMV35S promoter. The constructed SiWRKY89-GFP vector was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The transgenic Agrobacterium cells were injected into tobacco leaves. After 2 to 3 days, the injected leaves were pre-stained with 4′,6-diamino-2-phenylindole (DAPI) at 5 mg/mL for 10 min. Then the fluorescence signals in the leaves were observed under the Leica TCS SP8 laser scanning confocal microscope (Mannheim, Germany).
RNA isolation and qRT-PCR analysis
The total RNA was isolated from S. italica and Arabidopsis using a Plant Total RNA Isolation Kit (Tiangen, Beijing, China) and treated with RNase-free DNase I (RQ1, Promega, Madison, WI, USA). The cDNA was synthesised using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) with oligo(dT)18 (Promega) following the manufacturer’s protocol. The cDNA samples were analysed using quantitative real-time polymerase chain reaction (qRT-PCR) on a StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermocycling conditions were as follows: 95 °C for 5 min, followed by 40 cycles (95 °C for 15 s and 60 °C for 1 min). Three biological replicates were used. The primers used for qRT-PCR are listed in Table S1. A constitutive SiACTIN (Seita.8G043100) was used as an internal reference gene for F. millet (Pan et al. 2018), and AtTUBULIN for Arabidopsis (Xu et al. 2017). Relative gene expression values were determined using the 2−ΔΔCt method (Livak and Schmittgen 2001).
35S:SiWRKY89 vector construction and Arabidopsis transformation
To create the 35S:SiWRKY89 construct, the coding sequence of SiWRKY89 with restriction sites NcoI and BstEII was amplified from the cDNA of Yugu1 with the specific primers 35SSiWRKY89-F and 35SSiWRKY89-R (Table S1). The amplified fragment was digested with NcoI and BstEII, and cloned into the corresponding sites of the pCAMBIA1302 vector to generate the recombinant construct 35S:SiWRKY89 driven by the CaMV35S promoter. The 35S:SiWRKY89 vector was transformed into A. tumefaciens strain GV3101 by electroporation, and then transformed into Arabidopsis using the floral dipping method (Clough and Bent 1998). The T0 transformants were screened on 1/2 MS medium with hygromycin (50 μg/mL). Ten T1 transgenic lines were generated and confirmed using semi-quantitative PCR with the primers of SiWRKY89 CDS-F and SiWRKY89 CDS-R. The AtACTIN2 gene was used as the reference gene (Xu et al. 2020). The seeds of T1 generation were screened on 1/2 MS medium containing hygromycin. (50 μg/mL) to select the T2 generation. The same process was repeated with seeds of T2 generation to select T3 generation. Three T3 generation transgenic lines (W1-3, W3-5, and W6-3) with 100% resistance to hygromycin were considered homozygous lines and used in the subsequent experiments.
Drought treatments of transgenic Arabidopsis lines
For drought treatments in the seedling stage, the seeds of wild-type (WT), W1-3, W3-5, and W6-3 were sown onto 1/2 MS medium. After growing for 7 days, the seedlings were transferred to square petri dishes containing 1/2 MS as controls and to some dishes containing 1/2 MS medium with 300 mM mannitol (to induce osmotic stress). After 7 days of culture, the root length of the seedlings was recorded. The seedlings were transplanted into pots containing soil and vermiculite, at four seedlings per pot, under normal watering conditions in three replicates. After 3 weeks, the plants were dehydrated by withdrawing water for 7 days. At 3 days after re-watering the plants, we evaluated the survival rates of the transgenic and WT plants.
Measurement of water loss rate, proline content, and peroxidase and superoxide dismutase activities
The water loss rate, proline content, superoxide dismutase (SOD) activity, and peroxidase (POD) activity of the drought-treated transgenic plants were measured. Arabidopsis leaves were collected during drought treatment. To measure the water loss rate, 20 rosette leaf from five plants were detached and weighed at four-time points over a 2 h period (He et al. 2016). The mixture of rosette leaves derived from 10 WT individuals or transgenic plants (0.1 g) under drought stress were used to evaluate proline content, and SOD and POD activities (Huang et al. 2014). The experiments were performed using appropriate kits following the manufacturer’s instructions (A107-1-1, A001-3-2, and A084-3-1; Jiancheng, Nanjing, China). Three biological replicates were performed for each experiment.
Yeast one-hybrid assay
For the protein–DNA-binding experiment, an oligonucleotide sequence containing triple tandem copies of the W-box and mW-box was synthesised, annealed, and cloned into the pHIS2.1 vector forming W-box- and mW-box-specific reporter vectors. The full-length CDS sequence of SiWRKY89 was cloned and fused with the transcription activating domain (GAL4) of the pGADT7 vector (Clontech, Santa Clara, CA, USA). To determine the interaction between DNA and protein, pGADT7 and pGADT7-SiWRKY89 were co-transformed with the reporter vectors, pHIS2.1-Wb and pHIS2.1-mWb, into the Y187 strain following the manufacturer’s instructions (Clontech). All co-transformed yeast cells grown on SD/-Trp/-Leu were re-streaked on SD/-Trp/-Leu/-His with 30 mM 3-AT (3-amino-1,2,4-triazole) to examine protein–DNA interaction.
Statistical analysis
All experiments were repeated thrice and experimental data of all parameters were analysed using one-way analysis of variance in SPSS version 17.0 software (SPSS, Chicago, IL, USA). The data are shown as mean ± standard deviation (SD). Significant differences between the means were identified using Duncan’s multiple range test at P ≤ 0.05.
Results
Bioinformatic analysis of SiWRKY89
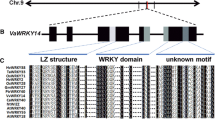
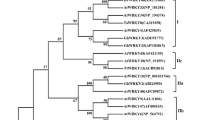
The transcript sequence of SiWRKY89 is 1300 bp long, and SiWRKY89 is composed of 207 amino acids. Phylogenetic analysis revealed that SiWRKY89 was classified into the same phylogenetic clade with AtWRKY57 (Fig. 1A). SiWRKY89 and AtWRKY57 showed a 22.3% identity at the amino acid level, however, they both contained the conserved WRKY domain WRKYGQK at N-terminal and the zinc-finger-like motif at C-terminal (Fig. 1B). In addition, promotor cis-elements analysis revealed that drought response cis-elements, such as ABA-responsive element, ARE, and MBS, and elements for hormone response, such as CGTCA-box, TGA-element, and TGACG-motif, were identified in the 2000 bp SiWRKY89 promoter (Table 1). The results indicate that SiWRKY89 regulates plant drought response and development possibly through hormone regulation.
Setaria italica and Arabidopsis’s WRKY gene family’s phylogenetic tree (A) and alignment study of AtWRKY57 and SiWRKY89 (B). SiWRKY89 and AtWRKY57 are denoted with a red arrow in the phylogenetic tree while the zinc-finger-like motif (Eulgem et al. 2000) is denoted by a red circle, and the conserved WRKY domain WRKYGQK of SiWRKY89 and AtWRKY57 is shown in a red box
SiWRKY89 subcellular localisation
The subcellular localization of SiWRKY89 was detected with a confocal microscope. The green fluorescence signal of SiWRKY89-GFP was detected in the nucleus, which was marked by DAPI staining of the nucleus-targeted control (Fig. 2), which suggests that SiWRKY89 is a nuclear protein.
Root length of 35S:SiWRKY89 transgenic lines under drought stress
The function of SiWRKY89 was investigated by ectopically expressing the gene under the CaMV35S promoter in Arabidopsis. The semi-quantitative PCR results revealed that the SiWRKY89 gene was successfully integrated into the genome of transformed Arabidopsis (Fig. 3C). The expression of SiWRKY89 in three homozygous transgenic lines (W1-3, W3-5, and W6-3) was significantly higher than the background level in the Yugu1 leaves (Fig. 3D). The root growth of WT and transgenic Arabidopsis plants were comparable on 1/2 MS medium (Fig. 3A). Although the root length of W1-3 (2.47 cm) was slightly longer than that of the WT (2.43 cm) on the medium containing mannitol, W3-5 (3.15 cm) and W6-3 (2.69 cm) plants developed significantly longer roots on mannitol (Fig. 3B, E).
Root length of 35S:SiWRKY89 transgenic lines under drought stress. A 1/2 MS medium; B 1/2 MS medium supplemented with 300 mM mannitol; C semi-quantitative PCR analysis of SiWRKY89 in WT and transgenic lines (W1-3, W3-5, and W6-3); D qRT-PCR expression analysis of SiWRKY89 in Yugu1 leaves (CK) and transgenic lines (W1-3, W3-5, and W6-3). E Root length of 35S:SiWRKY89 transgenic lines under drought stress. The data are shown as mean ± SD of three replicates
Survival rate and physiological indices of 35S:SiWRKY89 transgenic lines under drought
The survival rate of transgenic and WT plants was compared in order to look at the survival rate of 35S:SiWRKY89 transgenic plants under drought stress. The findings demonstrated that after 1 week of no watering, the leaves of the WT showed chlorosis and withered, whereas just a few leaves of the transgenic plants withered (Fig. 4A). Furthermore, after resuming watering, only 30% of the WT plants resumed growth, whereas over 90% of the transgenic plants resumed growth (Fig. 4B).
Analysis of drought tolerance of 35S:SiWRKY89 transgenic lines under drought stress. WT and transgenic lines (W1-3, W3-5, and W6-3) were A under drought stress for 1 week and B 3 days after re-watering; C The water loss rate of WT and transgenic lines (W1-3, W3-5, and W6-3). D The proline content in WT and transgenic lines (W1-3, W3-5, and W6-3). E SOD activity in WT and transgenic lines (W1-3, W3-5, and W6-3); F POD activity in WT and transgenic lines (W1-3, W3-5, and W6-3). The data are shown as mean ± SD of three replicates
The physiological parameters of the WT and transgenic plants under drought stress, including the water loss rate, proline content, and SOD and POD activities, were compared in order to understand the mechanism behind the increased drought tolerance of 35S:SiWRKY89 transgenic plants. After detaching over a 2-h period, the leaves of the WT plants showed a higher water loss rate than those of the transgenic lines (Fig. 4C). Additionally, transgenic plants had proline content and SOD and POD activities that were 2–3fold higher than those of WT plants (Fig. 4D-F), demonstrating the critical role played by osmotic substances and the antioxidant system in the 35S:SiWRKY89 transgenic plants’ responses to drought stress.
Expression of drought-responsive genes in 35S:SiWRKY89 transgenic lines under drought stress
To further understand the molecular mechanism underlying enhanced stress tolerance in 35S:SiWRKY89 transgenic plants, the expression of four genes, AtSOD1, AtPOD, AtP5CS, and AtNCED3 was evaluated in the WT and transgenic Arabidopsis plants under drought stress. In transgenic lines compared to WT plants, the transcript levels of AtSOD1, AtPOD, and AtP5CS were 2–6 fold higher. In particular, the expression of AtP5CS was significantly increased in W1-3 (Fig. 5A–C), which is consistent with the high levels of proline and antioxidants accumulated in transgenic plants. In addition, we found that the three transgenic lines had considerably higher levels of the AtNCED3 gene, a crucial enzyme in the ABA biosynthesis pathway (Fig. 5D), suggesting that SiWRKY89 may be controlling this gene.
qRT-PCR expression analysis of four stress-responsive genes. The expression level of four stress-responsive genes AtNCED3, AtSOD1, AtPOD and AtP5CS under drought stress was analysed in WT and three homozygous transgenic lines (W1-3, W3-5, and W6-3). Relative gene expression values were determined using the 2−ΔΔCt method. The data are shown as mean ± SD of three replicates. AtTUBULIN was used as an internal control to normalise the data. Values are mean ± SD
Determination of DNA-binding activity of SiWRKY89 using the yeast one-hybrid assay
To determine the DNA-binding activity of SiWRKY89, the yeast one-hybrid assay was performed. All yeast cells harbouring the two kinds of plasmids grew well on SD/-Trp/-Leu medium (Fig. 6) suggesting the success of co-transformation. On SD/-Trp/-Leu/-His medium plus 30 mM 3-AT, the cells containing pHIS2.1-Wb plus pGADT7-SiWRKY89 vectors grew well, and pHIS2.1-mWb plus pGADT7-SiWRKY89 did not grow (Fig. 6), suggesting that SiWRKY89 could interact with W-box rather than mW-box to induce the transcription of HIS3. In addition, the negative control (pHIS2.1-Wb/pGADT7 and pHIS2.1-mWb/pGADT7) did not grow on SD/-Trp/-Leu/-His medium plus 30 mM 3-AT.
DNA-binding assay of SiWRKY89. A The sequence of the triple tandem copies of the W-box and mW-box binding elements. B Yeast one-hybrid assay using the W-box or mW-box as bait. Yeast cells were co-transformed with the plasmid combination of pHIS2.1-Wb/pGADT7, pHIS2.1-Wb/pGADT7-SiWRKY89, pHIS2.1-mWb/pGADT7, and pHIS2.1-mWb/pGADT7-SiWRKY89 on SD/-Trp/-Leu or SD/-Trp/-Leu/-His + 30 mM 3-AT plates
Discussion
Abiotic factors, especially drought stress, severely limit crop production worldwide (Godfray et al. 2010; Gupta et al. 2020). WRKY transcription factors are a large family of transcription factors in plants (Thomas et al. 2000), and the functions of WRKYs in growth and development have been extensively explored in a variety of plants (He et al. 2016). Numerous earlier investigations also reported on the various functions that WRKY transcription factors play in response to biotic and abiotic stressors (Wu et al. 2009; Ren et al. 2010; Tao et al. 2011; Chen et al. 2012; Niu et al. 2012; Luo et al. 2013; Rabara et al. 2014; Jiang et al. 2016; Yang et al. 2020). The role of the Setaria italica WRKY genes in the response to drought stress hasn’t, however, received much thorough research. SiWRKY89 was revealed to be up-regulated under drought stress when we previously did genome-wide identification and expression profiling of SiWRKY (Zhang et al. 2017).In the present study, the role of SiWRKY89 in drought tolerance was examined by exploring the effects of overexpressing the gene in Arabidopsis. The 35S:SiWRKY89 transgenic seedlings displayed longer roots under mannitol stress (Fig. 3B, E), and presented a higher survival rate than WT plants under drought stress conditions (Fig. 4A, B). The ABA biosynthesis was activated with the AtNCED3 accumulation (Fig. 5D), and the antioxidant system was also triggered (Fig. 4D-F) in the transgenic plants.
The functions of the WRKY genes in response to drought stress have been extensively established. For instance, Jiang et al. (2012) demonstrated that overexpression of AtWRKY57 enhances Arabidopsis’ ability to withstand drought. Also, by altering the root architecture’s length, sorghum SbWRKY30 has been reported to make rice more tolerant to drought stress (Yang et al. 2020). In this study, foxtail millet SiWRKY89 over-expression plants showed longer roots under mannitol stress in comparison with wild type (Fig. 3B, E). AtWRKY57 was highly expressed in rosette leaves (Jiang et al. 2012), but the expression of SiWRKY89 was much higher in roots than in leaves and flowers (Zhang et al. 2017), and SbWRKY30 is highly expressed in sorghum taproots (Yang et al. 2020). Notably, sorghum and foxtail millet are both natural stress tolerant crops. However, further studies are required to determine the mechanism by which SiWRKY89 regulates root length.
A great deal of evidence has shown that abiotic stresses such as drought, cold, salinity, heat, and light could induce ROS generation in plant cells (Huang et al. 2019). As signalling molecules, ROS trigger signal transduction pathways in response to abiotic stresses and play key roles in the acclimation process of plants to abiotic stresses (Sarvajeet et al. 2010). However, more ROS accumulation could cause irreversible cellular damage through their strong oxidative properties. Antioxidant enzymes such as SOD, POD, CAT, and APX, et al. were regarded as the enzymatic system to scavenge excess ROS (Huang et al. 2019). The function of antioxidants in ROS scavenging were reviewed well in many reviews (Huang et al. 2019; Sarvajeet et al. 2010). In our study, the antioxidants SOD and POD, and the encoding genes AtSOD1, AtPOD were all up-regulated in transgenic plants (Fig. 4E, F; Fig. 5A, B), which was consistent with previous reports (Huang et al. 2019; Sarvajeet et al. 2010). Proline is an important osmolyte accumulated in plants under drought stress (Yoshiba et al. 1997; Kavi Kishor et al. 2014). It has been reported that an increased proline content could enhance drought tolerance in various plants overexpressing WRKY genes. For example, higher accumulation of proline has been shown to enhance the drought tolerance of AtWRKY57 transgenic rice (Jiang et al. 2016). Overexpression of MuWRKY3 from groundnut led to the accumulation of higher proline levels under drought stress (Kiranmai et al. 2018). In line with these earlier findings, we found that SiWRKY89 overexpression led to a large increase of the proline-encoding gene AtP5CS (Fig. 5C) and proline content (Fig. 4D), and that these OX-plants had high drought tolerance. Thus, we came to the conclusion that SiWRKR89 overexpressing plants’ high drought tolerance was likely brought on by an abundance of antioxidant enzymes and proline, which lessened ROS damage. This is further supported by Nishizawa et al. (2008) who reported that proline performs the same role as antioxidant enzymes in scavenging reactive oxygen species.
By attaching to the W-box cis-element in the target gene promoter, WRKY TFs can control the expression of target genes (Rushton et al. 2010). The key enzyme gene in the ABA biosynthesis pathway is AtNCED3 (Woo et al. 2011). According to a previous study, AtWRKY57 could directly bind to the W-box of the AtNCED3 promoter sequences, which led to high levels of AtNCED3 expression and ABA content in transgenic plants with good drought tolerance (Jiang et al. 2012). SiWRKY89 and AtWRKY57 share the same WRKY conserved domain, although having an amino acid identity of only 22.30 percent (Fig. 1). Importantly, the yeast one-hybrid assay revealed that SiWRKY89 could bind to the W-box elements of AtNCED3 (Fig. 6), indicating that SiWRKY89 could bind to the promoter of AtNCED3 and regulate the plants’ response to drought stress in a manner similar to that of AtWRKY57 (Jiang et al. 2012).
Conclusions
We elucidated the role of SiWRKY89 in drought tolerance by exploring the consequences of overexpressing this gene in Arabidopsis. The functional analysis revealed that overexpressing SiWRKY89 enhanced drought tolerance in Arabidopsis by regulating the downstream gene of AtNCED3 and activating the antioxidant system. Our study provides valuable information for analysing the basis of drought tolerance of S. italica and a candidate gene for breeding drought tolerant crops.
Data availability
Not applicable.
References
Bencke-Malato M, Cabreira C, Wiebke-Strohm B, Bücker-Neto L, Mancini E, Osorio MB, Homrich MS, Turchetto-Zolet AC, De Carvalho MC, Stolf R, Weber RL, Westergaard G, Castagnaro AP, Abdelnoor RV, Marcelino-Guimarães FC, Margis-Pinheiro M, Bodanese-Zanettini MH (2014) Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol 14:236
Ceasar SA (2022) Foxtail millet (Setaria italica) as a model system to study and improve the nutrient transport in cereals. Plant Growth Regul. https://doi.org/10.1007/s10725-022-00878-x
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:163
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812–818
GuoY HD, Wang X, Wang H, Wu Z, Yang P, Zhang B (2022) Comparative transcriptomics reveals key genes contributing to the differences in drought tolerance among three cultivars of foxtail millet (Setaria italica). Plant Growth Regul. https://doi.org/10.1007/s10725-022-00875-0
Gupta A, Rico-Medina A, Caño-Delgado AI (2020) The physiology of plant responses to drought. Science 368:266–269
He G, Xu J, Wang Y, Liu J, Li P, Chen M, Ma Y, Xu Z (2016) Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol 16:116
Huang X, Wang W, Zhang Q, Liu J (2014) A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162:1178–1194
Huang H, Farhan U, Zhou D, Yi M, Zhao Y (2019) Mechanisms of ROS regulation of plant development and stress responses. Front Plant Sci 10:800
IPCC Climate Change Synthesis Report Contribution of Working Groups I. II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC, Geneva. 151 2014
Jiang YJ, Liang G, Yu DQ (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5:1375–1388
Jiang Y, Qiu Y, Hu Y, Yu D (2016) Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front Plant Sci 7:145
Kavi Kishor PB, Sreenivasulu N (2014) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37:300–311
Kiranmai K, Lokanadha Rao G, Pandurangaiah M, Nareshkumar A, Amaranatha Reddy V, Lokesh U, Venkatesh B, Anthony Johnson AM, Sudhakar C (2018) A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front Plant Sci 9:346
Lata C, Sahu PP, Prasad M (2010) Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem Biophys Res Commun 393:720–727
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Luo X, Bai X, Sun X, Zhu D, Liu B, Ji W, Cai H, Cao L, Wu J, Hu M, Liu X, Tang L, Zhu Y (2013) Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot 64:2155–2169
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263
Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ 35:1156–1170
Okay S, Derelli E, Unver T (2014) Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol Genet Genomics 289:765–781
Pan J, Li Z, Wang Q (2018) Comparative proteomic investigation of drought responses in foxtail millet. BMC Plant Biol 18(1):315
Qie L, Jia G, Zhang W, Schnable J, Shang Z, Li W, Liu B, Li M, Chai Y, Zhi H, Diao X (2014) Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica×Setaria viridis. PLoS ONE 9:e101868
Qin L, Chen E, Li F, Yu X, Liu Z, Yang Y, Wang R, Zhang H, Wang H, Liu B, Guan Y, Ruan Y (2020) Genome-wide gene expression profiles analysis reveal novel insights into drought stress in foxtail millet (Setaria italica L.). Int J Mol Sci 21:8520
Rabara RC, Tripathi P, Rushton PJ (2014) The potential of transcription factor-based genetic engineering in improving crop tolerance to drought. OMICS 18:601–614
Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63:417–429
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Sarvajeet S, Narendra Tu (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Tang S, Li L, Wang Y, Chen Q, Zhang W, Jia G, Zhi H, Zhao B, Diao X (2017) Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses). Sci Rep 7:10009
Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62:4863–4874
Thomas E, Paul J, Silke R, Imre E (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Tom H, Ibis B, Carlsbad Ca (2011) BioEdit: an important software for molecular biology. GERF Bull Biosci 2(1):60–61
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Wang H, Hao D, Wang X, Zhang H, Yang P, Zhang L, Zhang B (2021) Genome-wide identification and expression analysis of the SNARE genes in Foxtail millet (Setaria italica) reveals its roles in drought stress. Plant Growth Regul 95:355–369
Woo D, Park H, Kang I, Lee S, Moon B, Lee C, Moon Y (2011) Arabidopsis lenc1 mutant displays reduced ABA accumulation by low AtNCED3 expression under osmotic stress. J Plant Physiol 168:140–147
Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K (2009) Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep 28:21–30
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189
Xu Z, Wang M, Shi D, Zhou G, Niu T, Hahn G, O’Neill M (2017) DGE-seq analysis of MUR3-related Arabidopsis mutants provides insight into how dysfunctional xyloglucan affects cell elongation. Plant Sci 258:156–169
Xu Z, Marowa P, Liu H, Du H, Zhang C, Li Y (2020) Genome-wide identification and analysis of P-type plasma membrane H+-ATPase sub-gene family in sunflower and the role of HHA4 and HHA11 in the development of salt stress resistance. Genes 11:361
Yang Z, Chi X, Guo F, Jin X, Luo H, Hawar A, Chen Y, Feng K, Wang B, Qi J, Yang Y, Sun B (2020) SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J Plant Physiol 246–247:153142
Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol 38:1095–1102
Zandalinas SI, Mittler R, Balfagón D, Arbona V, Gómez-Cadenas A (2018) Plant adaptations to the combination of drought and high temperatures. Physiol Plant 162:2–12
Zhang W, Zhi H, Liu B, Peng H, Li W, Wang Y, Li H, Li Y, Diao X (2010) Indexes screening for drought resistance test of foxtail millet. J Plant Genet Resour 11:560–565
Zhang L, Shu H, Zhang AY, Liu BL, Xing GF, Xue JA, Yuan LX, Gao CY, Li RZ (2017) Foxtail millet WRKY genes and drought stress. J Agric Sci 155:777–790
Acknowledgements
This work was financially supported by the National Key Research and Development Program (2020YFD1000803-2); State Key Laboratory of Sustainable Dryland Agriculture (202105D121008-2-5); the Natural Science Foundation of China (NSFC) (31860409; 32060509); Natural Science Foundation of Shanxi Province (201901D111221); Key Research and Development Program of Shanxi Province (201803D221019-1); Research Program Sponsored by Shanxi Key Laboratory of Minor Crops Germplasm Innovation and Molecular Breeding, Shanxi Agricultural University (202105D121010-06); Demonstration and Guidance Program for Technology People-Benefit in Qingdao (20-3-4-7-nsh), and the Agricultural Science and Technology Innovation Program (ASTIP No. CAAS-ZDRW202201).
Funding
The State Key Laboratory of Sustainable Dryland Agriculture, 202105D121008-2-5, Li Zhang, Natural Science Foundation of Jilin Province,31860409, Li Zhang,32060509, Li Zhang, Key Technologies Research and Development Program,2020YFD1000803-2, AiYing Zhang, Natural Science Foundation of Shanxi Province, 201901D111221, Li Zhang
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Communicated by Zhen Liang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, A., Zhang, L., Guo, E. et al. Setaria italica SiWRKY89 enhances drought tolerance in Arabidopsis. Plant Growth Regul 99, 125–135 (2023). https://doi.org/10.1007/s10725-022-00916-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-022-00916-8