Abstract
Key message
Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes, including COR15A, COR15B, COR413, KIN2, and RD29A.
Abstract
The WRKY family is one of a largest transcription factors in plants, and it is a key component of multiple stress responses. In this study, the drought- and cold-induced WRKY family gene VaWRKY14 was isolated and characterized. Phylogenetic analysis indicated that VaWRKY14 belongs to the WRKY IIa subfamily, of which several members participate in biotic and abiotic stress responses in plants. Fluorescence observation from Arabidopsis mesophyll protoplasts transformed with the VaWRKY14::eGFP fusion vector suggested that VaWRKY14 was localized in the nucleus. The VaWRKY14 in yeast cells did not display any transcriptional activity. The expression of VaWRKY14 could be induced by exogenous phytohormones, including salicylic acid (SA) and abscisic acid (ABA). Overexpression of VaWRKY14 enhanced the drought tolerance of transgenic Arabidopsis. Compared with wild-type Arabidopsis, the VaWRKY14-OE lines exhibited higher water content and antioxidant enzyme activities in leaves after drought treatment. RNA sequencing analysis revealed that several stress-related genes, including COR15A, COR15B, COR413, KIN2, and RD29A, were upregulated in transgenic plants relative to their expression in wild-type Arabidopsis under normal conditions. Several genes (3 upregulated and 49 down-regulated) modulated by VaWRKY14 were also affected by drought stress in wild-type plants. These data suggest that VaWRKY14 responds to drought and cold stresses and that drought tolerance may be enhanced by regulating the expression of stress-related genes in Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought is an adverse factor that restricts plant growth, survival, and distribution. To adapt to drought stress, plants have evolved various physiological and biochemical processes, including regulation of stomatal closure, accumulation of osmolytes (e.g., proline, soluble sugar, and glycine betaine), and scavenging of reactive oxygen species (Shinozaki et al. 2003). Transcriptional regulation mechanisms also play critical roles in plant response to drought stress. Transcriptome modifications that occur during drought stress in plants have been extensively identified. Transcription factors (TFs), such as DREB, NAC, ERF, MYB, and WRKY, play important roles during drought stress response in plants (Dubouzet et al. 2003; Fang et al. 2016; Pan et al. 2012; Cominelli et al. 2005; Wang et al. 2015).
WRKY is one of the largest TF family in plants. Members of this gene family contain a DNA-binding region (WRKY domain) of approximately 60 amino acids in length, which comprises a conserved heptad WRKYGQK amino acid motif adjacent to a zinc-finger motif (Ülker and Somssich 2004). In view of these features of the WRKY domain, genes in this family were classified into three groups (Eulgem et al. 2000). The WRKY II group can be further divided into five subgroups (IIa, IIb, IIc, IId, and IIe) on the basis of their primary amino acid sequences. Electrophoretic mobility shift assay confirmed that WRKY preferentially binds to a markedly conservative DNA motif named the W box (T/CTGACC/T, Rushton et al. 2010). Since the first WRKY TF (SPF1) was cloned from sweet potato (Ipomoea batatas), an increasing number of WRKY family genes have been identified in several species, including Arabidopsis, rice, and grape (Ishiguro and Nakamura 1994; Dong et al. 2003; Wu et al. 2005; Wang et al. 2014).
WRKY genes play pivotal roles in stress responses (Pandey and Somssich 2009, 2010) and developmental processes, such as senescence, flowering, seed dormancy, and biosynthetic pathway regulation (Xu et al. 2004; Guillaumie et al. 2010; Besseau et al. 2012; Ding et al. 2014; Yu et al. 2016). Recent evidence has suggested that WRKY genes play important roles in abiotic stress responses. In Arabidopsis, abscisic acid overly sensitive 3 (ABO3), which is a WRKY TF, mediates ABA-related signaling pathways and negatively regulates drought tolerance (Ren et al. 2010). GmWRKY27 interacts with GmMYB174 to reduce the expression of GmNAC29, which is a negative factor regulating drought tolerance in soybean (Wang et al. 2015). FcWRKY70 from Fortunella crassifolia functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene (Gong et al. 2015). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice (Shen et al. 2012). In addition, WRKY TFs may serve as nodes of different stress-mediated signaling pathways. In Arabidopsis, WRKY46 plays dual roles of regulating plant responses to drought and salt stress as well as facilitating light-dependent stomatal opening in guard cells (Ding et al. 2014). OsWRKY76 in rice acts as a transcriptional repressor that plays a negative role in blast disease resistance and a positive role in cold stress tolerance (Yokotani et al. 2013). Overexpression of OsWRKY45 enhances salt and drought tolerance while increasing disease resistance (Qiu and Yu 2009), demonstrating the multiple roles of WRKY genes during the cross talk among different stress responses in plants.
In view of the key roles of WRKY genes in plant stress responses, the WRKY gene family has been extensively analyzed, and 59 members have been identified in grapevine (Wang et al. 2014). Based on the previous study in our lab (Wang et al. 2014), a WRKY gene that responds to drought and cold stresses was cloned from Vitis amurensis and named as VaWRKY14. In the present study, we analyzed the transcription patterns of VaWRKY14 in response to abiotic stresses (drought and cold) and under exogenous phytohormone (SA and ABA) treatments. Yeast assay and Arabidopsis mesophyll protoplasts were used to analyze the transcriptional activity and subcellular location of VaWRKY14, respectively. Moreover, VaWRKY14-overexpressing lines were constructed in Arabidopsis to examine the effects of VaWRKY14 during cold and drought stresses. In addition, RNA sequencing (RNA-seq) analyses were performed on transgenic lines and wild-type (WT) plants under normal and drought conditions to elucidate the molecular mechanisms underlying the enhanced stress tolerance by VaWRKY14. The results of this study suggest that VaWRKY14 plays a positive role in plant drought responses and can be an important candidate gene in breeding new cultivars with excellent drought tolerance.
Materials and methods
Plant materials and growth conditions
V. amurensis was collected from Changbai Mountain in Jilin Province in China and micropropagated on half-strength (1/2) Murashige and Skoog (MS) medium containing 3% sucrose and 0.7% agar in a growth chamber at a constant temperature of 26 °C under a 16 h light/8 h dark photoperiod and 100 µmol m− 2 s− 1 light intensity. Seeds of Arabidopsis (ecotype Columbia Col-0) were vernalized at 4 °C in the dark for 2 days and sown in the same pots with equiponderate substrates in a growth chamber at 22 °C under a 16 h light/8 h dark cycle and 60% relative humidity.
ORF cloning and phylogenetic analysis
The coding region of VaWRKY14 was amplified from the cDNA of V. amurensis using a specific primer (5ʹ-GCTGAGTTTGATGGCTATGGAT-3ʹ/5ʹ-CCGTCTTAACATTGGACTTGG-3ʹ). Homologous proteins were searched in the NCBI database utilizing the BLASTp program, and multiple alignments were performed using Clustal X with the deduced amino acid sequences. A phylogenetic tree was constructed through the neighbor-joining method on MEGA5 with 1000 bootstrap replicates (Hall 2013).
Subcellular location of VaWRKY14 in Arabidopsis protoplasts
The open reading frame (ORF) of VaWRKY14 was amplified by conducting polymerase chain reaction (PCR) using primers (5′-CGCTCTAGAATGGCTATGGATAGTTCTAATTGG-3′ and 5′-GGGGGTACCCCATTTTTCAGTTTGATTGCG-3′) without the stop codon. The amplified fragment was ligated into the pEZS-NL-EGFP vector to construct the plasmid pEZS-NL-VaWRKY14-EGFP, which was introduced into the Arabidopsis protoplasts cells by the PEG-mediated protocol (Yoo et al. 2007; Wu et al. 2009). After culturing in the dark at 25 °C, the GFP was localized with a confocal laser scanning microscope (TCS SP8, Leica, Germany) at an excitation wavelength of 488 nm and an emission wavelength of 507 nm.
Transactivation assay of VaWRKY14
The coding region of VaWRKY14 was sub-cloned into the pGBKT7 vector fused with the GAL4 DNA-binding domain. VaNAC26 was used as the positive control to test the yeast system (Fang et al. 2016). The VaWRKY14/pGBKT7, VaNAC26/pGBKT7, and pGBKT7 vectors were transformed to Y2HGold yeast cells using the Yeastmaker™ yeast transformation system 2 in accordance with the manufacturer’s protocol (Clontech, CA, USA). Y2HGold yeast cells harboring VaWRKY14-pGBKT7, VaNAC26-pGBKT7, and pGBKT7 were cultivated on SD/-Trp, SD/-Trp-His, and SD/-Trp/x-α-gal media at 30 °C for 3 days.
Cold, drought, and exogenous phytohormone treatments
For drought treatment, 40-day-old grapevine plantlets were transferred into liquid medium with an additional 6% polyethylene glycol (PEG) 6000. For cold treatment, the grapevine plantlets were transferred into a growth chamber with a constant temperature of 4 °C. Shoot apexes with one well-developed leaf were harvested at 0, 2, 4, 8, 24, and 48 h after initiating treatments. For exogenous phytohormone treatment, the plantlets were sprayed with 100 µM ABA or 200 µM SA. The samples were harvested at 0, 0.5, 1, 2, 4, and 8 h for ABA treatment and at 0, 2, 4, 8, 24, and 48 h for SA treatment. All samples were frozen in liquid nitrogen and stored at − 80 °C for subsequent total RNA isolation and quantitative real-time PCR (qRT-PCR) analyses.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted using the TIANDZ Column Plant RNAout 2.0 Kit (Tiandz, Beijing, China). Approximately 1 µg of the total RNA per sample was used for cDNA synthesis using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). An SYBR Green-based real-time PCR assay was performed in a total volume of 10 µL reaction mixture containing 5.0 µL of 2 × SYBR Green Master Mix (Roche, Basel, Switzerland), 0.4 µL of 10 mM primer mix, and 1.0 µL of template cDNA. The VvACT gene (GenBank accession no. XM_002282480) was used as the reference gene for normalizing the gene expression in V. amurensis, whereas AtACT2 (GenBank accession: BAH20120.1) was used in Arabidopsis. Three biological and technical replicates were performed, and the relative expression was calculated through the relative quantization method (2− ΔΔCT). All primers used in this study are listed in Table S1.
Arabidopsis transformation
A pair of primers, 5ʹ-GGTACCGCTGAGTTTGATGGCTATGGAT-3ʹ/5ʹ-TCTAGACCGTCTTAACATTGGACTTGG-3ʹ, was designed to amplify the entire coding region of VaWRKY14. The forward and reverse primers contained Kpn I and Xho I sites at the 5ʹ end, respectively. The PCR product was digested with Kpn I and Xho I and inserted into the Kpn I/Xho I-digested pCAMBIA1301. Arabidopsis transformation was performed through the floral dip method (Clough and Bent 1998). Transgenic seedlings were identified through hygromycin resistance screening. Homozygous T3 transgenic lines were used for stress tolerance assessment.
Drought and cold stress tolerance assays of transgenic Arabidopsis
For drought tolerance assessment, seeds of transgenic and WT Arabidopsis were sowed and adequately watered for 18 days. Then, phenotypes were observed after an additional 14 days without watering. When the WT plants exhibited lethal effects of dehydration, watering was resumed, and the plants were allowed to grow for another 5 days. For cold tolerance assessment, 3-week-old Arabidopsis seedlings were exposed to − 1 °C for 8 h and then cooled at a rate of 1 °C h− 1 up to – 11 °C in the dark. After exposure to – 11 °C for 3 h, the seedlings were recovered at 22 °C for 7 days. The survival rate was calculated based on three replicates. To measure the leaf water content (LWC), we weighed (fresh weight, FW) the transgenic and WT Arabidopsis plants after 12 days of drought treatment. Then, the dry weight (DW) was measured after 16 h of incubation at 80 °C, and the LWC was calculated as follows: LWC (%) = (FW − DW)/FW × 100 (Yin et al. 2017).
Measurement of the antioxidant enzyme activities
To measure the antioxidant enzymes activities, we ground 0.1 g of leaf samples with 2 mL of ice-cold sodium phosphate buffer (50 mM, pH 7.8) containing 1% polyvinylpyrrolidone to extract the total enzymes. The activities of catalase (CAT), peroxidase (POD) and superoxide dismutase (SOD) were assessed using the CAT Assay Kit (A007-1, Nanjing Jiancheng Bioengineering Institute, China), POD Assay Kit (A084-3, Nanjing Jiancheng Bioengineering Institute, China) and SOD Assay Kit (A001-1, Nanjing Jiancheng Bioengineering Institute) in accordance with the manufacturers’ instructions, respectively.
RNA-Seq analysis of transgenic Arabidopsis
Leaves from WT and transgenic Arabidopsis plants were collected under normal and drought stress (without watering for 5 days) conditions. Three overexpressed lines were pooled as OE lines, and four samples (WT-N and W14-N under normal conditions; WT-D and W14-D under drought conditions) were used for RNA-Seq analysis, with two replicates for each sample. Total RNA was extracted using TRIzol Reagent (Invitrogen, Life Technologies, USA) and treated with DNase I (Invitrogen, Life Technologies, USA). The cDNA library for each sample was constructed using the BGI RNA-Seq kit. Then, the libraries were purified using the AMPure XP system (Beckman Coulter, Beverly, USA) and validated with the Agilent Technologies 2100 bioanalyzer. Finally, the libraries were sequenced using the BGISEQ-500 platform (BGI, Wuhan, China, http://www.seq500.com) (Wang et al. 2009; Mortazavi et al. 2008). Differentially expressed genes were identified based on fold change ≥ 2 and diverge probability ≥ 0.8. The functional categories of the differently expressed genes and the pathway were obtained using MapMan (http://bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) as the classification source (Thimm et al. 2004).
Results
VaWKRY14 belongs to the WRKY IIa subgroup
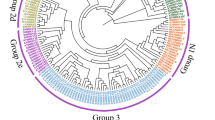
VaWRKY14 is homologous to the gene GSVIVT01015952001, which is mapped to chromosome 9 of V. vinifera cv. Pinot Noir clone P40024 and both comprises five exons and four introns (Fig. 1a, b). The ORF of VaWRKY14 is 933 bp in length, and it encodes a putative protein of 310 amino acids with a calculated molecular mass of 34.3 kDa and an isoelectric point of 8.99. Thus, a phylogenetic tree was constructed to clarify the relationship of VaWRKY14 to other WRKYs proteins from different plants. The results revealed that VaWKRY14 belongs to the WKRY IIa subfamily and displays a 60 and 52% protein sequence similarity to GmWRKY27 and AtWRKY40 protein, respectively (Fig. 2). GmWRKY27 could positively regulate the drought tolerance of soybean (Wang et al. 2015). This result indicates that VaWRKY14 probably involved in the regulation of drought tolerance in grapes. In addition, we conducted multiple sequence alignments of VaWRKY14 with WRKY IIa members from different plants and showed that apart from the WRKY domain, two new conserved motifs/structures, namely, a leucine zipper (LZ) structure in the N-terminal and an unknown motif in the C-terminal, were identified (Fig. 1b, c). The functions of these new structures are worthy to be elucidated in future studies.
Phylogenetic tree constructed by VaWRKY14 and WRKY proteins from different plants. GenBank accession numbers are AtWRKY75 (At5g13080)—Pi starvation; GmWRKY21 (DQ322691)—cold stress, GmWRKY54 (DQ322698)—salt and drought, GmWRKY13 (DQ322694)—salt; OsWRKY45 (AY870611)—salt and drought; AtWRKY18/40/60 (AT4G31800, AT1G80840, and AT2G25000)—abiotic and biotic stress; CmWRKY17 (AJF11725)—salt stress; AtWRKY46 (At2g46400)—down-regulated osmotic (drought) stress; MrWRKY30—abiotic (cold and salt) stress and downy mildew; FcWRKY70 (FcWRKY70)—drought stress; GmWRKY27 (ABC26917)—drought stress; AtWRKY8 (AT5G46350)—salt stress; AtWRKY63 (At1g66600)—drought stress; OsWRKY30 (DQ298180)—drought stress; OsWRKY11 (AK108745)—drought stress; GhWRKY17(ADW82098)—drought and salt Stress; OsWRKY76 (ABC02813)—cold stress; HvWRKY38 (AAS48544)—cold and drought; CaWRKY40 (AAX20040)—heat stress; TaWRKY33 (ALR88711)—drought stress; OsWRKY71 (BAF80893)—cold stress; OsWRKY28 (DAA05093) and OsWRKY62 (ABC02810)—biotic stress; FcWRKY40 (AIZ94612)—oxidative stress; PtoWRKY60 (AIA66985.1) and PtrWRKY40 (XP_002332076)—biotic stress; NtWIZZ (AB028022); and GaWRKY1(AAR98818)
VaWRKY14 is localized in the nucleus
To investigate the subcellular localization of VaWRKY14, the vectors carrying GFP or GFP-fused full-length VaWRKY14 (VaWRKY14-GFP) under the control of the cauliflower mosaic virus 35S promoter were transformed into Arabidopsis protoplasts through the PEG-Ca2+ method. As shown in Fig. 3, GFP was detected in the nucleus and the cytosol, whereas VaWRKY14-GFP was detected only in the nucleus. Therefore, the VaWRKY14 protein is targeted to the nucleus and possibly functions as a TF.
VaWRKY14 does not exhibit transactivation activity in yeast
To analyze the transcriptional activity of VaWRKY14, the ORF was fused in-frame with the GAL4 DNA-binding domain in the pGBKT7 vector and subjected to yeast two-hybrid assay using the yeast strains carrying the dual reporter genes HIS3 and LacZ. VaNAC26, which has been identified as an active transcriptional activator, was used as the positive control to test the yeast system (Fang et al. 2016). As shown in Fig. 4, all cells grew well in the SD/-Trp medium. Only transformants harboring the recombinant plasmids VaNAC26/pGBKT7 (positive control) grew well on the SD/-Trp/-His medium and presented strong β-galactosidase activity, whereas transformants containing VaWKRY14/pGBKT7 and the negative control pGBKT7 could not grow on the SD/-Trp/-His medium and exhibited no β-galactosidase activity. This result indicates the absence of transactivation activity of VaWRKY14 in yeast cells.
Transcriptional activity assays of VaWRKY14. pGBKT7-VaNAC26, an active transcriptional activator, was used as positive control (Fang et al. 2016). pGBKT7 was used as negative control
Expression of VaWRKY14 is induced by abiotic stresses and exogenous SA and ABA
The expression profiles of VaWRKY14 under time-course cold and drought stresses were determined by conducting qRT-PCR. The transcription of VaWKRY14 was quickly induced by drought stress; it showed a strong and gradual increase from 0 to 4 h and a decrease from 4 to 24 h (Fig. 5a). During the cold treatment, the expression of VaWKRY14 was upregulated, and the highest expression level was detected at 8 and 24 h (Fig. 5b). The results indicate that VaWRKY14 participates in the drought and cold stresses of grapevine.
Transcription levels of VaWRKY14 in response to exogenous phytohormones treatment and abiotic stresses. a Drought stress (6% PEG600). b 4 °C cold stress. c 200 mM SA. d 100 µM ABA. Error bars refer to the standard deviations of three biological replicates. * and ** indicate significant differences compared with WT at P < 0.05 and P < 0.01 level (t test), respectively
Phytohormones, such as ABA and SA, play important roles in plant tolerance against abiotic stresses (Hubbard et al. 2010; Lindemose et al. 2013; Miura and Tada 2014). Phytohormones could change the expression patterns of TFs and regulate plant response to abiotic stresses. To determine the signaling pathway that VaWRKY14 may involve in, the transcription patterns of VaWRKY14 under exogenous phytohormones were investigated. The transcript level of VaWRKY14 was upregulated under both ABA and SA treatments. A rapid response of VaWRKY14 to SA treatment was detected at 2 h, whereas the highest transcript level of VaWRKY 14 under ABA treatment was detected at 4 h (Fig. 5c, d). These results suggest that VaWRKY14 is involved in the ABA and SA signaling pathways during cold and drought stress response in grapes.
Overexpression of VaWRKY14 enhances the drought stress tolerance in Arabidopsis
Three transgenic lines with overexpressed VaWRKY14 (OE1, OE2, and OE3) were examined for their performance under cold and drought stresses (Fig. 6d). For the cold treatment, no differences were observed between the OE lines and the WT plants (data not shown). For the drought stress, the transgenic plants showed better growth with higher survival rates, whereas the WT plants displayed severe symptoms (Fig. 6a). Under drought stress, the transgenic lines achieved a survival rate of over 60%, which was significantly higher than that of the WT plants (35%, Fig. 6b). Moreover, the LWCs of the OE plants, especially the OE2 and OE3 lines were significantly higher than that of the WT plants after 12 days of drought treatment. These data suggest that overexpression of VaWRKY14 enhances the drought tolerance of Arabidopsis.
Drought tolerance of VaWRKY14-OE lines and WT Arabidopsis. a 18-day plants (upper panel) were used to drought treatment for 14 days and its drought tolerance phenotypes were recorded after re-watering for 5 days. b Survival rate of WT and transgenic plants was analyzed. c Leaf water content of WT and transgenic plants. d Relative expression of VaWRKY14 in WT and transgenic OE lines. * and ** indicate significant differences compared with WT at P < 0.05 and P < 0.01 level (t test), respectively
Overexpression of VaWRKY14 improved the drought tolerance of transgenic Arabidopsis plants by increasing the antioxidant enzyme activities
Drought stress can induce the production of reactive oxygen species (ROS) that causes oxidative damage to plants. Antioxidant enzymes such as CAT, POD and SOD are involved in the ROS-scavenging system to protect plants from oxidative damage (Mittler et al. 2004). In the present study, we measured the activities of CAT, POD and SOD in the OE lines and WT plants under normal and drought conditions. As show in Fig. 7a, the CAT activity of OE1 was significantly higher than that of the WT plants in normal conditions. The CAT activities of both OE1 and OE3 were significantly higher than that of the WT plants under drought stress. No significant differences were observed in POD and SOD activities between the OE lines and WT plants in normal conditions (Fig. 7b, c). However, the POD activities in the three OE lines were significantly higher than that in the WT plants after 5-day drought treatment (Fig. 7b). In addition, the SOD activities in OE3 were significantly higher than in WT plants (Fig. 7c). These results suggest that overexpression of VaWRKY14 improve the drought tolerance of transgenic Arabidopsis plants by increasing the antioxidant enzyme activities.
Activities of antioxidant enzymes in WT and transgenic OE lines. The CAT (a), POD (b) and SOD (c) activities of WT and three transgenic lines both at 5 days after drought treatments and normal conditions. Error bars refer to the standard deviations of three replicates. * and ** indicate significant differences compared with WT at P < 0.05 and P < 0.01 level (t test), respectively
Expression of stress-related genes is affected by VaWRKY14 overexpression in Arabidopsis
Analysis of the subcellular location of VaWRKY14 suggests that VaWRKY14 functions as a TF. To further examine the regulatory roles of VaWRKY14, the total RNAs extracted from the WT and transgenic Arabidopsis plants under normal and drought stress conditions were used for RNA sequencing (RNA-seq) analysis.
Results showed that the expression of 113 genes (16.8% upregulated and 83.2% down-regulated) was significantly changed by the overexpression of VaWRKY14 under normal conditions (W14-N vs WT-N) (Fig. 8, Table S2). The drought treatment changed 484 genes (65.9% upregulated and 34.1% downregulated) in WT plants (WT-D vs WT-N) and 554 genes (82.6% upregulated and 17.4% downregulated) in VaWRKY14-OE lines (W14-D vs W14-N) (Fig. 8, Table S2). Under drought stress treatment, 77 genes were differently expressed in VaWRKY14-OE lines when compared with those in WT plants (W14-D vs WT-D) (Fig. 8, Table S2). Overlapping analysis revealed that three out of the 19 upregulated and 49 out of the 94 downregulated genes were changed by the overexpression of VaWRKY14 (W14-N vs WT-N) were also induced by drought stress in WT plants (WT-D vs WT-N) (Fig. 8, Table S3). This result indicates that VaWRKY14 affects the expression of drought-related genes. Among them, several stress-related genes include COR15A, PR, PDF, and OSM34 reportedly play important roles in abiotic stress (Thalhammer et al. 2010; Lcvan and Eavan 2002).
Furthermore, pathway enrichment analyses were performed on the differently expressed genes of four comparisons (WT-D vs WT-N, W14-N vs WT-N, W14-D vs WT-D and W14-D vs W14-N). After NF values were selected with P values < 0.05, the pathways are shown in Table 1. The results showed that various pathways, including secondary metabolism, amino acid metabolism, stress, and hormone metabolism, were severely over-represented in OE lines compared with those in WT plants under normal conditions. Among them, the stress pathway was consistently enriched in all four comparisons together with the miscellaneous pathway. The pathways of the cell wall were also enriched in OE lines.
Among the genes significantly changed by the overexpression of VaWRKY14 under normal conditions, the upregulated genes in OE lines compared to WT plants, including COR15A, COR15B, COR413, RD29A, and KIN2, were directly involved in response to abiotic stresses, such as drought and cold (Table 2). In addition to COR15A, another stress-induced protein and F-box family protein were also induced by drought stress in WT plants (Table 2 and S3). The downregulated genes affected by both VaWRKY14 and drought stress included defense response proteins, such as OSM34, PDF, MAPKKK19, and PR (Table 2 and S3) (Singh et al. 1987; Kitajima and Sato 1999; Zhang and Klessig 2001). In addition, the expression levels of PDF1.2, PDF1.2b, PDF1.2c and PR4 were down-expressed in the OE lines under drought condition compared with those under normal condition. These results suggest that changes in the stress-related genes caused by VaWRKY14 may be responsible for the increased drought tolerance in OE plants.
qRT-PCR analysis was performed on nine significantly changed genes to validate the data obtained by RNA-sEq. As shown in Fig. S1, all of the tested genes showed similar expression trends at the transcript level between the qRT-PCR and RNA-seq data.
Discussion
In recent years, increasing evidence has supported the essential roles of WRKY TFs not only in biotic stress responses but also in abiotic stresses, such as drought (Ren et al. 2010), low and high temperatures (Yokotani et al. 2013; Dang et al. 2013), and osmotic stress (Gong et al. 2014). In this study, VaWRKY14, which is a drought-response gene, was identified as a member of the WRKY IIa subgroup (Fig. 2). Several studies have already identified the pivotal roles of this WRKY subgroup in abiotic stress response in plants. GmWRKY27 in soybean positively regulates drought tolerance (Wang et al. 2015). HvWRKY38 from barley enhances the drought tolerance in turf and forage grass (Paspalumnotatum Flugge) (Xiong et al. 2010). TaWRKY33 confers drought and heat tolerance in Arabidopsis (He et al. 2016). WRKY76 and WRKY71 play positive roles in cold stress tolerance in rice (Yokotani et al. 2013; Kim et al. 2016). In the present study, we found that the ectopic overexpression of VaWRKY14 could also enhance the drought stress tolerance in Arabidopsis (Fig. 6). These results suggest that members of the WRKY IIa group participate in abiotic stress pathways in plants.
To illustrate the molecular mechanisms underlying the enhanced drought tolerance of VaWRKY14-overexpressing plants, RNA-seq was performed on OE lines and WT plants. The results revealed that many stress-related genes, such as COR15A, COR15B, COR413, RD29A, and KIN2 were upregulated in transgenic plants relative to their levels in WT plants under normal conditions. Among them, AtCOR15A and AtRD29A are considered marker genes for cold, drought, and salt stress responses and are regulated by various upstream TFs (Msanne et al. 2011; Thalhammer et al. 2014; Yamaguchi-Shinozaki and Shinozaki 2006). COR15B is also involved in dehydration tolerance (Kang et al. 2009). The upregulation of these stress-related genes may partly explain the increased tolerance to drought stress in VaWRKY14 transgenic Arabidopsis.
Phytohormones are central to the integration of diverse environmental cues in plants (Santner and Estelle 2009). An increasing number of studies are focusing on the signaling pathways activated by phytohormones, such as ABA, SA, jasmonic acid, and ethylene, as well as their roles during stress response in plants. In the present study, the transcription of VaWRKY14 could be induced by SA and ABA. Similar results were obtained for the WRKY IIa subgroup in Arabidopsis (Chen et al. 2010; Schön et al. 2013). The WRKY proteins may be key components in ABA signaling, acting as activators or repressors (Xie et al. 2005; Chen et al. 2012). The ABA response genes, such as RD29A, COR15A, COR15B, COR413, RD29A, and KIN2 were upregulated in transgenic plants relative to their levels in WT plants, suggesting that VaWRKY14 may be involved in ABA-related signaling pathways. SA is an important signaling molecule under stress conditions, particularly in defense against pathogens (Dempsey et al. 1997; Hamada 2001; Korkmaz et al. 2007). The results of RNA-seq showed that many pathogen-related genes that were regulated in Arabidopsis overexpressed VaWRKY14 compared to WT (Table S2). Therefore, the putative roles of VaWRKY14 during pathogen defense must be demonstrated through SA signaling pathways in grapevines.
An increased expression of VaWRKY14 against cold stress was detected, but the cold tolerance of its transgenic Arabidopsis was unaffected. This result disagrees with the finding of a previous study on OsWRKY76, which also belongs to the WRKY IIa subgroup and could enhance cold tolerance of rice (Yokotani et al. 2013). The possible explanation for this phenomenon is that VaWRKY14 may not functional solely for cold stress response. Mare et al. (2004) showed that two W-box motifs in the promoter region are required to ensure the binding of HvWRKY38 to W box. This finding suggests that the transcriptional regulations require an interaction between HvWRKY38 and another WRKY protein (Mare et al. 2004). Proteins, particularly regulatory proteins, rarely act alone (Chi et al. 2013). Protein interaction is an important mechanism by which TFs function effectively. In addition, the WRKY TFs physically interact with a wide range of proteins with roles in signaling, transcription, and chromatin remodeling (Chi et al. 2013). Two bulky hydrophobic residue domains, namely, LZ structure in the N-terminal and an unknown motif in the C-terminal (Fig. 1c), were found in VaWRKY14 and its homolog genes in other plants. The LZ structure could mediate the interaction of WRKY with other proteins (Eulgem et al. 2000). In Arabidopsis, three WRKY proteins from group IIa (AtWRKY18, AtWRKY40, and AtWRKY60) interact with themselves and with one another through the LZ motifs present at its N-terminal (Xu et al. 2006). Thus, the identification of interacting proteins for VaWRKY14 is necessary to explain its role during cold stress response in grapevines.
In summary, we identified a Group IIa stress-responsive WRKY gene (VaWRKY14) from wild grapevine species V. amurensis and found that this gene acts as a positive regulator of drought tolerance. Transcriptome analysis of Arabidopsis plants revealed that the expression levels of several stress-related genes, such as COR15A, COR15B, COR413, RD29A, and KIN2, were upregulated in transgenic plants relative to those in WT plants under normal conditions. Therefore, VaWRKY14 may mediate drought tolerance by regulating the expression levels of stress-related genes.
Author contribution statement
LZ and JC carried out most of the experiments and drafted the manuscript. XS performed phenotypes observation and statistical analysis. TZ participated in experiment of subcellular localization. ML, QW, SL and HX supervised the studies and revised the manuscript.
References
Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63:2667–2679
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:443–462
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819:120–128
Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6:287–300
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Dang FF, Wang YN, Lu YU, Eulgem T, Lai Y, Liu ZQ, Wang XU, Qiu AL, Zhang TX, Lin J (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36:757–774
Dempsey DA, Shah J, Klessig DF (1997) Salicylic acid and disease resistance in plants. Crit Rev Plant Sci 18:547–575
Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ (2014) Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J 79:13–27
Dong JX, Chen CH, Chen ZX (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51:21–37
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Fang L, Su L, Sun X, Li X, Sun M, Karungo SK, Fang S, Chu J, Li S, Xin H (2016) Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J Exp Bot 67:2829–2845
Gong XQ, Hu JB, Liu JH (2014) Cloning and characterization of FcWRKY40, a WRKY transcription factor from Fortunella crassifolia linked to oxidative stress tolerance. Plant Cell Tiss Org 119:197–210
Gong X, Zhang J, Hu J, Wang W, Wu H, Zhang Q, Liu JH (2015) FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ 38:2248–2262
Guillaumie S, Mzid R, Méchin V, Léon C, Hichri I, Destrac-Irvine A, Lauvergeat V (2010) The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol Biol 72:215–234
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
Hamada AM (2001) Salicylic acid versus salinity-drought-induced stress on wheat seedlings. Rostl Vyr 47:444–450
He GH, Xu JY, Wang YX, Liu JM, Li PS, Chen M, Ma YZ, Xu ZS (2016) Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol 16:116
Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Gene Dev 24:1695–1708
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet 244:563–571
Kang GZ, Zhu ZH, Guo TC, Ren JG (2009) Isolation and expression pattern of COR15b and KIN1 genes in watermelon and pumpkin. Afr J Biotechnol 8:5666–5672
Kim CY, Vo KTX, Cong DN, Jeong DH, Lee SK, Kumar M, Kim SR, Park SH, Kim JK, Jeon JS (2016) Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol Rep 10:13–23
Kitajima S, Sato F (1999) Plant Pathogenesis-Related Proteins: Molecular Mechanisms of Gene Expression and Protein Function. J Biochem 125:1–8
Korkmaz A, Uzunlu M, Demirkiran A (2007) Treatment with acetyl salicylic acid protects muskmelon seedlings against drought stress. Acta Physiol Plant 29:503–508
Lcvan L, Eavan S (2002) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant P 55:85–97
Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878
Mare C, Mazzucotelli E, Crosatti C, Francia E, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold-and drought-response in barley. Plant Mol Biol 55:399–416
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Miura K, Tada Y (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front Plant Sci 5:4
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Methods 5:621–628
Msanne J, Lin J, Stone JM, Awada T (2011) Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234:97–107
Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G (2012) An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep 31:349–360
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Pandey SP, Roccaro M, Schon M, Logemann E, Somssich IE (2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 64:912–923
Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65:35–47
Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63:417–429
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nat 459:1071–1078
Schön M, Töller A, Diezel C, Roth C, Westphal L, Wiermer M, Somssich IE (2013) Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol Plant Microbe Interact 26:758–767
Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, Wang X (2012) OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol 80:241–253
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Singh NK, Bracker CA, Hasegawa PM, Handa AK, Buckel S, Hermodson MA, Pfankoch E, Regnier FE, Bressan RA (1987) Characterization of Osmotin. Plant Physiol 85:529–536
Thalhammer A, Hundertmark M, Popova AV (2010) Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. BBA-Biomembranes 1798:1812–1820
Thalhammer A, Bryant G, Sulpice R, Hincha DK (2014) Disordered cold regulated 15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in Vivo. Plant Phyol 166:190–194
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for Transcriptomics. Nat Rev Genet 10:57–63
Wang L, Zhu W, Fang L, Sun X, Su L, Liang Z, Wang N, Londo JP, Li S, Xin H (2014) Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol 14:103
Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, Man WQ, Zhang JS, Chen SY (2015) GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J 83:224–236
Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12:9–26
Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis sandwich-a simpler Arabidopsis protoplast isolation method. Plant Meth 5(1):16
Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189
Xiong X, James VA, Zhang H, Altpeter F (2010) Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalumnotatum Flugge). Mol Breeding 25:419–432
Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol 135:507–515
Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310–1326
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yin MZ, Wang YP, Zhang LH, Li JZ, Quan WL, Yang L, Wang QF, Chan ZL (2017) The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J Exp Bot 68:2991–3005
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64:5085–5097
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, Xiang F (2016) WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J 85:96–106
Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6:520–527
Acknowledgements
This work was supported by the National Science Foundation of China (NSFC Accession No.: 31471857), Youth Innovation Promotion Association of CAS (2015281), Science and Technology Service Network Initiative of CAS (KFJ-STS-ZDTP-025), Grape Breeding Project of Ningxia (NXNYYZ201502) and Open Project Program of State Key Laboratory of Crop Stress Biology for Arid Areas (NWAFU, CSBAA2016009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability statements
The resulting RNA-Seq data were deposited in The National Center for Biotechnology Information (NCBI) GEO database repository (https://www.ncbi.nlm.nih.gov/geo/).
Additional information
Communicated by Ying-Tang Lu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Cheng, J., Sun, X. et al. Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep 37, 1159–1172 (2018). https://doi.org/10.1007/s00299-018-2302-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2302-9