Abstract
Drought stress is a severe environmental factor that greatly restricts plant distribution and crop production. Transgenic breeding offers new opportunities for developing drought-resistant varieties. The WRKY transcription factors have been reported to be involved in various plant physiological and biochemical processes. In this study, we report the impact of TaWRKY46 on abiotic tolerance in wheat (Triticum aestivum L.). The transcription levels of the TaWRKY46 gene were differentially regulated by diverse abiotic stresses and hormone treatments, including PEG-induced stress (20% polyethylene glycol 6000), cold (4 °C), salt (100 mM NaCl), abscisic acid (100 μM ABA) and hydrogen peroxide (10 mM H2O2). The TaWRKY46-GFP fusion protein was localized to the nucleus of wheat protoplast. The N-terminal of TaWRKY46 showed transcriptional activation activity. Overexpression of TaWRKY46 in wheat resulted in enhanced drought stress tolerance. TaWRKY46-overexpressing plants exhibited increase survival rate, soluble sugar, proline and superoxide dismutase (SOD), as well as higher activities of catalase (CAT) and peroxidase (POD), but lower contents of malondialdehyde (MDA) and H2O2 content. Taken together, our results indicate that TaWRKY46 functions as a positive factor under drought stress by regulating the osmotic balance and ROS scavenging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants constantly encounter various abiotic and biotic stresses throughout their life cycle (Abrahám et al. 2003). These stresses can simultaneously affect plant growth and development and/or alter the distribution of plant species. Although sessile plants lack a circulating, somatically adaptive immune system, they have developed specific mechanisms to respond to diverse environmental signals that may cause stress and restrict growth and development (Bechtold and Field 2018). Among these mechanisms, a number of stress-related genes directly protect the plants against stress, or either induce or repress the downstream target genes to regulate plant development and defense responses synergistically or antagonistically (Skriver and Mundy 1990). Transcriptional modulation is vital for the complex genetic and biochemical networks to respond to stress (Zhang et al. 2012). So far, a large number of transcription factors (TFs) belonging to different protein families (such as MYB, DREB, bZIP, NAC and WRKY families) have been identified in plants. Among them, the plant WRKY transcription factors, comprising a large family of regulatory proteins, are shown to play an important role in response to various stresses (Chen and Zhu 2004). Since the first member was isolated in sweet potato, an increasing number of WRKY members have been identified, including 74 members in Arabidopsis thaliana, 109 members in rice, 197 in soybean and 43 in wheat (Eulgem et al. 2000). All the WRKY proteins contain one or two conserved WRKY domains composed of about 60 amino acids with the conserved amino acid sequence WRKYGQK at N-terminus and a zinc-finger motif (C-X4-5-C-X22-23-H-X1-H or C-X7-C-X23-H-X1-C) at C-terminus (Ulker and Somssich 2004). The WRKY proteins act as transcription factors and bind the W box (TTGACC/T) via the WRKY domain to modulate the transcription of target gene. Based on the number of conserved WRKY domains and the structure of the zinc-finger motif, WRKY members can be subdivided into three major groups (I–III) (Wang et al. 2013). Furthermore, group II can be further classified into five distinct subgroups (IIa–IIe) based on additional conserved structural motifs outside the WRKY domain (Rushton et al. 2010).
Previous studies have examined the roles of plant WRKY proteins in response to pathogens, and WRKY proteins can function as either positive or negative regulators of the defense response (Zhou et al. 2015). For example, overexpression of ZmWRKY33 in Arabidopsis induced expression of RD29A that is known to be involved in stress signaling, and enhanced salt tolerance of the transgenic plants (Wang et al. 2018). Overexpression lines of OsWRKY11 showed significant heat and drought tolerance, as indicated by slower leaf-wilting and increased survival rate of green parts of plants (Wu et al. 2009). GmWRKY21 transgenic plants were tolerant to cold stress, whereas GmWRKY54 conferred salt and drought tolerance. Transgenic plants overexpressing GmWRKY13 exhibited sensitivity to salt and mannitol stress (Zhou et al. 2008). Although evidence that WRKY proteins are involved in abiotic stress is increasing, the understanding of the roles of these proteins in the responses to abiotic stress is progressing relatively slowly, and the challenge to elucidate the molecular mechanisms of defense remains (Xu et al. 2014). Wheat is the dominant crop for human food and livestock feed. Current and future concerns include improving wheat yield and quality under hostile environments. Understanding the molecular mechanism of WRKY genes in wheat is important for breeding of this important crop. To date, 28 WRKY genes have been reported to be expressed under abiotic stress, among which 21 genes were up-regulated under drought, oxidative stress and pathogen stress. In wheat, TaWRKY2 and TaWRKY19 are associated with abiotic tolerance in wheat (Niu et al. 2012). Together, the above findings suggest that the WRKY transcription factors play a crucial role in plant development. However, there is still little information about the function of WRKY transcription factors in biotic and abiotic stresses and hormone signaling in wheat. Therefore, understanding the underlying roles of WRKY proteins in the tolerance of biotic and abiotic is an important global problem for breeding programs. In the present study, a cDNA clone, TaWRKY46, encoding a putative III WRKY gene, was isolated and characterized, and the gene expression patterns under various biotic and abiotic stresses were investigated. TaWRKY46 can be induced by abiotic stresses and multiple defense-related signaling molecules. The overexpression of TaWRKY46 in wheat significantly enhanced resistance to drought. Hence, this study was conducted with the aim of understanding the mechanism by which the WRKY transcription factors regulate plant responses to drought stress.
Materials and methods
Plants materials and stress treatments
Wheat (Triticum aestivum L. cv. Bainong 207) was used in this study. Seeds were germinated and grown in greenhouse conditions at 25 °C with a 16 h light/8 h dark cycle. For salt and PEG treatments, 2-week-old seedlings were treated with 100 mM NaCl or 20% PEG-6000, respectively, and sampled at 0, 1, 3, 6, 12 and 24 h after treatment. For signaling molecule treatment, the seedlings were treated with 100 μM ABA and 10 mM H2O2, respectively, and then sampled at 0, 1, 3, 6, 12 and 24 h after treatment. For cold treatment, seedlings were transferred to low temperature room (4 °C) and sampled at 0, 1, 3, 6, 12 and 24 h. Leaves from sterile water treatment were taken as a control. For the tissue-specific expression assay, roots, stems and leaves were collected from sterile seedlings, while pistils and stamens were collected from wheat plants in the growth chamber. All organs and tissues were quickly frozen in liquid nitrogen, ground to powder by adding liquid nitrogen, and then stored at − 80 °C for use.
Amplification and sequence analysis of TaWRKY46
Total RNA was extracted from young wheat leaves using Plant RNA Reagent (TIANGEN, China). First-strand cDNA was obtained by using 1 μg of total RNA, Oligo (dT) 12–18 primers and reverse transcriptase according to the manufacturer's instruction. PCR amplification was done using ES Taq DNA Polymerase (CWBIO, Beijing, China) with proofreading activity. The temperature cycles were: 4 min at 94 °C, 40 s at 94 °C, 40 s at 58 °C, 30 s at 72 °C for 35 cycles; and 7 min at 72 °C. PCR products were purified by agarose gel recovery kit (TIANGEN, Beijing), subcloned into pMD-19T vector and sequenced. Nucleotide and deduced amino acid sequences were analyzed using DNAMAN 6.0. Theoretical isoelectric point (PI) and molecular weight were predicted in an internet server, ExPASy (Expert Protein Analysis System, http://www.expasy.org/tools). Multiple sequence alignments of translated gene sequence was carried out with the program DNAMAN 6.0. A phylogenetic tree, based on the genetic distance of the protein sequences as determined using the MEGA-5.0 program, was generated.
Quantitative real-time PCR
Organ-specific expression patterns and gene expression patterns of TaWRKY46, after various stress treatments, were determined by qRT-PCR analysis. qRT-PCR was performed using the SYBR Premix Ex Taq (TaKaRa, Dalian, China) and the CFX96TM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The wheat TaActin gene was used as a control. During the qRT-PCR analysis, each sample was analyzed using three technical replicates, and data analyzed by analysis software based on the comparative 2−ΔΔCt method of relative gene quantification.
Subcellular localization analysis
To observe the subcellular localization, the full open reading frame (ORF) of TaWRKY46 was amplified with primers as shown in Table 1. The PCR product was inserted in frame into the pBI121-GFP vector. The resultant construct p35S:TaWRKY46-GFP and the control vector p35S:GFP were genetically introduced into wheat protoplasts. Following pre-incubation at 22 °C for 24 h, the green fluorescence protein (GFP) signal was detected with a laser confocal microscope (Zeiss LSM510 Meta, Jena, Germany).
Transcriptional activation assay
For transcriptional activation analysis, the full-length coding region and deletions fragments of TaWRKY6 were generated by PCR using specific primers (primers are provided in Table 1). The PCR products were ligated to the yeast expression vector pGBKT7 and were named pBD-TaWRKY, pBD-TaWRKY-N1, pBD-TaWRKY-N2, pBD-TaWRKY-C1, pBD-TaWRKY-C2, respectively. The plasmid pGBKT7 (pBD) was used as the negative control. These plasmids were transformed into yeast strain AH109. The transformed yeast cells were selected on SD/-Trp and were verified by PCR. The positive colonies were transferred onto the SD/-His plates with or without X-α-D-Galactoside (X-α-D-gal). The plates were incubated at 30 °C for 3 days.
Generation of transgenic wheat plants
To obtain transgenic wheat plants, the coding sequence of the TaWRKY46 containing termination codon was amplified by RT-PCR and cloned into BamH I/Sma I restriction sites of the pBI121 vector under the control of the 35S promoter of cauliflower mosaic virus. The primers containing the BamH I/Sma I restriction sites are listed in Table 1. Agrobacterium strain GV3101 harboring the recombinant plasmid was used for wheat transformation through (Zhao et al. 2006). To isolate positive transgenic lines, RNA was isolated from leaves of transgenic wheat seedlings, and RT-PCR analysis was conducted. Primers used in these studies are shown in Table 1.
Analysis of transgenic plants under drought condition
Drought tolerance assays were performed using 4-week-old plants. Three independent transgenic homozygous T3 lines seedlings (Transgenic lines TG1, TG4, and TG8) and wild-type (WT) seeds were transplanted in the same pot and treated with drought stress by withholding water for 21 days. Three independent pots were repeated at the same time and a representative result displayed. Three independent experimental replications were conducted.
NBT staining and antioxidant enzyme activities
Histochemical staining with NBT has been proposed as a common technique to localize O2− in plants. Therefore, accumulation of O2− in the drought-treated leaves were examined by histochemical staining, as has been described by Jiang et al. (2016). In addition, the H2O2 concentration and MDA content were determined using a hydrogen peroxide test kit and a maleic dialdehyde assay kit (Nanjing Jiancheng Bioengineering Institute), respectively, according to the manufacturer's instructions. Soluble carbohydrate contents were determined by the phenol reaction method (Niu et al. 2012). The activity of three antioxidant enzymes, CAT, POD, and SOD was spectrophotometrically measured with kits produced by the Nanjing Jiancheng Institute.
Statistical analysis
The results are expressed as the mean standard deviation (SD) of triplicate experiments (n = 3). Statistical significance was determined by Duncan’s multiple range test with an analysis of variance (ANOVA) use Statistical Analysis System (SAS) version 9.1. The significance was set at P < 0.05.
Results
Identification and sequence analysis of TaWRKY46
The full-length cDNA of TaWRKY46 isolated from wheat leaves was obtained by RT-PCR. The sequence, which comprised 872 bp, contained a 669 bp ORF encoding 222 amino acid residues with an estimated molecular mass of 24.49 kDa and a theoretical isoelectric point of 8.57. Analyzing the evolutionary relationships among the various species of WRKYs would provide an insight into the evolution of their function. Multi-alignment analysis revealed that the deduced WRKY protein was closely related to other plant WRKY proteins, sharing 57.14, 57.01, 51.90 and 57.32% homology with PtWRKY48 (XP_002301524), PtWRKY23 (ABK41486.1), VvWRKY48 (XP_002279385), and PtWRKY13 (ACV92015), respectively (Fig. 1a). Similar to the other WRKY transcription factors, TaWRKY46 has one WRKY domain that contains the highly conserved amino acid sequence WRKYGEK and a C2HC zinc-finger motif. Phylogenetic analysis further revealed the evolutionary relationship to other WRKYs from various plant species, suggesting that TaWRKY46 belongs to Group III of the WRKY family (Fig. 1b).
Sequence and phylogenetic analyses of TaWRKY46. a Sequence alignment of the deduced TaWRKY46 protein with BdWRKY55, SbWRKY48, SiWRKY48, and OsWRKY70, Identical amino acids are shaded black. The approximately 60-amino acid WRKY domain is marked by the two-headed arrow. The C and H residues in the zinc-finger motif are marked by triangles. The highly conserved amino acid sequence WRKYGEK in the WRKY domain is boxed. b Phylogenetic tree of the TaWRKYs domains from various plants
Expression pattern of TaWRKY46 under various stress conditions
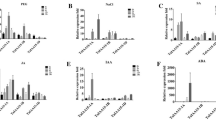
To clarify the tissue expression patterns of TaWRKY46, mRNA isolated from different wheat tissues were using as the templates for qRT-PCR. As shown in Fig. 2a, TaWRKY46 mRNA abundance was ubiquitously but differentially detected in all tissues, with the highest expression level being observed in leaves and roots and the lowest in stems, pistils and stamens. Transcriptional modulation is a vital aspect of the complex signal transduction pathway that enables plants to respond to biotic and abiotic stresses. To investigate the response of TaWRKY46 to abiotic stresses, TaWRKY46 expression levels were examined following NaCl, PEG600, H2O2, ABA and low temperature treatments (Fig. 2b–e). qRT-PCR analysis revealed that TaWRKY46 was obviously up-regulated after treatments with PEG and NaCl. During ABA and H2O2 treatments, the expression of TaWRKY46 was gradually increased by 3.2-fold at 6 h and 2.9-fold at 12 h, respectively. Low temperature treatment led to a slight up-regulation. These results indicate that TaWRKY46 expression was induced under various stress conditions.
Tissue-specific expression assay of TaWRKY46 and expression patterns of TaWRKY46 under PEG, NaCl, 4, ABA and H2O2 in wheat leaves. a Tissue-specific expression assay of TaWRKY46 in wheat. The tissues (root, stem, leaf, pistil and stamen) are represented by R, S, L, P, and ST, respectively. Expression analysis of TaWRKY46 in 10-day-old wheat seeding leaves under different treatments by qRT-PCR. b 20%PEG6000 treatments; c 200 mM NaCl treatment; d 4 treatment; e 100 μM ABA treatment; f 10 mM H2O2 treatment. Values are means ± SD of three replicates. *p < 0.05;**p < 0.01
The TaWRKY46 is localized in the nucleus
To investigate the subcellular localization of TaWRKY46, a TaWRKY46-GFP gene fusion construct was transformed into wheat mesophyll protoplasts. After 48-h incubation at room 28, free GFP protein was distributed throughout the cells while the TaWRKY46-GFP fusion protein had strong fluorescence signals in the nucleus (Fig. 3a). Therefore, TaWRKY46 likely function in the nucleus.
Analysis of the subcellular localization and transcriptional activation activity of TaWRKY34. a Subcellular localization of TaWRKY46. The TaWRKY46-GFP fusion and the GFP plasmids were transformed into wheat mesophyll protoplasts and examined by a confocal microscope (Leica). b Transactivation activity of the TaWRKY46 protein in yeast. Schematic diagrams of fused vectors illustrating the different portions of TaWRKY46 that were fused to the yeast vector pGBKT7. Yeast strain AH109 was used in the transactivation activity analysis of TaWRKY46. The transformants were incubated on the SD/-Trp or SD/-His medium and subjected to X-α-gal assay
Transcription activation activity of TaWRKY46
The yeast expression system was used to investigate whether TaWRKY46 possesses transcription activation activity. The ORF of TaWRKY46 gene was combined with the DNA-binding domain to identify transcriptional activation activity. Yeast strain AH109 was transformed with fusion plasmids pBD-TaWRKY, pBD-TaWRKY-N1, pBD-TaWRKY-N2, pBD-TaWRKY-C1, pBD-TaWRKY-C2, and pGBKT7 as a control. As shown in Fig. 1, the yeast cells transformed with pBD-WRKY, pBD-WRKY-N1 and pBD-WRKY-N2 grew well in the SD/-His medium and SD/-Trp medium. Meanwhile, yeast cells transformed with pBD-WRKY-C1, pBD-WRKY-C2 and pBD could only survive in the SD/-Trp medium. The staining result showed that the yeast cells turned blue in the presents of X-a-D-gal. These results indicated that the His and LacZ reporter genes were activated and N-terminal domain and full-length TaWRKY46 have transcription activation activity (Fig. 3b).
Overexpression of TaWRKY46 enhances the drought tolerance of transgenic plants
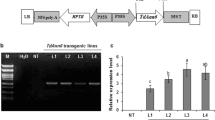
The differential expression patterns analysis suggested that TaWRKY46 may play a role in multiple stress defense responses, especially in the osmotic stress response. Further functional analyses of TaWRKY46 in stress tolerance, transgenic wheat plants constitutively overexpressing TaWRKY46 was produced by Agrobacterium-mediated transformation. The transgene overexpression in the T3 lines was analyzed by qRT-PCR, which indicated that TaWRKY46 expressed differentially in the ten transgenic lines, with higher level being detected in line TG1, TG4 and TG8 than in other lines (Fig. 4a).
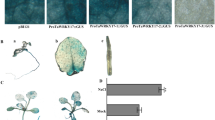
Analysis of the enhanced drought tolerance in transgenic lines. a TaWRKY46 transcript levels of WT and ten TG wheat lines determined by RT-PCR. b Phenotype of WT and TG wheat lines after 21 days of drought treatment. c Survival rate. d, e Fresh weight (d) and dry weight (e) of WT and TG wheat lines under drought stress. f Proline content. g Relative electrolyte leakage. h MDA content. g Tissue localization of O2− generation by NBT staining. Values are means ± SD of three replicates. *p < 0.05;**p < 0.01
To further assess the effect of TaWRKY46 overexpression on drought tolerance at the vegetative growth stage, we performed a drought stress assay using T3 plants of the three lines (TG1, TG4 and TG8). Four-week-old WT and TG plants were grown in the same pot without water for 21 days. After 21-day treatment, the WT showed obvious drought stress symptoms (Fig. 4b). When they were re-watered, the survival of the WT plants only 21.2%; however, all of the transgenic wheat plants survived (Fig. 4c). In addition, the fresh weight and dry weight of TG plants were significantly higher than WT plants (Fig. 4d, e). The phenotypic characterization suggested that overexpression of TaWRKY46 enhanced drought stress tolerance.
To investigate the physiological differences between WT and TG plants, some important physiological indices were measured. The accumulation of proline in plant is associated with adaptation to environmental stress through metabolic adjustments (Ju et al. 2020). We checked the proline contents of transgenic lines and control plants under normal growth and drought stress conditions to characterize the physiological basis for the improved stress tolerance. The proline content was higher in the TG plants after 21 days of drought stress (Fig. 4f). Electrolyte leakage, an indicator of membrane damage, was also measured following drought stress. The results showed that the leaves of TG plants exhibited significantly lower electrolyte leakage levels, compared to those of WT plants leaves (Fig. 4g). In addition, NBT staining showed that the accumulated the lower O2− than the WT plants (Fig. 4h). These results demonstrate that the transgenic lines possess more powerful resistance to hyperosmotic stresses compared to control lines. The activities of antioxidant enzymes (SOD, POD, CAT) in TG plants were higher than that in WT plants after drought treatment (Fig. 5a–c). Moreover, the contents of MDA and H2O2 in TG plants were significantly lower than those in WT plant under drought (Fig. 5d, e). In addition, the soluble sugar levels exhibited a profile similar to that of proline (Fig. 5f). In conclusion, the physiological characterization results suggested that overexpression of TaWRKY46 increased proline and sugar content and decreased MDA and ROS accumulation under drought conditions.
Discussion
Combination of abiotic and biotic stresses used to limit the production. Drought is the most important abiotic stress, which seriously restricts crop yield. Numerous studies have shown that the regulation of TFs is highly complex, involving transcription and protein levels, DNA binding, subcellular localization, and other characteristics achieved through post-translational mechanisms (Chi et al. 2013). WRKY gene is involved in a series of physiological and biochemical processes, including plant growth and development and resistance to external environment (Chen et al. 2012). Although some researchers indicated that plant WRKY transcription factors are involved in biotic and abiotic stresses, the most of these studies mainly focus on the model plants, such as A. thaliana and rice, and little is known about the role of WRKY transcription factors in wheat (Tripathi et al. 2014). To explore the function of the WRKY transcription factors in wheat, we isolated a WRKY gene from wheat, which is the foremost staple food crop in the world, and provides both calories and proteins to over 35% of the human population. In this study, multiple sequence alignment showed that TaWRKY46 possessed the conserved WRKYGQK domain and a C2HC (C-X7-C-X23-H-X-C) motif (Fig. 2d). The subcellular localization analysis demonstrated that the TaWRKY46 protein localized to the nucleus (Fig. 3a), which is consistent with previous studies on WRKY transcription factors from other species (Zou et al. 2010). Transcriptional activation analysis illuminated that the sequence of TaWRKY46 has transcriptional activation activity (Fig. 3b). These data conclude that the TaWRKY46 is a member of the WRKY family in wheat and may serve as a transcription activator, and suggest that TaWRKY46 may activate the expression of target genes in the nucleus and may participate in various plant processes.
The structural conservation of WRKY protein determines the functional specificity of regulating gene expression. Many studies have showed most WRKY proteins of the same group have similar functions in many plants. The dynamic expression patterns of TaWRKY46 were affected by stressors, including cold (4 °C), NaCl, PEG, ABA and H2O2 (Fig. 2). The WRKY transcription factors mediate signal transduction by activating adaptive responses and regulating downstream genes. During responses to multiple stresses, a single WRKY gene often participates various signaling pathways, indicating its diverse regulatory mechanism. The expression of some stress-induced genes has been proved to be related to stress tolerance. The response of TaWRKY46 to a broad range of environmental stresses implied that TaWRKY46 may be involved in the cross talk of the multi-environmental stress response in wheat. The drought tolerance phenotype of TaWRKY46 transgenic wheat plants was the result of a collection of physiological indexes observed in the overexpressing plants. Under drought stress conditions, the TG wheat displayed higher survival rates most likely because the water loss was reduced in these plants compared to control plants (Fig. 4c). Our findings were consistent with previous results reporting that BcWRKY46, TaWRKY2, TaWRKY19, and HvWRKY38 conferred drought and salt stresses, and enhanced stress tolerance, in transgenic plants (Niu et al. 2012; Wang et al. 2012; Xiong et al. 2010). Our results suggest that overexpression of TaWRKY46 in wheat remarkably enhances the plants’ tolerance to drought stress.
Drought first causes osmotic stress to plants and then causes secondary damage such as maladjustment of oxygen metabolism. Under osmotic stress, plants usually maintain water balance by accumulating organic matters. In this study, TaWRKY46 could regulated many biochemical and physiological processes to resist drought, e.g., increase the activities of antioxidant enzymes (SOD, CAT and POD) (Fig. 5a–c) to minimize oxidative damage during drought stress, reduced the cell membrane that functions as an osmoprotectant to reduce the effects of osmotic stress. These results showed that TaWRKY46 can maintain cell water absorption, prevent oxidative damage and improve plant resistance to drought stress by enhancing plant osmotic regulation and antioxidant capacity.
In conclusion, a wheat Group II WRKY gene, TaWRKY46, was up-regulated by PEG, NaCl, ABA, H2O2 and cold treatments. Overexpression of TaWRKY46 enhanced the tolerance to drought stress in transgenic wheat with increased proline and soluble sugar accumulation, decreased MDA and IL, improved antioxidant system and up-regulated transcription levels of ROS-related and stress responsive genes under various stresses. These results demonstrated that TaWRKY46 is a potential candidate gene for crop improvement. In our study, TaWRKY46 transgenic plants underwent normal development compared with controls, but we have not statistically analyzed the effects on productivity. Further studies should focus on the grains yield of transgenic plants under drought stress conditions.
References
Abrahám E, Rigó G, Székely G, Nagy R, Koncz C, Szabados L (2003) Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol Biol 51:363–372
Bechtold U, Field B (2018) Molecular mechanisms controlling plant growth during abiotic stress. J Exp Bot 69:2753–2758
Chen W, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9:591–596
Chen L, Yu S, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Acta Biochim Biophys Sin 1819:121–128
Chi Y, Yang Y, Zhou Y, Jie Z, Fan B, Yu J, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6:287–300
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Jiang Y, Qiu Y, Hu Y, Yu Q (2016) Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front Plant Sci 7:154
Ju Y, Yue X, Zhao M, Wang X, Fang Y, Zhang J (2020) VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol Biochem 146:98–111
Niu C, Wei W, Zhou Q, Tian A, Chen S (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ 35:1156–1170
Rushton PJ, Somssich IE, Ringler P, Shen Q (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2:503–512
Tripathi P, Rabara RC, Rushton P (2014) A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239:255–266
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Wang F, Hou X, Tang J, Wang Z, Wang S, Jiang F, Ying L (2012) A novel cold-inducible gene from Pak-choi (Brassica campestris ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol Biol Rep 39:4553–4564
Wang C, Deng P, Chen L, Wang X, Ma H, Hu W, Yao N, Feng Y, Chai R, Yang G, He G (2013) A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 8:e65120
Wang C, Ru J, Liu Y, Li M, Zhao D, Yang J, Fu J, Xu Z (2018) Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int J Mol Sci 19:3046
Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K (2009) Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep 28:21–30
Xiong X, James VA, Zhang H, Altpeter F (2010) Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalumnotatum Flugge). Mol Breed 25:419–432
Xu Q, Feng W, Peng H, Ni Z, Sun Q (2014) TaWRKY71, a WRKY transcription factor from wheat, enhances tolerance to abiotic stress in transgenic Arabidopsis thaliana. Cereal Res Commun 42:47–57
Zhang L, Zhao G, Jia J, Liu X, Kong X (2012) Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot 63:203–214
Zhao T, Zhao S, Chen H, Zhao Q, Hu Z, Hou B, Xia G (2006) Transgenic wheat progeny resistant to powdery mildew generated by Agrobacterium inoculum to the basal portion of wheat seedling. Plant Cell Rep 25:1199–1204
Zhou Q, Tian A, Zou H, Xie Z, Lei G, Huang J, Wang C, Wang H, Zhang J (2008) GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503
Zhou L, Wang N, Gong S, Lu R, Li Y, Li X (2015) Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol Biochem 96:311–320
Zou C, Jiang W, Yu D (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61:3901–3914
Acknowledgements
This work was financially supported by Henan Provincial Department of Science and Technology Research Project (192102110031).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu, Y., Zhang, L. Overexpression of TaWRKY46 enhances drought tolerance in transgenic wheat. CEREAL RESEARCH COMMUNICATIONS 50, 679–688 (2022). https://doi.org/10.1007/s42976-021-00215-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00215-4