Abstract
Main conclusion
A novel broad-spectrum powdery mildew resistance gene PmPB74 was identified in wheat- Agropyron cristatum introgression line Pubing 74.
Development of wheat cultivars with broad-spectrum, durable resistance to powdery mildew has been restricted by lack of superior genetic resources. In this study, a wheat-A. cristatum introgression line Pubing 74, originally selected from a wide cross between the common wheat cultivar Fukuhokomugi (Fukuho) and Agropyron cristatum (L.) Gaertn (2n = 4x = 28; genome PPPP), displayed resistance to powdery mildew at both the seedling and adult stages. The putative alien chromosomal fragment in Pubing 74 was below the detection limit of genomic in situ hybridization (GISH), but evidence for other non-GISH-detectable introgressions was provided by the presence of three STS markers specific to A. cristatum. Genetic analysis indicated that Pubing 74 carried a single dominant gene for powdery mildew resistance, temporarily designated PmPB74. Molecular mapping showed that PmPB74 was located on wheat chromosome arm 5DS, and flanked by markers Xcfd81 and HRM02 at genetic distances of 2.5 and 1.7 cM, respectively. Compared with other lines with powdery mildew resistance gene(s) on wheat chromosome arm 5DS, Pubing 74 was resistant to all 28 Blumeria graminis f. sp tritici (Bgt) isolates from different wheat-producing regions of northern China. Allelism tests indicated that PmPB74 was not allelic to PmPB3558 or Pm2. Our work showed that PmPB74 is a novel gene with broad resistance to powdery mildew, and hence will be helpful in broadening the genetic basis of powdery mildew resistance in wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Powdery mildew (caused by Blumeria graminis f. sp. tritici, Bgt) is a destructive wheat disease all over the world, causing not only significant yield loss but also severe quality deterioration (Bowen et al. 1991; Everts and Leath 1992). Resistant cultivars are the most economical and environmental strategy to reduce the prevalence of powdery mildew, considering that fungicide application can cause environmental problems and likely future acquisition of fungicide tolerance by the pathogen (Huang et al. 1997; Paillard et al. 2000; Huang and Roder 2004).
More than 70 powdery mildew resistance alleles have been identified, a number of which were introduced from wild relatives of common wheat (Friebe et al. 1996; Gill et al. 2011; Mohler et al. 2013; McIntosh et al. 2014; Petersen et al. 2015), such as Triticum monococcum (2n = 2x = 14; genome AA) (Shi et al. 1998; Yao et al. 2007; Schmolke et al. 2012), Aegilops tauschii (2n = 2x = 14; genome DD) (Miranda et al. 2006, 2007; Schneider et al. 2008), Secale cereale L. (2n = 2x = 14; genome RR) (Friebe et al. 1994; Mohler et al. 2001), Haynaldia villosa L. (2n = 2x = 14; genome VV) (Chen et al. 1995, 2013; Xie et al. 2012; Zhang et al. 2012), and Thinopyrum intermedium (2n = 6x = 42; genome JJJsJsSS) (He et al. 2009; Liu et al. 2014; Shen et al. 2015). However, some powdery mildew resistance genes, such as Pm8 from S. cereale L. have become ineffective due to changes in the pathogen population (Hsam and Zeller 2002; McDonald and Linde 2002; Parks et al. 2008). Consequently, researchers are constantly seeking novel germplasms with broad-spectrum resistance to the disease.
Various molecular markers, including restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and single nucleotide polymorphism (SNP), have been used to map powdery mildew resistance genes in wheat. SSR markers including genomic SSR or EST-derived SSR markers have most commonly been used (Röder et al. 1998; Paillard et al. 2003; Sourdille et al. 2004; Somers et al. 2004; Yu et al. 2004). Currently, SNP markers are becoming increasingly popular (Akhunov et al. 2009; Berard et al. 2009; Chao et al. 2010; Lai et al. 2012; Allen et al. 2013), followed by the development of the wheat 9 and 90 K SNP chip platforms (Cavanagh et al. 2013; Avni et al. 2014; Wang et al. 2014). Upon SNP identification, various systems have been devised for SNP profiling, such as single strand conformation polymorphism (SSCP), kompetitive allele specific PCR (KASPar) assay, and fluorescent high resolution DNA melting (HRM) analysis. HRM analysis, a powerful tool for discrimination of a single SNP, has been successfully used in wheat (Matsuda et al. 2012; Tan et al. 2013; Terracciano et al. 2013).
Agropyron cristatum (L.) Gaertn (2n = 4x = 28; genomes PPPP), a perennial species of the Triticeae, harbors many favorable traits that can be exploited for wheat genetic improvement (Dewey 1984; Dong et al. 1992), such as enhanced fertile tiller number per plant (Ye et al. 2015), high grain number per spike (Wu et al. 2006; Luan et al. 2010), and high resistance to powdery mildew and other diseases (Han et al. 2014; Lu et al. 2015). Various wheat-A. cristatum derivative lines with elite traits including addition, translocation and introgression lines have been produced (Wu et al. 2006; Han et al. 2014; Ye et al. 2015; Lu et al. 2015; Zhang et al. 2015a). Pubing 74 is a putative wheat-A. cristatum introgression line, which displays a high level of resistance to powdery mildew at both the seedling and adult stages. However, the genetic basis of the resistance was still uncharacterized. We determined the chromosomal location of the resistance gene and also evaluated its effectiveness against a comprehensive set of Bgt isolates from a wide range of wheat-producing regions in China.
Materials and methods
Materials
Pubing 74 was originally selected from a wide cross between the common wheat cultivar (cv.) Fukuhokomugi (Fukuho) and A. cristatum (accession No. Z559). Common wheat cv. Mingxian 169 highly susceptible to powdery mildew was crossed with Pubing 74 to generate F2 and F2-derived F3 family populations. Common wheat cv. Zhongzuo 9504 was used as the susceptible control in the powdery mildew assessment. Wheat lines with known powdery mildew resistance genes on wheat chromosome arm 5DS, including wheat landrace derivative Ulka/8*Cc (Briggle 1966; Qiu et al. 2006), German wheat cv. Tabasco (Gao et al. 2012), Chinese wheat cv. Liangxing 66 (Huang et al. 2012), wheat-A. cristatum introgression line PB3558 (Lu et al. 2015), common wheat line D57 (Ma et al. 2011), indigenous germplasm X3986-2 (Ma et al. 2014), and Chinese breeding line KM2939 (Ma et al. 2015), were used in this study. Tabasco was provided by Dr. Shibin Cai (Jiangsu Academy of Agricultural Sciences), D57 by Dr. Zhengqiang Ma (Nangjing Agricultural University), X3986-2 and KM2939 by Dr. Diaoguo An (Chinese Academy of Sciences, Shijiazhuang, Hebei province). The other wheat lines are maintained by our laboratory. All 28 single spore-derived Bgt isolates used in this study were kindly provided by Dr. Hongjie Li (Institute of Crop Science, Chinese Academy of Agricultural Sciences).
Genomic in situ hybridization and meiosis observation
Genomic in situ hybridization (GISH) was conducted according to Luan et al. (2010). Agropyron cristatum and Fukuho genomic DNA were used as the probe and blocker, respectively. Agropyron cristatum genomic DNA was labeled with Dig-Nick-Translation Mix (Roche, Mannheim, Germany). Wheat and A. cristatum chromosomes were pseudo-colored as blue and red, respectively. The procedure used for meiotic studies was described by Jauhar and Peterson (2006). Young spikes from Pubing 74 with pollen mother cells (PMCs) at the metaphase I (MI) stage were fixed in Carnoy solution (6-ethanol: 3-chloroform: 1-acetic acid, by vol.) for 24 h and stored at 4 °C until used. All cytological images were taken under a Nikon Eclipse E600 fluorescence microscope and captured with a CCD camera (Diagnostic Instruments, Sterling Heights, MI, USA).
Evaluation of powdery mildew response
The prevailing Bgt isolate E09 was used to test Pubing 74 × Mingxian 169 F1 hybrids, an F2 population, and F2-derived F3 families at the seedling stage. Seedlings at the one-leaf-stage were inoculated by dusting conidiospores of Bgt isolate E09 from susceptible cv. Zhongzuo 9504. Infection types (ITs) were scored on the first leaf using a 0–4 scale around 15 days after inoculation (Liu et al. 2002). Scores of 0–2 were classified as resistant and 3–4 as susceptible. Twenty seedlings of each line in the F2:3 population were tested against Bgt isolate E09 to determine the genotypes of the F2 individuals. We used 20 seedlings for each line, since this was adequate to reduce the probability of erroneously determining a heterozygous plant as homozygous resistant to 0.3 %, and to reduce the probability of determining a heterozygous plant as homozygous susceptible to 9e−13.
Twenty-eight Bgt isolates originating from different wheat-producing regions of Northern China (Sun et al. 2015) were used to compare the reaction patterns of Pubing 74 and lines with known alleles on chromosome arm 5DS. The reactions to 28 Bgt isolates were determined using detached leaf segments as described by Limpert et al. (1988). Three leaf segments from different plants of each genotype were examined and the tests were repeated three times. Besides, a mixture of Bgt isolates mainly composed of E09 was employed to inoculate the populations as well as two parents in the field at the adult stage. All plants were sown in 2.0 m rows, spaced 0.3 m apart. The susceptible control cv. Zhongzuo 9504 was planted in every fifth row to ensure that all plants were evenly infected. Disease reactions were scored using a 0–9 scale at the ear emergence and milky ripe stages.
DNA extraction and bulked segregant analysis
Genomic DNA was isolated from leaves of young seedlings following the CTAB method (Allen et al. 2006). To detect A. cristatum chromosomal fragments in Pubing 74, three sequence-tagged-site (STS) markers (Agc2970, Agc6287 and Agc21686) designed according to the expressed sequence tags (EST) of the A. cristatum transcriptome (Zhang et al. 2015b). Bulked segregant analysis (BSA) was applied as described by Michelmore et al. (1991). Briefly, resistant and susceptible DNA bulks were constructed by separately mixing equal amounts of DNA from 20 homozygous resistant (IT = 0) and 20 homozygous susceptible (IT = 4) F2 plants, homozygosity being established on the basis of progeny testing.
Genotyping with publicly available and new developed molecular markers
SSR markers evenly distributed across all the wheat chromosomes were used for a polymorphism survey on the two parents and two DNA bulks, and polymorphic markers were subsequently used for genotyping the segregating population. A series of Xbwm SSR markers, which were developed recently and located on wheat chromosome arm 5DS (Lu et al. 2015), were chosen for genotyping. Three HRM markers were designed based on the flanking sequences of SNPs, taking the sequences and their physical locations on wheat chromosome arm 5DS as reference (Jia et al. 2013). PCR was performed in 10 µL volumes containing 0.1 U Taq DNA polymerase, 2 µM forward and reverse primers, and approximately 30 ng of template DNA. The PCR amplification conditions were: 35 cycles at 95 °C for 30 s, 55–65 °C for 30 s and 72 °C for 30 s with an initial denaturation at 95 °C for 2 min and a final extension step at 72 °C for 5 min.
When PCR was carried out with HRM markers, 1 µL 10 x LC Green (Idaho Technology, Salt Lake City, UT, USA) was included in 10 µL PCR volumes. The resulting PCR products were analyzed using a light scanner (Idaho Technology), by ramping the temperature from 55 to 95 °C at 0.1 °C per second. Data were analyzed using the analytical light scanning software, and the amplification patterns of the HRM markers were shown in two different ways: normalized melting peaks and normalized melting curves.
Statistical analysis and linkage map construction
Chi squared (χ 2) tests were used to compare the observed and theoretically expected ratios. Polymorphic markers were used for constructing a linkage map flanking the powdery mildew resistance gene in Pubing 74, and a linkage map was established with Mapmaker 3.0 and Mapdraw (Lincoln et al. 1993; Liu and Meng 2003). The LOD score threshold was set at 3.0 and a maximum genetic distance at 50 cM. The Kosambi mapping function was used to estimate genetic distances between linked markers and the powdery mildew resistance gene based on recombination values.
Results
Pubing 74 is a novel wheat-A. cristatum introgression line
Pubing 74 was produced from a wide cross between A. cristatum and wheat cv. Fukuho, and then selected over five selfing generations. Pubing 74 displayed full fertility and normal agronomic performance, such as 56–73 grains per spike, and 15–20 fertile tillers per plant (Fig. 1a). GISH was performed to determine whether Pubing 74 is a wheat-A. cristatum introgression line. No visible hybridization signal was detected in Pubing 74 (Fig. 1b), despite strong hybridization signals in the positive control (Fig. 1c). However, evidence for the presence of A. cristatum chromatin was provided by STS markers specific to the A. cristatum P genome. STS markers Agc2970, Agc6287 and Agc21686 were amplified in A. cristatum and Pubing 74, but not in Fukuho or Mingxian 169 (Fig. 1d). These results indicated that A. cristatum chromatin was present in Pubing 74, but was presumably too small to be detected by the standard cytological methods. Meiotic metaphase PMC cells showed normal 21 bivalent chromosome pairing in Pubing 74 (Fig. 1e), indicating that Pubing 74 could be a stable wheat-A. cristatum introgression line.
Identification of wheat-A. cristatum introgression line Pubing 74. a Normal appearance of a Pubing 74 plant. b, c GISH detection of Pubing 74 (b) and one wheat-A. cristatum translocation line WAT12-9 as the positive control (c). d PCR patterns of three STS markers specific to A. cristatum (Agc2970, Agc6287 and Agc21686). Lanes M DNA ladder; 1 A. cristatum; 2 Fukuho; 3 Pubing 74; 4 Mingxian 169. e PMC from Pubing 74 with 21 bivalents at meiotic metaphase I (MI)
Inheritance of powdery mildew resistance in Pubing 74
Pubing 74 was highly resistant under natural field epidemic conditions over several years. It was also resistant at the seeding and adult plant stages under controlled conditions (Fig. 2). The inheritance data presented in Table 1 showed that resistance to Bgt race E09 was conferred by a single dominant gene, which was tentatively designated PmPB74. Results from seedling and adult plant tests were identical.
Chromosomal location of PmPB74 in Pubing 74
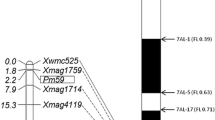
Three hundred and seventy-eight wheat SSR markers distributed randomly throughout the wheat genome were screened for polymorphisms between the parents and DNA bulks, and nine markers showed polymorphisms. Among these nine markers, only Xcfd81 showed evidence of linkage with PmPB74. Since Xcfd81 was known to be on chromosome arm 5DS, additional SSR markers on that chromosome arm were surveyed for polymorphisms. Four SSR markers (Xgpw5201, Xwmc805, Xgpw302 and Xcfd40) were polymorphic and linked to PmPB74 (Fig. 3). Four of 25 Xbwm SSR markers, and three of 15 HRM markers were also polymorphic and linked to PmPB74 (Fig. 3; Table 2). The linkage map based on the powdery mildew response and marker data indicated that PmPB74 was flanked by markers Xcfd81 and HRM02 at genetic distances of 2.5 and 1.7 cM, respectively (Fig. 3). Xcfd81 was earlier reported to be located on the bin C-5DS1-0-0.63, hence PmPB74 is also likely to be located on this bin. However, the three STS markers specific to A. cristatum were not linked with PmPB74, nor were they linked to each other, suggesting that there were multiple small chromosome segments from A. cristatum distributed at different chromosomal locations in Pubing 74. Amplification patterns of two SSR markers (Xcfd81 and Xbwm25) and two HRM markers (HRM01 and HRM02) are shown as examples in Figs. 4, 5, respectively.
Amplification profiles of SSR markers Xcfd81 (a) and Xbwm25 (b). Lanes M DNA ladder; PR resistant parent Pubing 74; PS susceptible parent Mingxian 169; BR resistant DNA bulk, BS susceptible DNA bulk; HR homozygous resistant F2 individual; HZ heterozygous resistant F2 individual; HS homozygous susceptible F2 individual. Polymorphic PCR bands are indicated by arrows
Amplification profiles of HRM markers: HRM01 (a) and HRM02 (b). Left and right panels display normalized melting peaks and normalized melting curves for the same amplicons, respectively. Lines with different colours represent different genotypes: red lines represent Mingxian 169 and the homozygous susceptible F2 plants; blue lines represent Pubing 74 and the homozygous resistant F2 plants; gray lines represent heterozygous F2 plants
Comparative reactions of Pubing 74 and other lines with known powdery mildew resistance genes on chromosome arm 5DS to 28 Bgt isolates
To determine the relationships of PmPB74 with other genes located on chromosome 5DS, 28 Bgt isolates including E09 from different wheat-producing regions of Northern China were used in inoculations. As shown in Table 3, Pubing 74 and A. cristatum were resistant to all the 28 isolates tested. However, Ulka/8*Cc (Pm2) was susceptible to 12 isolates, Tabasco (Pm48) was susceptible to eight isolates, Liangxing 66 (PmLX66) was susceptible to 12 isolates, and PB3558 (PmPB3558) was susceptible to seven isolates. The disease reactions of seven lines to six Bgt isolates as examples are shown in Fig. 6. Besides, seven isolates were used to compare the reactions of Pubing 74 and three lines (D57, X3986-2 and KM2939). As shown in Table 4, both D57 and X3986-2 were each susceptible to two Bgt isolates, and KM2939 was susceptible to E21. Therefore, Pubing 74 displayed a broader spectrum of disease resistance than any other wheat line tested above. Finally, when 20 F2:3 lines homozygous resistant and 20 F2:3 lines homozygous susceptible to isolate E09 were tested with the other 27 isolates at the seedling stage, all these lines showed disease reactions identical to those inoculated with E09, thus showing that PmPB74 conferred resistance to all the isolates.
PmPB74 and PmPB3558 were both derived from A. cristatum accession Z559. To determine whether PmPB74 was PmPB3558, the Bgt isolate Bg44-6, which was virulent to PmPB3558 (and Mingxian 169) but avirulent to PmPB74, was used to inoculate the Pubing 74 × Mingxian 169 F2:3 population at the seedling stage. A segregation pattern identical with that of E09 was obtained and PmPB74 was assigned to the same chromosomal location, indicating that PmPB74 was indeed not identical to PmPB3558. To further determine whether PmPB74 and PmPB3558, and also PmPB74 and Pm2 were different alleles in a single locus, crosses between Pubing 74 and Ulka/8*Cc, and between Pubing 74 and PB3558, respectively, were made. When large F2 seedling populations were tested with isolate E09, which was avirulent to all three host lines, two of 6986 F2 plants from Pubing 74 × Ulka/8*Cc were susceptible to powdery mildew (IT = 4), and just one of 2260 F2 plants from Pubing 74 × PB3558 displayed susceptibility to powdery mildew (IT = 4). Thus, PmPB74 was not allelic to Pm2 nor to PmPB3558. To confirm the conclusion that PmPB74 represents a novel locus, we inoculated Pubing 74 × PB3558 F2:3 lines with E09, and two lines showing 3 resistant:1 susceptible segregation were identified from 168 F2:3 lines. Similarly, we tested the relationship between PmPB74 and Pm2 in Pubing 74 × Ulka/8*Cc F2:3 lines, and two segregating lines were identified from 392 F2:3 lines.
Discussion
Introgression lines are valuable sources of resistance genes in common wheat
The wild relatives of wheat have been used effectively as donors of desirable genes conferring superior agronomic traits. However, large alien chromosomal fragments may carry additional genes that confer undesirable traits in wheat, a phenomenon known as ‘linkage drag’. Therefore, smaller alien chromosomal fragments are generally preferred. Nevertheless, small alien chromosomal fragments with desirable genes are sometimes too small to be detected by standard cytological methods. Examples include the alien introgression of small chromosomal fragments from Dasypyrum villosum and Thinopyrum ponticum to wheat (Caceres et al. 2012; Chen et al. 2012), rust resistance genes introgressed from Ae. geniculata and Ae. triuncialis to wheat (Kuraparthy et al. 2007a, b), and powdery mildew and stripe rust resistance genes putatively derived from T. intermedium (He et al. 2009; Liu et al. 2013; Huang et al. 2014). Pubing 74, was obtained from distant hybridization between the common wheat cv. Fukuho and A. cristatum, displayed high resistance to powdery mildew at both the seedling and adult stages. The putative chromosomal fragment from A. cristatum was below the detection limit of GISH, but evidence for other non-GISH-detectable introgressions was provided by the presence of three STS markers specific to A. cristatum. However, these markers were genetically independent of PmPB74 and also independent of each other, suggesting that there were multiple small chromosome segments from A. cristatum distributed at different chromosomal locations in Pubing 74. Therefore, further studies are needed to develop more markers specific to A. cristatum and also linked to PmPB74. Nevertheless, the resistance in Pubing 74 was presumably derived from A. cristatum, considering that A. cristatum was the only parent highly resistant to powdery mildew.
PmPB74 is a novel resistance gene
PmPB74 occupied a different chromosomal position and conferred a different array of powdery mildew response compared with other genes at or near the Pm2 locus. PmPB74 and PmPB3558 are present in wheat-A. cristatum introgression lines, both of which display high levels of resistance to powdery mildew at both the seedling and adult stages (Lu et al. 2015). However, PmPB74 and PmPB3558 conferred different response spectra, and an allelism test indicated that they were not allelic. Similar results were also obtained for PmPB74 and Pm2. We also compared the response spectra of lines carrying PmPB74 and other resistance genes located on wheat chromosome 5DS. When inoculated with 28 Bgt isolates, the lines with Pm48 and PmLX66 was susceptible to 8 and 12 Bgt isolates, respectively (Table 3). When inoculated with seven out of 28 Bgt isolates, the lines with PmD57-5D and PmX3986-2 were susceptible to two isolates, and KM2939 was susceptible to one isolate (Table 4). By contrast, PmPB74 was highly resistant to all the 28 Bgt isolates, showing that the resistance spectrum of PmPB74 was broader than all of the other genes mentioned above. We concluded that PmPB74 is a novel gene.
PmPB74 is potentially valuable for resistance breeding
Most powdery mildew resistance sources derived from wild relatives of wheat are usually not directly applicable in wheat breeding because of linkage drag. Pubing 74 not only displayed a broad-spectrum of resistance against Bgt isolates from northern China, but also exhibited superior agronomic performance without linkage drag. Therefore, Pubing 74 was an ideal germplasm potentially useful to enhance the powdery mildew resistance at different wheat genetic backgrounds. In this study, no virulence to PmPB74 was found; therefore, identification of PmPB74 and its closely linked markers will be useful for breeders to combine it with other powdery mildew resistance genes. Actually, transferring PmPB74 into different commercial wheat varieties is currently being conducted in our group, and a range of powdery mildew resistant introgression lines have been obtained. In conclusion, the identification of PmPB74 reported here and its corresponding closely linked molecular markers will be beneficial to increasing the diversity of the genetic sources of powdery mildew resistance.
Author contribution statement
L. H. Li and Y. Q. Lu designed the research; M. M. Yao, Y. Q. Lu, J. P. Zhang, L. Q. Song, W. H. Liu, X. M. Yang and X. Q. Li performed technical work; Y. Q. Lu and M. M. Yao analyzed the data. Y. Q. Lu wrote the manuscript.
Abbreviations
- Fukuho:
-
cv. Fukuhokomugi
- GISH:
-
Genomic in situ hybridization
- HRM:
-
High resolution DNA melting
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
References
Akhunov E, Nicolet C, Dvorak J (2009) Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay. Theor Appl Genet 119:507–517
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325
Allen AM, Barker GL, Wilkinson P, Burridge A, Winfield M, Coghill J, Uauy C, Griffiths S, Jack P, Berry S, Werner P, Melichar JP, McDougall J, Gwilliam R, Robinson P, Edwards K (2013) Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol J 11:279–295
Avni R, Nave M, Eilam T, Sela H, Alekperov C, Peleg Z, Dvorak J, Korol A, Distelfeld A (2014) Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol Breed 34:1–14
Berard A, Le Paslier MC, Dardevet M, Exbrayat-Vinson F, Bonnin I, Cenci A, Haudry A, Brunel D, Ravel C (2009) High-throughput single nucleotide polymorphism genotyping in wheat (Triticum spp.). Plant Biotechnol J 7:364–374
Bowen KL, Everts KL, Leath S (1991) Reduction in yield of winter wheat in North Carolina due to powdery mildew and leaf rust. Phytopathology 81:503–511
Briggle LW (1966) Three loci in wheat involving resistance to Erysiphe graminis f. sp. tritici. Crop Sci 6:461–465
Caceres ME, Pupilli F, Ceccarelli M, Vaccino P, Sarri V, Depace C, Cionini PG (2012) Cryptic introgression of Dasypyrum villosum parental DNA in wheat lines derived from intergeneric hybridization. Cytogenet Genome Res 136:75–81
Cavanagh CR, Chao SM, Wang SC, Huang BE, Stephen S et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110:8057–8062
Chao S, Dubcovsky J, Dvorak J, Luo MC, Baenziger SP et al (2010) Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genom 11:727
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128
Chen G, Zheng Q, Bao YG, Liu SB, Wang HG, Li XF (2012) Molecular cytogenetic identification of a novel dwarf wheat line with introgressed Thinopyrum ponticum chromatin. J Biol Sci 37:149–155
Chen PD, You CF, Hu Y, Chen SW, Zhou B, Cao AZ, Wang X (2013) Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol Breed 31:477–484
Dewey DR (1984) The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP (ed) Gene manipulation in plant inprovement. Plenum Press, New York, pp 209–279
Dong YC, Zhou RH, Xu SJ, Li LH, Cauderon Y, Wang RRC (1992) Desirable characteristics in perennial Triticeae collected in China for wheat improvement. Hereditas 116:175–178
Everts KL, Leath S (1992) Effect of early season powdery mildew on development, survival, and yield contribution of tillers of winter wheat. Phytopathology 82:1273–1278
Friebe B, Heun M, Tuleen N, Zeller FJ, Gill BS (1994) Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci 34:621–625
Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87
Gao HD, Zhu FF, Jiang YJ, Wu JZ, Yan W, Zhang QF, Jacobi A, Cai SB (2012) Genetic analysis and molecular mapping of a new powdery mildew resistant gene Pm46 in common wheat. Theor Appl Genet 125:967–973
Gill BS, Friebe BR, White FF (2011) Alien introgressions represent a rich source of genes for crop improvement. Proc Natl Acad Sci USA 108:7657–7658
Han HM, Bai L, Su JJ, Zhang JP, Song LQ, Gao AN, Yang XM, Li XQ, Liu WH, Li LH (2014) Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PLoS One 9:e91066
He RL, Chang ZJ, Yang ZJ, Yuan ZY, Zhan HX, Zhang XJ, Liu JX (2009) Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor Appl Genet 118:1173–1180
Hsam SLK, Zeller FJ (2002) Breeding for powdery mildew resistance in common wheat (T. aestivum L.). In: Belanger RR, Bushnell WR, Dik AJ, Carver TLW (eds) The powdery mildews, a comprehensive treatise. APS Press, St Paul, pp 219–238
Huang XQ, Roder MS (2004) Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137:203–223
Huang XQ, Hsam SLK, Zeller FJ (1997) Identification of powdery mildew resistance genes in common wheat (Triticum aestivum L. em Thell).9. Cultivars, land races and breeding lines grown in China. Plant Breed 116:233–238
Huang J, Zhao ZH, Song FJ, Wang XM, Xu HX, Huang Y, An DG, Li HJ (2012) Molecular detection of a gene effective against powdery mildew in the wheat cultivar Liangxing 66. Mol Breed 30:1737–1745
Huang Q, Li X, Chen WQ, Xiang ZP, Zhong SF, Chang ZJ, Zhang M, Zhang HY, Tan FQ, Ren ZL, Luo PG (2014) Genetic mapping of a putative Thinopyrum intermedium-derived stripe rust resistance gene on wheat chromosome 1B. Theor Appl Genet 127:843–853
Jauhar PP, Peterson TS (2006) Cytological analyses of hybrids and derivatives of hybrids between durum wheat and Thinopyrum bessarabicum, using multi-colour fluorescent GISH. Plant Breed 125:19–26
Jia JZ, Zhao SC, Kong XY, Li YR, Zhao GY et al (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kuraparthy V, Chhuneja P, Dhaliwal HS, Kaur S, Bowden RL, Gill BS (2007a) Characterization and mapping of cryptic alien introgression from Aegilops geniculata with new leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor Appl Genet 114:1379–1389
Kuraparthy V, Sood S, Chhuneja P, Dhaliwal HS, Kaur S, Bowden RL, Gill BS (2007b) A cryptic wheat-Aegilops triuncialis translocation with leaf rust resistance gene Lr58. Crop Sci 47:1995–2003
Lai KT, Duran C, Berkman PJ, Lorenc MT, Stiller J, Manoli S, Hayden MJ, Forrest KL, Fleury D, Baumann U, Zander M, Mason AS, Batley J, Edwards D (2012) Single nucleotide polymorphism discovery from wheat next-generation sequence data. Plant Biotechnol J 10:743–749
Limpert E, Andrivon D, Felsenstein FG (1988) Influence of different benzimidazole concentrations in agar medium on senescence of wheat leaf segments and on growth and sporulation of the wheat powdery mildew pathogen. J Plant Dis Protect 95:301–306
Lincoln SE, Daly MJ, Lander ES (1993) Constructing linkage maps with MAPMAKER/Exp version 3. 0: a tutorial reference manual, 3rd edn. Whitehead Institute for Medical Res, Cambridge
Liu RH, Meng JL (2003) MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25:317–321
Liu ZY, Sun QX, Ni ZF, Nevo E, Yang TM (2002) Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica 123:21–29
Liu J, Chang ZJ, Zhang XJ, Yang ZJ, Li X, Jia JQ, Zhan HX, Guo HJ, Wang JM (2013) Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor Appl Genet 126:265–274
Liu ZH, Xu M, Xiang ZP, Li X, Chen WQ, Luo PG (2014) Registration of the novel wheat lines L658, L693, L696, and L699, which are resistant to Fusarium head blight, stripe rust, and powdery mildew. J Plant Regist 9:121–124
Lu YQ, Wu XY, Yao MM, Zhang JP, Liu WH, Yang XM, Li XQ, Du J, Gao AN, Li LH (2015) Genetic mapping of a putative Agropyron cristatum-derived powdery mildew resistance gene by a combination of bulked segregant analysis and single nucleotide polymorphism array. Mol Breed 35:1–13
Luan Y, Wang XG, Liu WH, Li CY, Zhang JP, Gao AN, Wang YD, Yang XM, Li LH (2010) Production and identification of wheat-Agropyron cristatum 6P translocation lines. Planta 232:501–510
Ma HQ, Kong ZX, Fu BS, Li N, Zhang LX, Jia HY, Ma ZQ (2011) Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor Appl Genet 123:1099–1106
Ma PT, Xu HX, Luo QL, Qie YM, Zhou YL, Xu YF, Han HM, Li LH, An DG (2014) Inheritance and genetic mapping of a gene for seedling resistance to powdery mildew in wheat line X3986-2. Euphytica 200:149–157
Ma PT, Xu HX, Xu YF, Li LL, Qie YM, Luo QL, Zhang XT, Li XQ, Zhou YL, An DG (2015) Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor Appl Genet 128:613–622
Matsuda R, Iehisa JC, Takumi S (2012) Application of real-time PCR-based SNP detection for mapping of Net2, a causal D-genome gene for hybrid necrosis in interspecific crosses between tetraploid wheat and Aegilops tauschii. Genes Genet Syst 87:137–143
McDonald BA, Linde C (2002) The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124:163–180
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC (2014) Catalogue of gene symbols for wheat: 2013–2014 supplement. Komugi-wheat genetic resources database. http://www.shigen.nig.ac.jp/wheat/komugi/
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Miranda LM, Murphy JP, Marshall D, Leath S (2006) Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor Appl Genet 113:1497–1504
Miranda LM, Murphy JP, Marshall D, Cowger C, Leath S (2007) Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor Appl Genet 114:1451–1456
Mohler V, Hsam SLK, Zeller FJ, Wenzel G (2001) An STS marker distinguishing the rye-derived powdery mildew resistance alleles at the Pm8/Pm17 locus of common wheat. Plant Breed 120:448–450
Mohler V, Bauer C, Schweizer G, Kempf H, Hartl L (2013) Pm50: a new powdery mildew resistance gene in common wheat derived from cultivated emmer. J Appl Genet 54:259–263
Paillard S, Goldringer I, Enjalbert J, Doussinault G, de Vallavieille-Pope C, Brabant P (2000) Evolution of resistance against powdery mildew in winter wheat populations conducted under dynamic management. I–Is specific seedling resistance selected? Theor Appl Genet 101:449–456
Paillard S, Schnurbusch T, Winzeler M, Messmer M, Sourdille P, Abderhalden O, Keller B, Schachermayr G (2003) An integrative genetic linkage map of winter wheat (Triticum aestivum L.). Theor Appl Genet 107:1235–1242
Parks R, Carbone I, Murphy JP, Marshall D, Cowger C (2008) Virulence structure of the Eastern US wheat powdery mildew population. Plant Dis 92:1074–1082
Petersen S, Lyerly JH, Worthington ML, Parks WR, Cowger C, Marshall DS, Brown-Guedira G, Murphy PJ (2015) Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor Appl Genet 128:303–312
Qiu YC, Sun XL, Zhou RH, Kong XY, Zhang SS, Jia JZ (2006) Identification of microsatellite markers linked to powdery mildew resistance gene Pm2 in wheat. Cereal Res Commun 34:1267–1273
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Schmolke M, Mohler V, Hartl L, Zeller FJ, Hsam SLK (2012) A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol Breed 29:449–456
Schneider A, Molnar I, Molnar-Lang M (2008) Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163:1–19
Shen XK, Ma LX, Zhong SF, Liu N, Zhang M, Chen WQ, Zhou YL, Li HJ, Chang ZJ, Li X, Bai GH, Zhang HY, Tan FQ, Ren ZL, Luo PG (2015) Identification and genetic mapping of the putative Thinopyrum intermedium-derived dominant powdery mildew resistance gene PmL962 on wheat chromosome arm 2BS. Theor Appl Genet 128:517–528
Shi AN, Leath S, Murphy JP (1998) A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology 88:144–147
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi LL, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of geneticphysical map relationships in wheat (Triticum aestivum L.). Funct Integr Genom 4:12–25
Sun YL, Zou JW, Sun HG, Song W, Wang XM, Li HJ (2015) PmLX66 and PmW14: new alleles of Pm2 for resistance to powdery mildew in the Chinese winter wheat cultivars Liangxing 66 and Wennong 14. Plant Dis 99:1118–1124
Tan YY, Fu HW, Zhao HR, Lu S, Fu JJ, Li YF, Cui HR, Shu QY (2013) Functional molecular markers and high-resolution melting curve analysis of low phytic acid mutations for marker-assisted selection in rice. Mol Breed 31:517–528
Terracciano I, Maccaferri M, Bassi F, Mantovani P, Sanguineti MC, Salvi S, Simkova H, Dolezel J, Massi A, Ammar K, Kolmer J, Tuberosa R (2013) Development of COS-SNP and HRM markers for high-throughput and reliable haplotype-based detection of Lr14a in durum wheat (Triticum durum Desf.). Theor Appl Genet 126:1077–1101
Wang SC, Wong D, Forrest K, Allen A, Chao SM et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wu J, Yang XM, Wang H, Li HJ, Li LH, Li XQ, Liu WH (2006) The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet 114:13–20
Xie WL, Ben-David R, Zeng B, Dinoor A, Xie CJ, Sun QX, Röder MS, Fahoum A, Fahima T (2012) Suppressed recombination rate in 6VS/6AL translocation region carrying the Pm21 locus introgressed from Haynaldia villosa into hexaploid wheat. Mol Breeding 29:399–412
Yao GQ, Zhang JL, Yang LL, Xu HX, Jiang YM, Xiong L, Zhang CQ, Zhang ZZ, Ma ZQ, Sorrells ME (2007) Genetic mapping of two powdery mildew resistance genes in einkorn (Triticum monococcum L.) accessions. Theor Appl Genet 114:351–358
Ye XL, Lu YQ, Liu WH, Chen GY, Han HM, Zhang JP, Yang XM, Li XQ, Gao AN, Li LH (2015) The effects of chromosome 6P on fertile tiller number of wheat as revealed in wheat-Agropyron cristatum chromosome 5A/6P translocation lines. Theor Appl Genet 128:797–811
Yu JK, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47: 805–818
Zhang RQ, Wang X, Chen PD (2012) Molecular and cytogenetic characterization of a small alien-segment translocation line carrying the softness genes of Haynaldia villosa. Genome 55:639–646
Zhang J, Zhang JP, Liu WH, Han HM, Lu YQ, Yang XM, Li XQ, Li LH (2015a) Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. Theor Appl Genet 128:1827–1837
Zhang JP, Liu WH, Han HM, Song LQ, Bai L, Gao ZH, Zhang Y, Yang XM, Li XQ, Gao AN, Li LH (2015b) De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics 106:129–136
Acknowledgments
This work was funded by the National Natural Science Foundation of China (No. 31271714), the National Key Technology Support Program of China (No. 2013BAD01B02), and the CAAS Innovation Team Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Y. Lu and M. Yao are contributed equally to this work.
Rights and permissions
About this article
Cite this article
Lu, Y., Yao, M., Zhang, J. et al. Genetic analysis of a novel broad-spectrum powdery mildew resistance gene from the wheat-Agropyron cristatum introgression line Pubing 74. Planta 244, 713–723 (2016). https://doi.org/10.1007/s00425-016-2538-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2538-y