Abstract
Key message
A new powdery mildew resistance gene conferring a wide spectrum of resistance to Bgt isolates in the USA, Pm63 , was identified in Iranian wheat landrace PI 628024 and mapped to the terminal region of the long arm of chromosome 2B.
Abstract
Powdery mildew is a globally important wheat disease causing severe yield losses, and host resistance is the preferred strategy for managing this disease. The objective of this study was to characterize a powdery mildew resistance gene in Iranian landrace PI 628024, which exhibited a wide spectrum of resistance to representative Blumeria graminis f. sp. tritici (Bgt) isolates collected from different regions of the USA. An F2 population and F2:3 lines derived from the cross PI 628024 × CItr 11349 were used in this study, and genetic analysis indicated that a single dominant gene, designated Pm63, conferred resistance to Bgt isolate OKS(14)-B-3-1. Linkage analysis located Pm63 to an interval of about 13.1 Mb on the long arm of chromosome 2B, spanning 710.3–723.4 Mb in the Chinese Spring reference sequence. Bin mapping assigned Pm63 to the terminal bin 2BL6-0.89-1.0, 1.1 cM proximal to STS marker Xbcd135-2 and 0.6 cM distal to SSR marker Xstars419. Allelism tests indicated that Pm63 is a new powdery mildew resistance gene, which differs from other genes in the terminal bin by origin, genomic location, and responses to a set of 16 representative US Bgt isolates. Pm63 can be widely used to enhance powdery mildew resistance in the Great Plains, western, and southeastern regions of the USA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is the second most important staple food crop after rice and is cultivated on approximately 220 million ha in diverse geographical regions, environments, and production systems (Singh et al. 2016). With the global population projected to exceed 9 billion by 2050, a 1.6% annual increase in wheat production was estimated to satisfy the demands of this increase in global population, such that wheat yields should be increased from the current 3 tons/ha to 5 tons/ha in 2050 (Tilman et al. 2002; Singh et al. 2016). Powdery mildew, a serious wheat disease caused by the biotrophic fungus Blumeria graminis f. sp. tritici, looms as a logical threat to this yield goal, as powdery mildew occurs globally in wheat-growing regions, especially in highly productive regions where modern wheat cultivation technologies, characterized by the use of semidwarf and high-yielding cultivars, irrigation, and high levels of nitrogen fertilizer, are utilized. A typical example is Hungary, where there were moderate or severe powdery mildew epidemics in 11 of the 14 years between 1986 and 1999, and yield losses of 5 to 8% were estimated for years of average infection and up to 30% in years of severe epidemics (Szunics et al. 2001). In the USA, yield losses inflicted by powdery mildew ranged from 5 to 34% (Conner et al. 2003; Griffey et al. 1993), mainly due to reductions in tiller number, grain number, and kernel weight, while grain protein content was also reduced (Parry 1990; Bowen et al. 1991).
The serious yield losses caused by powdery mildew have stimulated breeding of powdery mildew-resistant cultivars. A previous survey suggested that powdery mildew was one of the top four disease priorities in 115 winter and facultative wheat-breeding programs worldwide (Braun et al. 1997). A considerable number of powdery mildew resistance genes, including 62 permanently designated genes and over 20 temporarily named genes or QTL, have been identified (McIntosh et al. 2013, 2017), and some of them, especially those identified in bread wheat, have been widely used in wheat breeding. However, most of these genes are race-specific and confer immunity or high resistance to powdery mildew, but thereby exert strong selection on Bgt populations and subsequently lead to the buildup of virulent pathotypes with matching virulence genes (McDonald and Linde 2002). Recent studies indicated that Pm2, Pm3a, Pm3b, Pm3f, Pm4a, Pm6, Pm8, and Pm17 have been defeated in part or all of the USA, while Pm1a was defeated in Australia, China, and Egypt (Parks et al. 2008; Cowger et al. 2018). The ability of Bgt to overcome deployed race-specific resistance genes necessitates a continuous search for new resistance genes.
Wheat powdery mildew resistance gene pools include bread wheat and wheat relatives. A considerable number of these resistance genes originated from cultivated and wild relatives. Some of them, such as Pm6 from Triticum timopheevii and Pm8 from rye (Secale cereale L.) (Helmsjørgensen and Jensen 1973; McIntosh et al. 2011), have been widely used in wheat improvement. However, it is a daunting task to eliminate linkage drag associated with alien genes. For example, Pm21 was transferred to wheat in the early 1990s (Qi et al. 1996), but elite breeding lines with Pm21 were not released in the most important wheat-growing region in China, the Huang-Huai River Valley, until recently (Cao et al. 2015). On the other hand, powdery mildew resistance genes identified in landraces, which have experienced extreme environmental challenges, can be more easily introgressed and deployed in new cultivars. Several powdery mildew resistance genes, such as Pm2c, Pm3b, Pm5d, Pm5e, Pm24a, Pm24b, Pm45, Pm47, Pm59, and Pm61, have been identified in landraces and used in germplasm enhancement and wheat breeding (Huang et al. 2000, 2003; Hsam et al. 2001; Yahiaoui et al. 2004; Ma et al. 2011; Xue et al. 2012; Xiao et al. 2013; Xu et al. 2015; Sun et al. 2018; Tan et al. 2018).
Recently, a set of landraces exhibiting high resistance to Bgt isolates from the Great Plains of the USA were identified; many of them had been collected from Middle East, a major center of origin for cereal species (Li et al. 2016). Of these, PI 628024, a landrace collected from Iran, showed a wide spectrum of resistance to Bgt isolates collected in the USA. The objectives of this study were to characterize the resistance gene in PI 628024 using molecular markers and to determine its resistance spectrum.
Materials and methods
Plant materials
PI 628024, an Iranian landrace, was highly resistant to Bgt isolates collected from Oklahoma, including OKS (14)-B-3-1 [infection type (IT) = 0] and Bgt2015 (IT = 0; Li et al. 2016). An F2 population and a set of F2:3 lines derived from PI 628024 × CItr 11349 were used to map the powdery mildew resistance gene in PI 628024. CItr 11349, obtained from Bulgaria in 1929, is highly susceptible to both OKS (14)-B-3-1 (IT = 4) and Bgt2015 (IT = 4). The F2 population consisted of 243 plants, 212 of which produced sufficient F3 seeds for phenotypic analysis. PI 628024 and CItr 11349 were provided by the USDA-ARS National Small Grains Collection at Aberdeen, Idaho.
Evaluation of powdery mildew resistance
A protocol described previously by Tan et al. (2018) was used to determine powdery mildew responses. In brief, F2 plants were inoculated with Bgt isolate OKS(14)-B-3-1 at the two-leaf stage, and the inoculated plants were grown under natural light at 20 ± 2 °C in a greenhouse at the USDA-ARS Wheat, Peanut, and other Field Crops Research Unit in Stillwater, OK. ITs were recorded 7–10 days after inoculation when the susceptible check, Jagalene, was severely infected and re-assessed for confirmation after 2 days. A 0-to-4 infection type scale, representing highly resistant (IT = 0, 0, and 1), moderately resistant (IT = 2), moderately susceptible (IT = 3), and highly susceptible (IT = 4) responses, was employed (Tan et al. 2018).
After evaluation, all F2 plants were vernalized at 5 °C for 6 weeks and then transplanted to a greenhouse. The F2:3 lines were evaluated for powdery mildew responses in spring 2017 using a randomized complete block design with two replicates. For each replicate, 16 plants of each F2:3 lines were evaluated using the protocol described above. Assuming variation at a single locus, the genotype of each F2 plant was inferred from corresponding F3 phenotypic data.
PI 628024, CItr 11349, Coker747, Jimai 22, and Jagalene were evaluated for responses to a set of 16 Bgt isolates collected from different regions of the USA and maintained as pure cultures on detached leaves at the USDA-ARS Plant Science Research Unit at Raleigh, North Carolina (Table 1). Coker747 and Jimai 22 carry Pm6 and PmJM22, respectively, and Jagalene was used as a susceptible control. The detached-leaf method described previously by Cowger et al. (2018) was followed, and disease severities were assessed 10 days after inoculation using a 0-to-9 scale, which distinguished resistant (0–4), intermediate (5–6), and susceptible (7–9) reactions. The 0-to-9 scale was designed to differentiate phenotypes conferred by different powdery mildew resistance genes and to obtain accurate phenotypic data comparable to that based on the 0-to-4 scale. The 0-to-4 scale is suitable for evaluation of mapping or breeding populations.
Bulked segregant analysis
Bulked segregant analysis (BSA) (Michelmore et al. 1991) was used to map the powdery mildew resistance gene in PI 628024. In brief, the F2 genotypes were inferred from F3 phenotypic data, and a protocol described by Dubcovsky et al. (1994) was used to extract genomic DNA from two-week-old leaves. Equal amounts of genomic DNA from 10 highly resistant (IT = 0) F2 plants and 10 highly susceptible (IT = 4) F2 plants were pooled to construct the resistant and susceptible bulks, respectively. A set of over 400 simple sequence repeat (SSR) markers evenly distributed across the wheat chromosomes were initially surveyed for polymorphism between the two parents and the two contrasting bulks. Polymerase chain reaction (PCR) amplifications were performed on Applied Biosystems 2720 thermal cyclers (Applied Biosystems Inc., CA) using a previously described protocol (Xu et al. 2006), and denatured PCR products were separated in 6.5% polyacrylamide gels running in a Li-Cor DNA Analyzer. Electrophoresis conditions were set at 1500 V, 40 W, 35 mA, and 50 °C for 3 h in 1X Tris–borate-EDTA (TBE) buffer.
Informative SSR markers exhibiting polymorphism between the two contrasting bulks, as well as the parents, were used to genotype the F2 population, leading to the identification of SSR markers closely linked to the single powdery mildew resistance gene. Based on their genomic locations, molecular markers previously mapped in the target region were further used to genotype the population. In addition, two SSR loci, including STARS382 (forward primer: TGGGATGGAGGGAGTACTTG; reverse primer: TCATATCCATGGTGGGGAAC) and STARS419 (forward primer: GCCCTTGTCAGTTTCAGTCC; reverse primer: GTCGATCGCTCCACCTCTAC), were identified in the target region, and primers were designed to genotype the F2 population.
Date analysis
MAPMAKER 3.0 software (Lander et al.1987) was used to construct the genetic linkage map, and the Kosambi mapping function was used to convert recombination values to map distances (Kosambi 1943). A logarithm of the odds (LOD) score of 3.0 was set as the threshold for linkage. Chi-squared (χ2) tests were performed to test the hypothesis that a dominant powdery mildew resistance allele conferred resistance in the F2 and F3 populations.
Bin mapping
Based on linkage mapping results, molecular markers flanking the powdery mildew resistance gene in PI 628024 were used to genotype Chinese Spring nulli-tetrasomic lines, N2AT2B, N2BT2A, and N2DT2A, as well as five 2BL deletion lines, 2BL-3, 2BL-4, 2BL-5, 2BL-1, and 2BL-6, to determine the physical locations of markers close to the powdery mildew resistance gene. All aneuploid lines were provided by the Kansas State University Wheat Genetics Resource Center (Manhattan, Kansas).
Allelism tests
An F2 population derived from PI 628024 × Jimai 22 was used to determine the allelic relationships between gene PmJM22 and the gene in PI 628024. The protocol described above was used to determine responses of each F2 plant to Bgt isolate OKS(14)-B-3-1, which is avirulent to PI 628024 and Jimai 22. The recombination fraction was estimated, and genetic distance in centiMorgans (cM) was calculated (Kosambi 1943). A Chi-squared (χ2) test was conducted to test the hypothesis that two independent powdery mildew resistance genes segregated in the F2 population.
Results
Inheritance of powdery mildew resistance in PI 628024
The parental lines and F2 population derived from PI 628024 × CItr 11349 were evaluated for responses to Bgt isolate OKS(14)-B-3-1. PI 628024 and CItr 11349 were highly resistant (IT = 0) and highly susceptible (IT = 4), respectively. Among F2 plants, 186 were resistant, and 57 were susceptible (Table 1). The χ2 test indicated a single dominant allele conferred resistance (χ 23:1 = 0.31, df = 1, p = 0.58).
Of the 212 F2:3 lines evaluated for response to OKS(14)-B-3-1, 47 and 49 were rated as homozygous resistant and homozygous susceptible, respectively, and 116 segregated, confirming that PI 628024 carries a dominant resistance allele (χ 21:2:1 = 1.92, df = 2, p = 0.38) (Table 1).
Linkage mapping
BSA identified four SSR markers that distinguished the two parents and two contrasting bulks, Xgwm148, Xgwm374, Xwmc441, and Xwmc332. All these markers were previously mapped to the long arm of chromosome 2B. Therefore, we used them to genotype the F2 population. Linkage analysis indicated that the powdery mildew resistance gene in PI 628024, originally designated Pm628024, was 7.7 cM distal to Xwmc441.
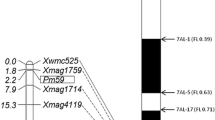
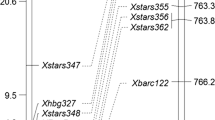
Xwmc441 was located at approximately 598.06 Mb in the Chinese Spring reference sequence IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr). Therefore, an additional 43 markers previously mapped in this region were tested for polymorphism, and 10 polymorphic markers were subsequently used to genotype the mapping population. Linkage analysis placed Pm628024 in an interval of 1.7 cM, flanked distally by STS marker Xbcd135-2 with a genetic distance of 1.1 cM and proximally by SSR marker Xstars419 with a genetic distance of 0.6 cM (Fig. 1).
Linkage (left) and physical bin map (right) of Pm63 (originally Pm628024). Marker names are shown at the right of the linkage map and genetic distances in cM on the left. The physical positions of some markers on the Chinese Spring reference assembly IWGSC RefSeq v1.0 are given in the following parentheses. Molecular markers flanking Pm63 are connected to their appropriate physical bins. The breakpoint of each Chinese Spring deletion line is shown with an arrow, and the corresponding fraction length (FL) value is given in the following parentheses
Physical location of Pm628024
Using the genomic sequence of the restriction fragment length polymorphism (RFLP) probe BCD135 (https://wheat.pw.usda.gov/GG3/), we located Xbcd135-2 to approximately 723.4 Mb in the Chinese Spring reference sequence IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr). Xstars419 was located at approximately 710.3 Mb. Therefore, Pm628024 resides in a genomic region of about 13.1 Mb, spanning 710.3–723.4 Mb of the Chinese Spring reference sequence.
We further located Pm628024 on the Chinese Spring bin map (Fig. 1). Five markers flanking Pm628024 (Xwmc175, Xstars419, Xcinau130, Xwmc332, and Xcinau139) were used to genotype three homoeologous group 2 nulli-tetrasomic lines. All target bands were amplified from N2AT2B and N2DT2A, but not from N2BT2A, confirming that all markers were located on chromosome 2B. We further used these markers to genotype deletion lines 2B-1, 2BL-3, 2BL-4, 2BL-5, and 2BL-6. All except Xwmc175 amplified the target bands from Chinese Spring, but not from any of these deletion lines, indicating that Xstars419, Xcinau130, Xwmc332, Xcinau139, as well as Pm628024, reside in terminal bin 2BL6-0.89-1.0. Xwmc175 amplified the target bands from Chinese Spring and 2BL-6, but not from the other four deletion lines, indicating that Xwmc175 was present in bin 2BL1-0.69-0.89 (Fig. 1). The target band of Xbcd135-2 was amplified from PI 628024, rather than Chinese Spring. Therefore, Xbcd135-2 was not used in physical mapping.
Responses of Pm628024, Pm6, and PmJM22 to Bgt isolates collected from different regions of the USA
A set of 16 representative Bgt isolates were used to determine the response spectrum of Pm628024. All isolates used in this study were virulent to CItr 11349 and the susceptible control Jagalene (Table 2). PI 628024 showed intermediate reactions to two isolates collected from North Carolina, NCF-D-1-1 and NCC-B-1-3, and a susceptible reaction to an isolate collected from New York, NYA-E-3-3. The other 13 isolates were avirulent to PI 628024. Pm6, introgressed from T. timopheevii into bread wheat, was also evaluated because it was also mapped in the terminal bin of chromosome 2BL. Pm6 was susceptible to 14 isolates and exhibited an intermediate reaction to the remaining two isolates, NYA-E-3-3 and MTG1-1a, confirming that Pm6 has been largely defeated in the USA (Cowger et al. 2018). Since PmJM22 was previously identified in bread wheat cultivar Jimai 22 and mapped to the terminal bin 2BL6-0.89-1.0 (Yin et al. 2009), Jimai 22 was included in the panel. Jimai 22 was resistant to 1, 2, 2 and 3 isolates from the Western, Mid-Atlantic, Great Lakes, and Great Plains regions, respectively, and exhibited a susceptible reaction to NYA-E-3-3. Additionally, it showed an intermediate reaction to another four isolates (Table 1).
Allelism test
PI 628024, Jimai 22, and 1557 F2 plants derived from PI 628024 × Jimai 22 were evaluated for responses to OKS(14)B-3-1. Both PI 628024 and Jimai 22 were immune (IT = 0), but 77 F2 plants were highly susceptible, indicating that Pm628024 was not allelic to PmJM22. A Chi-squared test of independence indicated that the two genes were loosely linked (χ 215:1 = 4.53, df = 1, p = 0.03), confirming that PmJM22 resides on chromosome 2B. The estimated recombination fraction and genetic distance between Pm63 and PmJM22 were 0.3145 and 37 cM, respectively.
Discussion
Pm628024, as well as Pm6, Pm33, Pm51, Pm52, PmJM22, MlZec1, and MlAB10, was mapped to the long arm of chromosome 2B; all but Pm52 were mapped to the terminal bin 2BL6-0.89-1.0 (Tao et al. 2000; Mohler et al. 2005; Zhu et al. 2005; Yin et al. 2009; Maxwell et al. 2010; Zhao et al. 2013; Zhan et al. 2014). Pm52 was identified in Lingxing99, a winter wheat cultivar widely grown in Northern China, and was mapped to bin 2BL2-0.36-0.5 (Zhao et al. 2013). Therefore, Pm628024 is not Pm52.
PmJM22 was identified in the Chinese winter wheat cultivar Jimai 22. It was 7.7 cM distal to SSR marker Xwmc149 (Yin et al. 2009), positioned at 779.1 Mb in the Chinese Spring reference assembly (https://urgi.versailles.inra.fr). Pm628024 was mapped to an interval spanning 710.3–723.4 Mb in the reference assembly. Thus, the physical distance between Pm628024 and PmJM22 is over 55.7 Mb. The allelism test further confirmed that Pm628024 and PmJM22 were not allelic, and the estimated genetic distance between them was 37 cM. The remaining five genes in the distal bin originated from wheat wild relatives, including Pm6 from T. timopheevii (Helmsjørgensen and Jensen 1973), Pm33 from T. carthlicum (Zhu et al. 2005), Pm51 from Thinopyrum ponticum (Zhan et al. 2014), and MlZec1 and MlAB10 from T. dicoccoides (Mohler et al. 2005; Maxwell et al. 2010). More recently, a new adult plant resistance gene, Pm62, was transferred from D. villosum into common wheat in the form of Robertsonian translocation T2BS.2VL#5 (Zhang et al. 2018). Given that PI 628024 is a landrace and should not carry any alien chromosomal segment, Pm628024 is not any of them.
A quantitative trait locus (QTL) for adult powdery mildew resistance, QPm.uga-2BL, was identified in the US soft red winter wheat cultivar 26R61 and mapped to a genomic region near SSR marker Xbarc332, positioned at about 576.0 Mb in the Chinese Spring reference sequence IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr) (Hao et al. 2015). QPm.uga-2BL is over 150 Mb away from Pm628024. Another minor QTL for adult powdery mildew resistance on chromosome 2B, QPm.vt-2B, was detected in the US winter wheat cultivar ‘Massey’ and was mapped to the terminal region of chromosome 2BL spanning 681.5 Mb (Xcdo244)–795.4 Mb (Xbcd1231) of the Chinese Spring reference sequence (Liu et al. 2011). Given that QPm.vt-2B only explained a small portion (11%) of the phenotypic variance in the population derived from a cross between Massey and ‘Becker,’ it is unlikely the same gene as the all-stage resistance gene Pm628024. Altogether, Pm628024 is a new powdery mildew resistance gene and has been designated Pm63.
Pm63 was mapped to a genomic region near Pm6, which has been widely used in wheat breeding and extensively characterized with molecular markers (Tao et al. 2000; Ji et al. 2008; Qin et al. 2011). In this study, Pm6 was susceptible to almost all representative US Bgt isolates, confirming that Pm6 has been defeated in the USA because of its extensive deployment in wheat cultivars (Cowger et al. 2018). A previous study indicated that Pm6 was 1.6 cM distal to RFLP marker Xbcd135 (Tao et al. 2000). Although two large populations were used to fine-map Pm6, its precise location is still elusive because of recombination suppression caused by the alien chromosome segment harboring Pm6 (Qin et al. 2011). Pm63 was 0.6 cM proximal to Xbcd135-2, an STS marker converted from Xbcd135. Given that recombination suppression is common between wheat and alien chromosomes (Qin et al. 2011), we cannot use an allelism test to determine whether Pm63 is allelic to Pm6.
Pm51 was identified in a wheat-Thinopyrum ponticum introgression line CH7086 and was 7 cM distal to Pm6 and 8.2 cM distal to Xbcd135 (Zhan et al. 2014). Therefore, Pm51 is more distal to Pm63 than Pm6. The remaining three genes, Pm33, MlZec1, and MlAB10, were mapped to the terminal region of 2BL, and they are either close or distal to PmJM22. Pm33 was introduced into bread wheat from T. carthlicum accession PS5. A previous study suggested that Pm33 was 1.1 cM proximal to SSR marker Xgwm317 (Zhu et al. 2005), while PmJM22 was 7.7 cM distal to SSR marker Xwmc149. Sequence alignment analysis located Xgwm317 and Xwmc149 to approximately 782.4 Mb and 779.1 Mb in the Chinese Spring reference assembly, respectively. Therefore, Pm33 is close to PmJM22. Both MlZec1 and MlAB10 were mapped to the end of chromosome 2BL. MlZec1 was 10 cM distal to Xwmc356, and MlAB10 was 7 cM distal to Xwmc445. Xwmc356 and Xwmc445 were located at approximately 796.7 Mb and 800.0 Mb in the reference assembly, respectively. Given that Pm63 is physically distant to these genes, Pm63 is unlikely allelic to any of them.
Pm63 exhibited high resistance to all Bgt isolates collected from the Great Plains and from western and southeast regions of the USA. Therefore, it can be used to enhance powdery mildew resistance in these regions. This is especially important because some powdery mildew resistance genes widely used in the Great Plains, such as Pm3a and Pm17, have been overcome in most of the USA (Cowger et al. 2018), and Pm63 is an ideal alternative to these genes in the Great Plains. PI 628024 was susceptible to one of four Bgt isolates collected from the Great Lakes region and exhibited an intermediate reaction to two isolates from the Mid-Atlantic region (Table 2). Thus, Pm63 must be combined with other genes in these mildew-prone regions. Gene pyramiding is a preferred strategy for prolonging powdery mildew resistance. Advances in wheat genomics have greatly facilitated gene stacking using marker-assisted selection. STS marker Xbcd135-2 and SSR marker Xstars419 were 0.6 cM distal and 1.1 cM proximal to Pm63, respectively, and have the potential to tag Pm63 in wheat-breeding populations.
Conclusions
Powdery mildew poses a persistent threat to wheat production worldwide, and plant host resistance is a cost-efficient and environmentally friendly alternative to chemical control. However, mutation to virulence and recombination in Bgt populations often lead to erosion of resistance, necessitating a continuous search for resistance genes. Pm63 is a dominant powdery mildew resistance gene identified in Iranian landrace PI 628024 and was mapped to an interval of approximately 13.1 Mb on the long arm of chromosome 2B, spanning 710.3–723.4 Mb in the Chinese Spring reference sequence. Pm63 was located in the terminal bin 2BL6-0.89-1.0 and is different from other genes in origin, genomic location, and response to differential Bgt isolates. Therefore, Pm63 should be considered a new powdery mildew resistance gene. Pm63 exhibited resistance to all representative Bgt isolates collected from the Great Plains, western, and southeastern regions of the USA and can be used to enhance powdery mildew resistance in these regions.
Author contribution statement
XX, BFC, CT, and GL designed the research; CT and GL performed the research; CC evaluated responses of differential lines to Bgt isolates; XX wrote the paper. All authors read, revised, and approved the manuscript.
Abbreviations
- Bgt :
-
Blumeria graminis f. sp. tritici
- cM:
-
Centimorgan
- RFLP:
-
Restrict fragment length polymorphism
- QTL:
-
Quantitative trait locus
- STS:
-
Sequence tag site
- SSR:
-
Simple sequence repeat
References
Bowen KL, Everts KL, Leath S (1991) Reduction in yield of winter wheat in North Carolina due to powdery mildew and leaf rust. Phytopathology 81:503–511
Braun HJ, Ekiz H, Eser V et al (1997) Breeding priorities of winter wheat programs. In: Braun HJ, Altay F, Kronstad WE, Beniwal SPS, McNab A (eds) Wheat: prospects for global improvement. Springer, Dordrecht, pp 553–560
Cao T, Chen Y, Li D et al (2015) Identification and molecular detection of powdery mildew resistance of new bred wheat varieties (lines) in Henan Province, China. Acta Agron Sin 41:1172–1182
Conner RL, Kuzyk AD, Su H et al (2003) Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can J Plant Sci 83:725–728
Cowger C, Mehra LK, Arellano C et al (2018) Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology 108:402–411
Dubcovsky J, Galvez AF, Dvořák J (1994) Comparison of the genetic organization of the early salt-stress-response gene system in salt-tolerant Lophopyrum elongatum and salt-sensitive wheat. Theor Appl Genet 87:957–964
Griffey CA, Das MK, Stromberg EL (1993) Effectiveness of adult-plant resistance in reducing grain yield loss to powdery mildew in winter wheat. Plant Dis 77:618–622
Hao Y, Parks R, Cowger C, Chen Z, Wang Y, Bland D, Murphy JP, Guedira M, Brown-Guedira G, Johnson J (2015) Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor Appl Genet 128:465–476
Helmsjørgensen J, Jensen CJ (1973) Gene Pm6 for resistance to powdery mildew in wheat. Euphytica 22:423
Hsam SLK, Huang XQ, Zeller FJ (2001) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.) 6. Alleles at the Pm5 locus. Theor Appl Genet 102:127–133
Huang XQ, Hsam SLK, Zeller FJ et al (2000) Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet 101:407–414
Huang X, Wang L, Xu M, Röder M (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Ji J, Qin B, Wang H et al (2008) STS markers for powdery mildew resistance gene Pm6 in wheat. Euphytica 163:159–165
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J et al (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li G, Xu X, Bai G, Carver BF, Hunger R, Bonman JM (2016) Identification of novel powdery mildew resistance sources in wheat. Crop Sci 56:1817–1830
Liu S, Griffey CA, Maroof MA (2011) Identification of molecular markers associated with adult plant resistance to powdery mildew in common wheat cultivar Massey. Crop Sci 41:1268–1275
Ma HQ, Kong ZX, Fu BS et al (2011) Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor Appl Genet 123:1099–1106
Maxwell JJ, Lyerly JH, Srnic G et al (2010) MlAB10: a Triticum turgidum subsp. dicoccoides derived powdery mildew resistance gene identified in common wheat. Crop Sci 50:2261–2267
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
McIntosh RA, Zhang P, Cowger C et al (2011) Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor Appl Genet 123:359–367
McIntosh RA, Yamazaki Y, Dubcovsky J et al (2013) Catalogue of gene symbols for wheat. In: Ogihara Y (ed) Proceeding of the 12th international wheat genetics symposium, Yokohama, Japan, 8–13 Sept 2013, pp 8–13
McIntosh RA, Dubcovsky J, Rogers WJ et al (2017) Catalogue of gene symbols for wheat: 2017 supplement. Annu Wheat Newslett 53:1–20
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Mohler V, Zeller FJ, Wenzel G et al (2005) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 9. Gene MlZec1 from the Triticum dicoccoides-derived wheat line Zecoi-1. Euphytica 142:161–167
Parks R, Carbone I, Murph JP et al (2008) Virulence structure of the eastern US wheat powdery mildew population. Plant Dis 92:1074–1082
Parry DW (1990) Plant pathology in agriculture. Cambridge University Press, Cambridge, pp 160–224
Qi L, Cao M, Chen P et al (1996) Identification, mapping, and application of polymorphic DNA associated with resistance gene Pm21 of wheat. Genome 39:191–197
Qin B, Cao A, Wang H et al (2011) Collinearity-based marker mining for the fine mapping of Pm6, a powdery mildew resistance gene in wheat. Theor Appl Genet 123:207–218
Singh RP, Singh PK, Rutkoski J et al (2016) Disease impact on wheat yield potential and prospects of genetic control. Annu Rev Phytopathol 54:303–322
Sun H, Hu J, Song W et al (2018) Pm61: a recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor Appl Genet 131:2085–2097
Szunics L, Szunics L, Vida G et al (2001) Dynamics of changes in the races and virulence of wheat powdery mildew in Hungary between 1971 and 1999. In: Bedo Z, Lang L (eds) Wheat in a global environment. Springer, Netherlands, pp 373–379
Tan C, Li G, Cowger C et al (2018) Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor Appl Genet 131:1145–1152
Tao W, Liu D, Liu J et al (2000) Genetic mapping of the powdery mildew resistance gene Pm6 in wheat by RFLP analysis. Theor Appl Genet 100:564–568
Tilman D, Cassman KG, Matson PA et al (2002) Agricultural sustainability and intensive production practices. Nature 418:671–677
Xiao M, Song F, Jiao J et al (2013) Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat andrace Hongyanglazi. Theor Appl Genet 126:1397–1403
Xu XY, Bai GH, Carver BF et al (2006) Molecular characterization of a powdery mildew resistance gene in wheat cultivar Suwon 92. Phytopathology 96:496–500
Xu H, Yi Y, Ma P et al (2015) Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor Appl Genet 128:2077–2084
Xue F, Wang C, Li C et al (2012) Molecular mapping of a powdery mildew resistance gene in common wheat landrace Baihulu and its allelism with Pm24. Theor Appl Genet 125:1425–1432
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Yin G, Li G, He Z et al (2009) Molecular mapping of powdery mildew resistance gene in wheat cultivar Jimai 22. Acta Agron Sin 35:1425–1431
Zhan H, Li G, Zhang X et al (2014) Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE 9:e113455
Zhang R, Fan Y, Kong L et al (2018) Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor Appl Genet 30:1–8
Zhao Z, Sun H, Song W et al (2013) Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor Appl Genet 126:3081–3089
Zhu Z, Zhou R, Kong X et al (2005) Microsatellite markers linked to 2 powdery mildew resistance genes introgressed from Triticum carthlicum accession PS5 into common wheat. Genome 48:585–590
Acknowledgements
We thank M. Hargrove and R. Whetten for excellent technical assistance and Dr. Robert McIntosh for critical reviewing the manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Communicated by Thomas Miedaner.
Rights and permissions
About this article
Cite this article
Tan, C., Li, G., Cowger, C. et al. Characterization of Pm63, a powdery mildew resistance gene in Iranian landrace PI 628024. Theor Appl Genet 132, 1137–1144 (2019). https://doi.org/10.1007/s00122-018-3265-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3265-5