Abstract
Pigeonpea is one of the most important pulse crops in the semi-arid tropical region, which is prone to several climatic uncertainties like unpredictable temperature, frequent drought and inconsistent rainfall. Additionally, during crop cycle pigeonpea also encounters a wide range of other biotic and abiotic constraints, ultimately leading to its fluctuating production and stagnant productivity. However, recently developed CGMS system has shown noteworthy impacts in enhancing pigeonpea productivity through exploitation of hybrid vigour. At present, A2-cytoplasm derived CGMS system has been well established in pigeonpea. Nevertheless, the commercial success of CGMS system relies largely on the continuous supply of genetically pure seeds of hybrids and corresponding parental lines. Traditionally, the genetic purity of seeds is guaranteed through conducting grow out test (GoT). In this context, DNA marker assays offer several advantages over conventional GoT especially in terms of time, space and money. Given its locus-specific and co-dominant nature, SSR or microsatellite marker is particularly suited for hybridity testing and purity assessment. Here we report a set of robust SSR markers, which could act as reliable molecular kit for ensuring the genetic purity of the CGMS-hybrid ‘IPH 09-5’ and its parental lines ‘PA 163A’ (A-or Male sterile-line) and ‘AK 261322’ (R- or Restorer-line).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pigeonpea (Cajanus cajan (L.) Millspaugh) is a warm-season grain legume grown primarily in tropical and semi-tropical regions. Worldwide, it is cultivated in about 5.83 m ha area with an annual production and an average productivity of 4.40 m t and 753 kg/ha, respectively [1]. With 4.42 m ha area and 2.86 m t production, pigeonpea remains the second most important pulse crop in India after chickpea [1]. The inherent properties like nitrogen fixation and drought tolerance make pigeonpea an integral component of rain-fed agro-ecosystem, thereby offering potential means for generating livelihood for the small-scale/marginal farmers along with ensuring nutritional security to the people inhabiting these areas [2, 3]. Concerted research efforts focusing on breeding and selection have facilitated the development and commercial cultivation of several pigeonpea varieties belonging to different maturity groups i.e. short-, medium- and long-duration [4]. Despite of meticulous attention paid towards pigeonpea improvement, limited success has been achieved in terms of productivity in pigeonpea [3].
Several impediments are responsible for its unstable production including biotic and abiotic constraints coupled with the cultivation of traditional less-responsive genotypes or landraces [2]. Recently, cytoplasmic genetic male sterility (CGMS)-based hybrid breeding system has emerged as a potential alternative to address the problem of yield stagnation that has been prevailing for several decades in pigeonpea [4]. To date, a total of seven different sources of sterile cytoplasm viz. A1 (C. sericeus), A2 (C. scarabaeoides), A3 (C. volubilis), A4 (C. cajanifolius), A5 (C. acutifolius), A6 (C. lineatus) and A7 (C. cajan) have been reported in pigeonpea [4, 5]. The classification is based on the wild progenitors acting as sources of cytoplasmic genetic male sterility [5]. Among all seven types, only two viz. A2 and A4 have been proven of commercial importance. Recently an A2-cytoplasm derived hybrid ‘IPH 09-5’ has been developed using lines ‘PA 163A’ (A-line) and ‘AK 261322’ (R-line) at Indian Institute of Pulses Research (IIPR) [6].
Commercial success of CGMS system largely depends on the adequate supply of genetically pure seeds of hybrids and its parental lines. Even a small fraction of impurity in the hybrid seed lot may lead to substantial reductions in the crop yield [7]. The genetic purity is tested conventionally using grow out test (GoT), which comprises of growing the crop in large fields in isolation followed by removal of the off-type plants based on visual inspection of morphological/floral characteristics [8]. Technically, the entire procedure is extremely cumbersome as it requires extensive field testing and most importantly, the test for fertility/sterility cannot be performed before the commencement of flowering [9]. Moreover, the morphological differences are often subjected to the influence by the external environment. Given the context, DNA marker assays offer an efficient and accurate system for assessing the genetic purity at very early stages of plant development, therefore considerably reducing the time, space and money invested in conducting GoT [8, 9].
Among the different kinds of marker systems available, simple sequence repeats (SSRs) are considered of great value due to their abundance in genome, multi-allelic nature, easily reproducible, user-friendly and co-dominant character [8–10]. These peculiar features make SSRs as the preferred class of DNA markers for estimating genetic purity especially in concern of detection of heterozygosity. With this view, here we have identified a robust set of SSR markers, which could be used in purity estimation of the CGMS based hybrid ‘IPH 09-5’ and its parental lines.
Materials and Methods
Plant Material and DNA Extraction
Hybrid and its parents were grown during Kharif 2012 at Indian Institute of Pulses Research (IIPR), Kanpur, India. Total genomic DNA was isolated from leaves of three-week old seedlings following the modified CTAB method [11]. Finally DNA samples were diluted to 10 ng/μl to perform polymerase chain reaction (PCR).
PCR Analysis
A total of 66 informative SSR primers were considered for the present investigation. PCR reactions were performed in G-STORM GS4 thermal cycler (G-Storm, United Kingdom) with a 20 μl reaction volume containing 2 μl of 10 × PCR buffer, 2 μl of 2 mM dNTPs (Bangalore Genei Pvt. Ltd., Bengaluru, India), 0.5 U of Taq DNA polymerase (Bangalore Genei Pvt. Ltd., Bengaluru, India) and 5 pM each of forward and reverse primers. A total of 4 μl of genomic DNA (10 ng/μl) was used for PCR reaction. A touch down PCR programme was chosen for SSR amplification. Following an initial denaturation for 5 min at 95 °C, five cycles were set as: denaturation for 20 s at 94 °C, annealing for 20 s at 56 °C (1 °C is reduced after each cycle) and extension for 30 s at 72 °C. The next 35 cycles were performed with 20 s at 94 °C, 30 s at 51 °C and 45 s at 72 °C and a final extension step for 20 min at 72 °C.
Gel Electrophoresis and Documentation
The PCR amplicons were resolved in 3 % agarose gel (Bangalore Genei Pvt. Ltd., India). The allelic sizes of the amplified fragments were determined using 100 bp DNA ladder (Fermentas). Amplified fragments were recorded and estimated with BioRad Gel Doc XR version 2.0.
Results and Discussion
In order to exploit heterosis or hybrid vigour, CGMS system has been established in several crop species like rice [12], sorghum [13], brassica [14], sunflower [9, 15] and so forth. However, assessment and maintenance of genetic purity is the most critical factor in the efficient utilization of CGMS technology at commercial scale. In addition to conventional GoT, PCR based DNA markers could be explored as potential tools to expedite the process of confirming the genetic purity of lines [8–10]. Various DNA markers are reported in pigeonpea including restriction fragment length polymorphisms (RFLPs) [16], random amplified polymorphic DNAs (RAPDs) [17], amplified fragment length polymorphisms (AFLPs) [18], SSRs [8, 10] and diversity arrays technologies (DArTs) [19]. In pigeonpea, these various marker assays were used for genetic diversity estimation [16], genetic linkage mapping [10] and quantitative trait loci (QTL) analysis [20]. However, remarkably low level of polymorphism in primary gene pool of pigeonpea was manifested by employing different marker assays [16–20].
Owing to its co-dominant nature, reproducibility and ease of scoring, SSR is the marker of choice for genetic purity testing [8]. SSRs can be further classified into two categories based on the entire length of repeat motif [21]. Class I includes SSRs with a total motif length of ≥20 bp while Class II consists of SSRs having repeat lengths of ≥12 < 20 bp. Among the two categories of SSRs mentioned herewith Class I SSRs (with longer repeat length) tend to be highly variable in plant species [21]. Besides, the polymorphism information content (PIC) values also provide an insight about the informativeness of a particular DNA marker that could be of immense importance [22], while discriminating genotypes at molecular level [23, 24]. More importantly, genomic-SSRs usually exhibit higher degree of polymorphism compared to genic- or expressed sequence tag (EST)-derived SSRs [25].
Taken the above considerations into account, here authors chose informative SSRs based on (i) the length of SSR tracts and (ii) PIC values (as reported in earlier studies). Consequently, a set of 66 genomic-SSRs was chosen to perform the genetic purity analysis. The SSR primer pairs were synthesized based on the primers information available from Burns et al. [26], Saxena et al. [24], Odeny et al. [23, 27] and Singh et al. [28]. Originally, these various SSRs were derived from bacterial artificial chromosome (BAC)-libraries [23–27] and through in silico SSR-mining of the whole genome sequence [28] of pigeonpea.
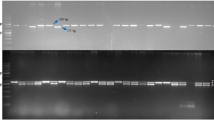
The parents (PA 163A and AK 261322) were screened for detection of marker polymorphism (Table 1). As a result, scorable amplicons with expected sizes were obtained for all the 66 SSRs. However, of the total 66 SSRs screened, 59 exhibited the similar SSR profiles between PA 163A and AK 261322, whereas the remaining seven SSRs viz. CCB9, HASSR3, HASSR9, HASSR23, HASSR35, HASSR37 and HASSR43 enabled the detection of polymorphic fragments (Table 1). All the seven polymorphic SSRs were further used to confirm the true hybrid status of the genotype IPH 09-5. Recovery of both paternal- and maternal-specific fragments/alleles in the SSR profiles of hybrid IPH 09-5 confirmed the true heterozygous nature of the hybrid (Fig. 1). Moreover, no residual heterozygosity was detected in the parental lines using these 66 SSRs thereby confirming their true pure-line nature harboring 100 % homozygosity (Table 1). It is important to note that absence of residual heterozygosity in the parental lines also contributes to the sustainability of CGMS system [8]. SSR-based molecular profiles of the hybrid and its parents have been shown in Fig. 1.
Similar instances of SSR-based hybridity/genetic purity testing were also reported for A4-cytolplasm derived hybrids in pigeonpea. For example, SSR profiles were constructed for hybrid ICPH 2438 and its parental lines (ICPA 2039 and ICPR 2438) using genomic-SSR markers [8]. Likewise, another hybrid ICPH 2671 and its parental lines (ICPA 2043 and ICPR 2671) were characterized with BAC-end sequence (BES) derived SSR markers (BES-SSRs) [10]. Besides pigeonpea, SSR markers were employed for DNA fingerprinting of CGMS based hybrids in several other crops including rice [12], maize [29] and so forth. In some instances, dominant markers such as RAPD were also employed for testing hybridity, if adequate number of paternal-specific fragments were available [30]. Nevertheless, meagre reproducibility and detection of multiple fragments offer potential impediments while dealing with marker systems like RAPD.
In summary, here authors report a set of seven robust SSR markers, which would facilitate rapid and accurate detection of the CGMS-hybrid and its parental lines. Accordingly, the SSR markers would contribute significantly to the commercial success of CGMS-hybrids in pigeonpea through avoiding any possibilities of contamination from self-inbred or other seeds. Moreover, mapping populations like F2 and Backcross are being developed at IIPR derived from the cross PA 163A × AK 261322 and hence, these polymorphic SSR markers might be of great use in future molecular tagging/mapping of gene(s)/QTL(s) responsible for fertility restoration using bulked segregants analysis (BSA) or quantitative trait loci (QTL) approach.
References
FAOSTAT (2011) http://www.faostat.fao.org
Odeny DA (2007) The potential of pigeonpea (Cajanus cajan (L.) Millsp.) in Africa. Nat Resour Forum 31:297–305
Varshney RK, Chen W, Li Y et al (2011) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89
Bohra A, Mallikarjuna N, Saxena K, Upadhyaya H, Vales MI, Varshney RK (2010) Harnessing the potential of crop wild relatives through genomics tools for pigeonpea improvement. J Plant Biol 37:1–16
Saxena KB, Sultana R, Mallikarjuna N, Saxena RK, Kumar RV, Sawargaonkar SL, Varshney RK (2010) Male-sterility systems in pigeonpea and their role in enhancing yield. Plant Breed 129:125–134
Pulses Newsletter IIPR, January–March 2013, pp8 (http://www.iipr.res.in/pdf/newsletter_2013_5june.pdf)
Yuan LP (1985) A concise course in hybrid rice. Hunan Science and Technology Publishing House, China, p 168
Saxena RK, Saxena KB, Varshney RK (2010) Application of SSR markers for molecular characterization of hybrid parents and purity assessment of ICPH 2438 hybrid of pigeonpea [Cajanus cajan (L.) Millspaugh]. Mol Breed 26:371–380
Yue B, Vick BA, Cai X, Hu J (2010) Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus L.) by SSR and TRAP markers. Plant Breed 129:24–28
Bohra A, Dubey A, Saxena RK et al (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol 11:56
Abdelnoor RV, Barros EG, Moreira MA (1995) Determination of genetic diversity within Brazilian soybean germplasm using random amplified polymorphic DNA techniques and comparative analysis with pedigree data. Braz J Genet 18:265–273
Sundaram RM, Naveenkumar B, Biradar SK et al (2008) Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163:215–224
Reddy BVS, Ashok Kumar A, Kaul SL (2008) Alternative cytoplasmic male sterility systems in sorghum and their utilization. In: Sorghum improvement in the new millennium. International Crops Research Institute for the Semi-Arid Tropics, Patancheru 502 324, Andhra Pradesh, India, pp. 132–144
Wan Z, Jing B, Tu J, Ma C, Shen J, Yi B, Wen J, Huang T, Wang X, Fu T (2008) Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napus. Theor Appl Genet 116:355–362
Rieseberg LH, Van Fossen C, Arias D, Carter RL (1994) Cytoplasmic male-sterility in sunflower-origin, inheritance, and frequency in natural populations. J Hered 85:233–238
Sivaramkrishnan S, Seetha K, Reddy LJ (2002) Diversity in selected wild and cultivated species of pigeonpea using RFLP of mtDNA. Euphytica 125:21–28
Ratnaparkhe MB, Gupta VS, Ven Murthy MR, Ranjekar PK (1995) Genetic fingerprinting of pigeonpea (Cajanus cajan (L.) Millsp) and its wild relatives using RAPD markers. Theor Appl Genet 91:893–898
Panguluri SK, Janaiah J, Govil JN, Kumar PA, Sharma PC (2005) AFLP fingerprinting in pigeonpea (Cajanus cajan L. Millsp.) and its wild relatives. Genet Resour Crop Evol 53:523–531
Yang S, Pang W, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A (2006) Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology (DArT). Theor Appl Genet 113:585–595
Bohra A, Saxena RK, Gnanesh BN, Saxena KB, Byregowda M, Rathore A, Kavi Kishor PB, Cook DR, Varshney RK (2012) An intra-specific consensus genetic map of pigeonpea [Cajanus cajan (L.) Millspaugh] derived from six mapping populations. Theor Appl Genet 125:1325–1338
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Odeny DA, Jayashree B, Ferguson M, Hoisington D, Crouch J, Gebhardt C (2007) Development, characterization and utilization of microsatellite markers in pigeonpea. Plant Breed 126:130–136
Saxena RK, Prathima C, Saxena K, Hoisington DA, Singh NK, Varshney RK (2010) Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.). Plant Breed 129:142–148
Varshney RK, Graner A, Sorrells E (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Burns MJ, Edwards KJ, Newbury HJ, Ford-Lloyd BV, Baggott CD (2001) Development of simple sequence repeat (SSR) markers for the assessment of gene flow and genetic diversity in pigeonpea (Cajanus cajan). Mol Ecol Notes 1:283–285
Odeny DA, Jayashree B, Gebhardt C, Crouch J (2009) New microsatellite markers for pigeonpea (Cajanus cajan (L.) Millsp.). BMC Res Notes 2:35
Singh NK, Gupta DK, Jayaswal PK et al (2011) The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol 21:98–112
Asif M, Rahman MU, Zafar Y (2006) Genotyping analysis of six maize (Zea mays L.) hybrid using DNA fingerprinting technology. Pak J Bot 38:1425–1430
Ali MA, Seyal MT, Awan SI, Niaz S, Ali S, Abbas A (2008) Hybrid authentication in upland cotton through RAPD analysis. Aust J Crop Sci 2:141–149
Acknowledgments
Financial support from the Indian Council of Agricultural Research (ICAR) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bohra, A., Singh, I.P., Yadav, A.K. et al. Utility of Informative SSR Markers in the Molecular Characterization of Cytoplasmic Genetic Male Sterility-Based Hybrid and its Parents in Pigeonpea. Natl. Acad. Sci. Lett. 38, 13–19 (2015). https://doi.org/10.1007/s40009-014-0288-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40009-014-0288-6