Abstract
Background
The cytoplasmic-genic male-sterility system has been extensively employed for the production of onion hybrids. Molecular marker-assisted characterization of the cytotypes and genotyping at the restorer-of male-fertility (Ms) locus is important for the accelerated breeding of onion hybrids. Indian onion breeding has focussed more on open-pollinated varieties than hybrids. To accelerate the breeding efforts, marker-assisted selection (MAS) plays a pivotal role.
Methods and results
This study aimed to characterize the Indian breeding lines, varieties, hybrids, and exotic accessions for cytotype and Ms locus. For cytoplasm, cytotype markers, accD, and MKFR and for Ms locus identification, PCR markers AcPMS1 and AcSKP1 were employed. Bulk strategy to identify cytoplasm and Ms locus was tested. Sequencing of PCR products amplified by accD was also tried. Both the accD and MKFR were synonymous in cytoplasm identification except in T821 where T cytoplasm was identified. AcPMS1 was more reliable than AcSKP1 for Ms locus identification. Sequencing proved that N and T cytoplasm are identical. Bulking strategy can be used for cytotype identification but not for Ms locus.
Conclusions
Indian onions have a predominance of normal (N) cytoplasm and homozygous recessive (msms) locus. This might be beneficial for hybrid development. S cytoplasm was identified in exotic varieties. For the first time, T cytoplasm has been reported from India. These findings will assist Indian onion breeders to develop MAS strategies for accelerating hybrid development programs. And for the release of onion hybrids with high productivity and uniformity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Onion (Allium cepa L.) is an extremely important and indispensable element in almost every culture as a vegetable crop. It is consumed year long and is not limited to a particular season, unlike other vegetables. Globally, amongst the top ten vegetable crops produced, onion is at second place and ranks third in the exported fresh vegetable crop category [1]. Onion dominates the local India export trade (> 50%) share with a foreign exchange value of US$ 336 million [2]. India leads in the area under cultivation and production with an area of 1.43 million hectares and a production of 26.74 million metric tonnes. But the productivity is less (18.6 t/ha) as compared to other countries like the Republic of Korea (79.6 t/ha), the USA, Australia, Spain, etc. [3]. Low productivity may be ascribed to the lack of hybrids (< 1%) in the Indian market compared to the 98% market share of hybrids in the USA, UK, Italy, Netherlands, etc. Studies in onion have some challenges owing to some of its features like its biennial nature, high inbreeding depression, the requirement of isolation distances to maintain genetic uniformity, and environmental factors.
Heterosis of hybrids for desirable utilitarian traits like yield, bulb weight, height, the diameter has been acknowledged and commercially exploited [4, 5]. In India, the public sector released two hybrids (Arka Kirtiman and Arka Lalima) but they were not high-yielding on the national level [6]. Hybrid onions are cherished by farmers and consumers throughout the world for their potential higher yield and uniformity in a bulb shape, size, color, and maturity. Research on onion hybrids was initiated with the discovery of ‘S’ cytoplasm [7] in cultivar ‘Italian Red’ and restorer of fertility (Ms) locus [8]. S cytoplasmic male sterility is favored worldwide due to its stable nature under multi-locations and maintenance by a single ‘Ms’ locus [9]. Since then, the phenomenon of male sterility has been reported in more than 140 species. Berninger [10] reported another type of male sterility, CMS-T, wherein the fertility restoration was controlled by three independent loci [11]. Male sterility in the CMS-S system is maintained by crossing a sterile plant with S cytoplasm and homozygous recessive (msms) loci to a plant with N cytoplasm and msms locus. Whereas the restoration of the fertility can be obtained by crossing the S cytoplasmic plants with a plant having either ‘Msms’ or ‘MsMs’ genotype. Conventionally, the genetic constitution of the parent plants can be identified by crossing a male sterile plant with a fertile plant via test crossing and then studying the F1 to determine the parentage also known as progeny testing [12].
Highly cross-pollinated nature coupled with high inbreeding depression and biennial generation time lengthens the period required for the development of onion inbreds with the known genetic constitution. Employment of PCR markers for the determination of cytoplasm and nuclear Ms locus is highly recommended for accelerated hybrid development. PCR markers identifying normal (N) and sterile (S) cytoplasm [13,14,15] and T cytoplasm [16,17,18] have been reported. Furthermore, Kim et al. [19] described the presence of cytotype X and Y based on the comparative analysis of chloroplast genome of male fertile and male-sterile cytoplasm. Marker-assisted selection of nuclear Ms locus was made possible with the development of RFLP marker [20], CAPS markers [21, 22], SCAR marker [23], SNPs [24] followed by other PCR markers [19, 25,26,27]. Till 2015, cytoplasm markers reported by Sato [14] and Engelke et al. [16] were mostly sought-after markers [28,29,30]. Recently, new primers for cytoplasm [15, 18] and Ms locus [19, 27] are being utilized for the identification of cytoplasm and Ms locus [6, 28, 31] (Table 1). Although reported to be in complete linkage disequilibrium with Ms locus, still new PCR markers linked to Ms locus are being reported. Khar and Saini [6] reported about the limitation of these PCR markers in open-pollinated populations and Khrustaleva et al. [32] hypothesized that limitations may be due to PCR markers assigned to the centromere of Chromosome 2 which is a region of lower recombination. New studies suggest a novel cytotype Y containing a unique stoichiometry of cox1 and orf725 [33], ‘T like’ designated as ‘R’ [34], and a novel haplotype of Ms locus [35] and a new Ms2 locus situated at 70 cM away from the Ms locus on the end of chromosome 2 [36]. To sum up, presently there are three male-sterile cytoplasms reported in onion. The first cytoplasm is the S cytoplasm with one [8] or possibly two [36] nuclear restorer loci. The second one is the T cytoplasm with a complex male-fertility restoration that is maintained possibly by three complementary loci [11]. And the third latest one is the R cytoplasm which appears to be restored by the Ms locus [34].
Our study aimed to characterize the commercially grown open-pollinated varieties (OPVs), breeding material, and hybrids marketed in India. Also, our purpose was to assign the genetic constitution to the breeding lines for the future hybrid breeding programs and to ascertain the genetic architecture of OPVs and hybrids available in the market so that a blueprint for heterosis breeding program can be outlined. For identification of cytoplasm, two PCR markers specific for onion cytoplasm viz., accD, an indel marker [15] and MKFR designed on a chimeric gene orf725 [18] were utilized. For the Ms locus determination, two PCR markers reported for the restorer-of-male-fertility (Ms) locus namely AcSKP1 [27] and AcPMS1 [19] were employed.
Materials and methods
Plant materials
Eighteen onion accessions including two commercially grown open-pollinated varieties (OPVs) (Pusa Red, Pusa Madhavi), four breeding lines (POS13, POS14, POS21, POS22), nine doubled haploid (DH) lines (PMDH2, PMDH3, PMDH7, Ha2hi3.1.2, Hap2hi3.1.4, Hap2hi3.1.5, Hap2hi3.1.6, Hap2hi3.1.7 and Hap2hi3.1.8), and three hybrids (Juni, Matahari, T821) were used for determining the onion cytotype and the genotype at Ms locus. Hybrids, popular among the farmers for high productivity, were purchased from the market. Additionally, twenty-five accessions of commercially grown OPVs including four 4 exotic accessions were also investigated for this study. All the onion bulb material was planted during the rabi (winter) season under net house conditions in the experimental field of the Division of Vegetable Science, Indian Agriculture Research Institute, New Delhi, India.
DNA extraction and PCR amplification
The genomic DNA from 626 individual plants of 18 onion accessions (Table 2) and 208 individuals from the OPVs and exotic lines (Table 3) was extracted individually from young leaves using the CTAB method [37]. DNA integrity was examined by agarose gel electrophoresis and quantified by a Nanodrop spectrophotometer. The DNA quantity of the samples was adjusted to a 10 ng/µl concentration and then stored at − 20 °C until PCR analysis. DNA amplification was performed in a programmable thermal controller (Eppendorf Vapo Protect Mastercycler Pro) in a 10 µl volume of reaction mixture containing 5.0 µl Dream Taq™ Green PCR (Thermo Fisher Scientific Inc., USA), 1.0 µl genomic DNA and 1.0 µl of the specific primers (10 pmol/µl) and volume made up with molecular grade water. PCR amplification for all the markers was done as per Khar and Saini [6]. The amplified PCR products were separated electrophoretically on 2% agarose gel, stained with ethidium bromide, and viewed and photographed under ultraviolet light using the gel documentation system (Cell Biosciences Alphaimager HP).
Suitability of accD in cytoplasm determination
To determine whether primer accD will be able to differentiate N, S, and T cytoplasm, a set of plants with known cytoplasm (N-, S-, and T-) were genotyped. PCR products amplified with accD were sequenced. PCR products were purified using the QIAquick PCR Purification kit (QIAGEN). The purified products were sequenced directly. The sequencing reactions were carried out using BigDye (Applied Biosystems, Foster City, CA, USA) and analyzed using an ABI 3700 Genetic Analyzer (Applied Biosystems) as per the manufacturer’s protocol. Differences between the N-, S- and T- cytoplasms were identified by comparing the aligned sequences using the software Clustal Omega [38].
Bulk DNA sampling to test the sensitivity of PCR markers
To reduce the burden of isolating genomic DNA from individual plants for cytoplasm and Ms locus determination, the efficiency of a bulking strategy was explored with the assumption that any allele present in a bulk will be detected by the given marker system. Each bulk had a combination of DNA extracted from several individuals from a given group or population. In this study, DNA from already validated male sterile and fertile plants was extracted and pooled in different combinations to assess their effectiveness. Normalized DNA was used for bulking wherein an equal quantity of DNA from individual plants was bulked. DNA from ten plants was pooled in the ratio of 1:9, 2:8, 3:7, 4:6, 5:5, 6:4, 7:3, 8:2, and 9:1. The cytoplasm type and the Ms allele genotype were assessed from these DNA pools using the accD and AcPMS1 primer respectively.
Results
Determination of cytotypes and Ms locus in breeding lines, doubled haploids (DHs), and hybrids
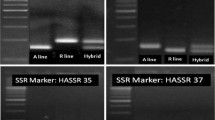
Based on the PCR profile of the marker accD, all the individual plants in the accessions POS13 (14 plants) and POS14 (22 plants) were detected to be S cytoplasmic and N cytoplasmic respectively (Table 1). These same observations were corroborated by the marker MKFR as well. In POS21 and POS22, multiplied as male sterile and maintainer lines, accD and MKFR markers identified all the individual plants as S and N cytoplasm, respectively. Two open-pollinated varieties, Pusa Red (67 plants) and Pusa Madhavi (20 plants), which have been in cultivation for the last 30 years, were also genotyped. In Pusa Red, both accD and MKFR unanimously identified 24 plants with S cytoplasm and 43 plants with N cytoplasm whereas, in Pusa Madhavi, 2 plants with S cytoplasm and 18 plants with N cytoplasm were identified. PMDH2, PMDH3, and PMDH7 are the doubled haploid (DH) lines derived from Pusa Madhavi whereas Hap2hi3.1.2, Haphi3.1.4, Hap2hi3.1.5, Hap2hi3.1.6, Hap2hi3.1.7, and Hap2hi3.1.8 are the DH plants derived from a commercial variety ‘Hisar 3’. All the DH lines were identified as N cytoplasmic and both the markers were in agreement. There was no ambiguity in the determination of cytoplasm in DH lines. Three hybrids viz., Juni, Matahari, and T821, commercially available in the market, were also analyzed. In Juni, 100% of the plants had S cytoplasm. In ‘Matahari’, out of 25 plants, twenty (80%) plants had S cytoplasm whereas five (20%) were having N cytoplasm. Both accD and MKFR gave the same results. In T821, primer accD identified all 47 plants as having N cytoplasm whereas based on MKFR primer, nine (19.1%) plants had S cytoplasm and thirty-eight (80.8%) had T cytoplasm.
Determination of Ms locus was carried out using two nuclear markers AcSKP1 and AcPMS1. All the plants of POS13 and POS14 carried recessive msms locus as determined by these two markers. All the plants of the male-sterile line POS21 and maintainer line POS22 were genotyped as carrying recessive msms locus as predicted. The two markers also congruently genotyped the nuclear Ms locus of all the plants of the doubled haploid lines derived from Pusa Madhavi as homozygous recessive (msms) with 100% efficiency. However, few discrepancies in the determination of the Ms locus by these two markers were observed in two of the Hisar-3 derived DH lines viz., Hap2hi3.1.6 and Hap2hi3.1.7. AcSKP1 determined these DH lines as having heterozygous Msms genotype whereas primer AcPMS1 identified them as having recessive msms genotype. The rest of the Hisar-3 derived DH lines were found to have recessive msms genotype as ascertained by both the nuclear markers. Similar discrepancies between the two markers were observed for Pusa Red and the hybrid lines Juni and Matahari. In Pusa Red, AcSKP1 determined that all the plants had msms genotype whereas AcPMS1 determined sixteen (23.9%) plants having Msms genotype and fifty-one plants (76.1%) were having msms genotype. In hybrid Juni, all 74 plants were detected to be homozygous recessive at Ms by AcSKP1 primer, but according to AcPMS1 primer, 88% of the plants (66) were homozygous recessive and 12% were heterozygous Msms. The results of the AcSKP1 marker in the Matahari hybrid line too revealed 100% plants with homozygous recessive (msms) genotype while AcPMS1 marker determined 72% of the plants with heterozygous (Msms) genotype and 28% with the recessive genotype (msms).
Determination of cytoplasm and nuclear Ms locus in commercial varieties
PCR markers accD and MKFR were synonymous in cytoplasmic identification. For Ms locus genotyping, AcPMS1 primer was used since this primer predicted more accurate results compared to AcSKP1, based on visual observations. Hence, accD and AcPMS1 were used for cytoplasm and Ms determination in 25 commercially released varieties of Indian onion and some exotic lines grown under short-day conditions (Table 2). Eight individual plants per genotype were used except VL Pyaz where 16 plants were used. Out of 25 accessions, twenty-one accessions had normal ‘N’ cytoplasm. The remaining four accessions had both N and S cytoplasm. Out of eight plants, Arka Kalyan had three ‘N’ and five ‘S’ cytoplasmic plants, Sel. 157 had two normal ‘N’ and six ‘S’ cytoplasmic plants, VL Pyaz had ten ‘N’ and six ‘S’ cytoplasmic plants whereas AVON1102 had seven normal ‘N’ and one ‘S’ cytoplasmic plant. Based on the nuclear Ms locus, eighteen accessions had all the plants with ‘msms’ constitution. In Arka Kalyan, one plant had ‘Msms’ whereas other seven plants were of ‘msms’ constitution. In Sel.157, one plant of ‘MsMs’, three of ‘Msms’, and four plants of ‘msms’ constitution were observed. Similarly, in VL Pyaz, four plants having MsMs, ten plants of Msms, and two plants of msms constitution were identified. In all the four exotic genotypes viz., NBPGR3, AVON1102, NBPGR2, and AKON067, plants of both ‘Msms’ and ‘msms’ genetic constitution were observed except NBPGR3 where in addition one plant of ‘MsMs’ was also observed.
Suitability of accD in cytoplasm determination
Chloroplast marker accD can identify S-cytoplasm, but cannot distinguish between the N- and T- cytoplasm. Hence a mitochondrial marker, MKFR was used which could differentiate between N, S, and T cytoplasm. In hybrid T821, primer accD classified all the plants (47) as N cytoplasmic. But MKFR differentiated plants into 9 sterile and 38 T cytoplasmic. To ascertain whether there is any SNPs present between N and T cytoplasm so that this marker (accD) can be used to distinguish between all three cytotypes, PCR amplification of already validated cytotypes (N, S, and T) was done using accD. The PCR product was sequenced directly and sequence similarity was conducted. It was observed that three SNPs and a 45 bp indel sequence were present among S and N and T cytoplasm whereas there was no difference between N and T cytoplasm (Fig. 1).
Sensitivity test of PCR markers
For the determination of cytoplasm and Ms locus through bulk strategy, DNA from individual plants having N and S cytoplasm was isolated, normalized, and then mixed in different proportions. Nine pools of 10 plants per sampling group and 9 combination ratios were constituted. This sampling allowed us to evaluate if the molecular markers can efficiently detect the cytoplasm and Ms locus in each particular sample size. The optimum sample size was found to be in the ratio of 8S:2 N where the primer accD was able to correctly determine the cytoplasmic nature of the individual plant present in the DNA bulk (Fig. 2). However, the remaining bulk of 9S:1 N was unable to detect N cytoplasm. A similar bulking study was done for determining the usability of bulk samples in male-sterile locus (Ms) identification using AcPMS1 primer. However, the data was inconclusive and only a single band could be observed Fig. (3)
Discussion
Onion is one of the most dominating vegetables in the global food economy along with tomatoes, chilies, and cucurbits [39]. Research and development of hybrid varieties, which are tailored to meet specific requirements like high-yielding or climate and disease resilient, that can match up to the global supply demands takes precedence. In the Indian onion production scenario, despite being one of the leading producers, the research studies on onion hybrids have not gained interest in either public institutions or private enterprises. There may be many reasons for the lack of enthusiasm among the researchers but the main reason is the time required for the identification of male-sterile and maintainer lines through conventional breeding. Further, the wrong selection of a genotype can contaminate the whole population which makes the identification of the maintainer line even more difficult. It is a well-known fact that male sterility in onion involves interaction between the cytoplasm and nuclear Ms locus. Through marker-assisted selection, identification of male sterile and maintainer lines in the segregating individuals of onion F1 hybrids has become a reality [16, 24, 40]. During the 1980s, work on male sterility in Indian onion was initiated leading to the identification of male sterile lines and the development of hybrid [6]. Pathak [41] highlighted the problems for hybrid development in India and further hypothesized a new source of male sterility in Indian onion using conventional breeding [42]. After a gap of 20 years, preliminary work on the identification of male sterility was initiated in India and it has gained momentum [30, 31, 43]. Validating the genetic purity of the parental lines and hybrids is indispensable in realizing the full potential of hybrids. So far, there is a lack of concerted effort towards the identification of PCR makers for distinguishing cytoplasm type and determining the Ms locus for the development of male sterile, maintainer lines, and consequently hybrid. Hence a step towards this direction was taken up in this study.
In this study, molecular markers that have been developed and validated in previous studies were adopted to determine the cytoplasm type (accD, MKFR) and nuclear genotypes (AcSKP1, AcPMS1). PCR markers for cytoplasm were in agreement for most of the onion material used. The only inconsistency was found with hybrid T821. MKFR marker designated 80% of the individual plants as having T cytoplasm whereas accD identified 100% of them as N cytoplasmic. It has been speculated that CMS-N and CMS-T might have emerged from a progenitor (M) cytoplasm as very few polymorphisms have been discovered between their chloroplast and mitochondrial DNA [33, 44, 45]. Holford et al. [44] detected no difference between the chloroplast and the organellar genomes of CMS-T and the fertile N-cytoplasm while studying the origin of male-sterile and male-fertile onions. Havey and Kim [34] reported that mitochondrial orf725 marker could not differentiate the inbred lines carrying the N cytoplasm and the bona fide T cytoplasm (proposed to be referred to as R cytoplasm) resulting in the production of only the 833-base pair amplicon. They have proposed that this bonafide CMS-T might be an additional CMS source that is more similar to N and T cytoplasm. Although accD characterized them with N cytoplasm, yet the orf725 marker produced both the 833 bp and 628 bp amplicons. Further analysis to determine if there is variation between the amplicons produced by the accD marker in genotypes carrying N, S, and T cytoplasm was performed. Sequencing data revealed that there was no difference between the N and T cytoplasm but a 45 bp indel and three SNPs at 208, 223, and 326 bp were observed in between the S, N, and T cytoplasm. This observation gives credence to the observations of Kim and Yoon [46] that CMS-T male sterility has recently developed from normal cytotype. It would be interesting to examine the restoration mechanism in these CMS-T lines to see if the dominant allele of Ms locus can restore male fertility. According to Kim [26], male fertility could be restored in male-sterile maternal plants carrying the CMS-T-like cytoplasm when crossed with male-fertile paternal plants as validated by the segregation of male-fertility phenotype and jnurf13 marker genotypes. Havey [9] on the other hand found that male-fertility restoration was not conditioned by the dominant Ms allele in CMS-T. He also suggested that the CMS-T-like cytoplasm might have emerged from the Japanese or Dutch population. The mechanism of male-fertility restoration in CMS-S and CMS-T cytoplasmic lines are different. Plants possessing S cytoplasm are restored by a dominant allele at a single locus [8] whereas male fertility restoration in plants with CMS-T cytoplasm is controlled by three independent loci [11]. However, Kim [26] propounded the idea that the three-gene inheritance model proposed for fertility restoration in the CMS-T system may not be the case. Instead, it might be due to the segregation distortion of a single Rf gene. From our study, we suspect the CMS-T plants to be most probably CMS-R (Rijnsburger) cytoplasmic and further examination of the phenotypic and genotypic segregation will allow better determination.
The fertility-restorer (Ms) locus was identified using two markers viz., AcPMS1 and AcSKP1. Male sterile (POS13, POS21) and maintainer line (POS14, POS22) possessed Smsms and Nmsms genotype, respectively. A similar nuclear genotype was confirmed for Pusa Madhavi-derived DH lines (PMDH2, PMDH3, PMDH7) and Hisar-3 derived DH lines viz. Hap2hi3.1.2, Hap2hi3.1.5, Hap2hi3.1.4, Hap2hi3.1.8) and T821 hybrid. Inconsistency among two markers was observed in Pusa Red, Hap2hi3.1.6, Hap2hi3.1.7, Juni, Matahari, and T821 which brings into question the reliability of these PCR markers for accurate prediction of Ms locus. Khar and Saini [6] reported the limitations of PCR markers accurately predicting Ms locus. Khrustaleva et al. [32] explained the reason for this phenomenon. The location of Ms locus is close to the centromere of chromosome2, a region of relatively low recombination, that gives false positives even though the tightly linked markers are physically distant from Ms locus. A thorough prediction of parents on morphological and molecular level followed by selfing and test crossing with the same PCR markers will give a clear picture of the linkage disequilibrium of Ms locus in Indian populations. Assessment of cytotypes in Indian varieties and exotic material led to the conclusion that 91.3% of the varieties had N cytoplasm whereas 8.7% had S cytoplasm. S-cytoplasm was present in Indian varieties viz., Arka Kalyan, Sel.157, and VL Pyaz and one exotic accession AVON 1102. Sel. 157 is derived from Early Grano, an exotic variety and VL Pyaz is an exotic introduction whereas the genealogy of Arka Kalyan is not clear. These observations support the hypothesis of Khar and Saini [6] that the source of male sterility in Indian material may be the exotic germplasm. Based on the AcPMS1 marker, homozygous recessive allele (msms) was the predominant Ms locus type in the Indian breeding lines and the DH varieties. According to Pike [47], the frequency of plants in a population possessing the Nmsms is less than 5% and a similar observation was made by Pathak [42] who stated that the possibility of the prevalence of germplasm lines involved in crossing program possessing the recessive ms gene is very rare. Pathak [42] postulated the possibility of a new source of male sterility in onion. He suggested that although the lines used in his crossing program were possessing dominant Ms alleles yet all the F1 hybrids were found to be male sterile. He hypothesized that the resulting sterility was due to a strong cytoplasmic factor. But, based on our observations, it is likely that the Ms locus of Indian germplasm used in his crossing programme might have been recessive ms alleles instead of assumed dominant alleles. Our data suggest that recessive msms alleles are more prevalent in the Indian onion germplasm. In India, mass selection is extensively followed and most of the improved varieties were developed through this method from a local collection of segregating populations. It is highly likely that the varieties that have been used in this selection process possessed recessive msms allele that might have been passed on to the germplasm for generations. There are around 45 onion varieties released by the public sector in India and all the varieties have been developed through mass selection. And most of these varieties may track their origin to Central India, Maharashtra, which is the largest grower and supplier of onion in India [48]. This explains the predominance of recessive Ms locus.
The suitability of accD primer to differentiate among the N, S, and T cytoplasm was also verified. Chloroplast markers can confidently distinguish N and S cytoplasm [9] but they are unable to identify T cytoplasm. Hence, the use of PCR marker (MKFR) distinguishing S, N, and T cytoplasm is important in a preliminary assessment of cytoplasms of a particular region. Use of 25–50 plants for DNA isolation [34] is possible in inbreds but in open-pollinated populations, individual plants should be used for DNA isolation. The use of bulk samples for detecting cytotype was correct only when the optimum sample size was 8Sterile:2Normal. But bulk sampling for Ms locus was not feasible. We believe this is the first DNA pool study done in onion and this type of study detecting the cytoplasm type from a bulk sample will not only reduce time and resource but can simplify the whole process of selection of the male-sterile, and maintainer line. The DNA pooling analysis study designed here can be adopted for screening numerous plants in a population and then identifying a subset for a more detailed study. The putative identification from the bulk study can be followed up by individual genotyping for identification of the cytoplasm and not for the Ms locus.
Conclusion
To conclude, it can be said that both accD and MKFR should be used for the first-time identification of N, S, and T cytotypes in the new accessions. Almost all the onion material grown in India has normal and sterile cytoplasm except T821 which recorded T cytoplasm. To the best of our knowledge, this is the first report of T cytoplasm from India. Research studies to evaluate the effect of maintainer/restorer loci on restoring fertility in this cytoplasm need to be undertaken. More than 95% of the Indian varieties possess homozygous recessive (msms) Ms locus which can be a good sign for heterosis breeding as the material can be directly used as a maintainer. Limitations of PCR markers to identify Ms locus still exist and need to be addressed by utilizing or identifying new PCR markers. The bulking strategy may be a good option for cytotype determination but not for the Ms locus. This study is a stepping stone for the heterosis breeding program in Indian short-day onion.
Data availability
All data needed to conduct this study is provided within the manuscript.
References
Rabobank (2018) The world vegetable map-Key global trends. https://www.rabobank.com/en/ raboworld/articles/the-world-vegetable-map.html
APEDA (2021) India export agro-food products. http://agriexchange.apeda.gov.in/indexp/product_profile/exp_f_india.aspx?category=0201. Accessed on 27th January 2022.
FAOSTAT (2022) Onion production, area, and productivity. http://www.fao.org/faostat/en /#data/QC. Accessed 27th Jan 2022.
Dowker BD, Gordon GH (1983) Heterosis and hybrid cultivars in onions, p. In: Frankel R (ed) Monographs on theoretical and applied genetics, vol 6. Heterosis. Springer-Verlag, Berlin, pp 220–233
Doruchowski RW (1986) Heterosis effect of some onion (Allium cepa L.) characteristics. Acta Agrobotanica 39:325–339
Khar A, Saini N (2016) Limitations of PCR-based molecular markers to identify male-sterile and maintainer plants from Indian onion (Allium cepa L.) populations. Plant Breeding 135:519–524. https://doi.org/10.1111/pbr.12373
Jones HA, Emsweller SL (1936) A male-sterile onion. Proc Am Soc Hortic Sci 34:582–585
Jones HA, Clarke AE (1943) Inheritance of male sterility in the onion and the production of hybrid seed. Proc Am Soc Hortic Sci 43:189–194
Havey MJ (2000) Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor App Genet 101:778–782
Berninger E (1965) Contribution à l’étude de la sterilité male de l’oignon (Allium cepa L.). Annales de l’Amelioration des Plantes 15:183–199
Schweisguth B (1973) A new type of male-sterility in onion, Allium cepa L. Ann Amélior Plant 23:221–233
Yen DE (1959) Pollen-sterility in Pukekohe Longkeeper onions. NZ J Agric Res 2:605–612
Havey MJ (1995) Identification of cytoplasms using the polymerase chain reaction to aid in the extraction of maintainer lines from open pollinated populations of onion. Theor Appl Genet 90:263–268. https://doi.org/10.1007/BF00222212
Sato Y (1998) PCR amplification of CMS-specific mitochondrial nucleotide sequences to identify cytoplasmic genotypes of onion (Allium cepa L.). Theor Appl Genet 96:367–370. https://doi.org/10.1007/s001220050750
Von Kohn C, Kiełkowska A, Havey MJ (2013) Sequencing and annotation of the chloroplast DNAs and identification of polymorphisms distinguishing normal male-fertile and male-sterile cytoplasm of onion. Genome 56:737–742. https://doi.org/10.1139/gen-2013-0182
Engelke T, Terefe D, Tatlioglu T (2003) A PCR-based marker system monitoring CMS-(S), CMS-(T) and (N)-cytoplasm in the onion (Allium cepa L.). Theor Appl Genet 107(1):162–167
Engelke T, Tatlioglu T (2000) Genetic analyses supported by molecular investigations provide evidence of a new genic (st1) and a new cytoplasmic male sterility (st2) in Allium schoenoprasum L. Theor Appl Genet 101:478–486. https://doi.org/10.1007/s001220051506
Kim S, Lee E, Cho DY, Han T, Bang H, Patil BS, Ahn YK, Yoon M (2009) Identification of a novel chimeric gene, orf725, and its use in development of a molecular marker for distinguishing three cytoplasm types in onion (Allium cepa L.). Theor Appl Genet 118:433–441. https://doi.org/10.1007/s00122-008-0909-x
Kim S, Kim CW, Park M, Choi D (2015) Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor Appl Genet 128:2289–2299. https://doi.org/10.1007/s00122-015-2584-z
Gokce AF, Havey MJ (2002) Linkage equilibrium among tightly linked RFLPs and the Ms locus in open-pollinated onion populations. J Am Soc Hortic Sci 127:944–946
Bang H, Kim S, Park SO, Yoo K, Patil BS (2013) Development of a codominant CAPS marker linked to the Ms locus controlling fertility restoration in onion (Allium cepa L.). Sci Hortic 153(4):42–49
Park J, Bang H, Cho D, Yoon MK, Patil B, Kim S (2013) Construction of high-resolution linkage map of the Ms locus, a restorer-of-fertility gene in onion (Allium cepa L.). Euphytica 192:267–278. https://doi.org/10.1007/s10681-012-0851-5
Yang YY, Huo YM, Miao J, Liu BJ, Kong SP, Gao LM, Liu C, Wang ZB, Tahara Y, Kitano H, Wu X (2013) Identification of two SCAR markers co-segregated with the dominant Ms and recessive ms alleles in onion (Allium cepa L.). Euphytica 190:267–277. https://doi.org/10.1007/s10681-012-0842-6
Havey MJ (2013) Single nucleotide polymorphisms in linkage disequilibrium with the male. -fertility restoration (Ms) locus of onion. J Amer Soc Hort Sci 138:306–309
Bang H, Cho DY, Yoo KS, Yoon MK, Patil BS, Kim S (2011) Development of simple PCR-based markers linked to the Ms locus: a restored-of-fertility gene in onion (Allium cepa L.). Euphytica 179:437–449. https://doi.org/10.1007/s10681-010-0342-5
Kim S (2014) A codominant molecular marker in linkage disequilibrium with a restorer-of-fertility gene (Ms) and its application in reevaluation of inheritance of fertility restoration in onions. Mol Breeding 34:769–778. https://doi.org/10.1007/s11032-014-0073-8
Huo YM, Liu BJ, Yang YY, Miao J, Gao LM, Kong SP, Wang ZB, Kitano H, Wu X (2015) AcSKP1, a multiplex PCR based co-dominant marker in complete linkage disequilibrium with the male-fertility restoration (Ms) locus, and its application in open pollinated populations of onion. Euphytica 204:711–722. https://doi.org/10.1007/s10681-015-1374-7
Ferreira RR, Santos CAF (2018) Partial success of marker-assisted selection of ‘A’ and ‘B’ onion lines in Brazilian germplasm. Sci Hortic 242:110–115. https://doi.org/10.1016/j.scienta.2018.08.002
Ragassi CF, Santos MDM, Fonseca MEN, Oliveira VR, Buzar AGR, Costa CP, Boiteux LS (2012) Genotyping of polymorphisms associated with male-sterility systems in onion accessions adapted for cultivation in Brazil. Hortic Bras 30:409–414. https://doi.org/10.1590/S0102-05362012000300009
Saini N, Hedau NK, Khar A, Yadav S, Bhatt JC, Agrawal PK (2015) Successful deployment of marker assisted selection (MAS) for inbred and hybrid development in long-day onion (Allium cepa L.). Ind J Genet 75:93–98. https://doi.org/10.5958/0975-6906.2015.00012.7
Manjunathagowda DC, Anjanappa M (2020) Identification and development of male sterile and their maintainer lines in short-day onion (Allium cepa L.) genotypes. Genet Resour and Crop Evolut 67(2):357–65
Khrustaleva L, Jiang J, Havey MJ (2016) High-resolution tyramide-FISH mapping of markers tightly linked to the male-fertility restoration (Ms) locus of onion. Theor Appl Genet 129:535–545. https://doi.org/10.1007/s00122-015-2646-2
Kim B, Kim C, Kim S (2019) Inheritance of fertility restoration of male-sterility conferred by cytotype Y and identification of instability of male fertility phenotypes in onion (Allium cepa L.). J Hort Sci Biotechnol 94:341–348. https://doi.org/10.1080/14620316.2018.1512383
Havey MJ, Kim S (2021) Molecular marker characterization of commercially used cytoplasmic male sterilities in onion. J Am Soc for Hortic Sci 146:351–355
Kim T, Kim S (2021) Identification of a novel haplotype of the Ms locus controlling restoration of male-fertility and its implication in origination of cytoplasmic male-sterility in onion (Allium cepa L.). J Hortic Sci Biotechnol 96:1–9. https://doi.org/10.1080/14620316.2021.1925598
Yu N, Kim S (2021) Identification of Ms2, a novel locus controlling male-fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L), and development of tightly linked molecular markers. Euphytica. https://doi.org/10.1007/s10681-021-02927-4
Murray MG, Thompson WF (1980) Rapid isolation of high-molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. https://doi.org/10.1093/nar/8.19.4321
Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins D (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Schreinemachers P, Simmons EB, Wopereis MCS (2018) Tapping the economic and nutritional power of vegetables Global. Food Security 16:36–45
Liu B, Huo Y, Yang Y, Gao L, Yang Y, Wu X (2019) Development of a genotyping method for onion (Allium cepa L) male fertility based on multiplex PCR. J Hortic Sci Biotechnol 95(2):203–210
Pathak CS, Gowda RV (1994) Breeding for the development of Onion hybrids in India: problems and prospects. Acta Hortic 358:239–242
Pathak CS (1997) A possible new source of male sterility in onion. Acta Hortic 433:313–316
Chaurasia AK, Adsul GG, Nair D, Subramaniam VR, Krishna B, Sane PV (2010) Diversity in Indian and some exotic onion cultivars as revealed by genomic and mitochondrial DNA. Acta Hortic 859:207–220
Courcel AGL, De Vedel F, Boussac JM (1989) DNA polymorphism in Allium cepa cytoplasms and its implications concerning the origin of onions. Theor Appl Genet 77:793–798. https://doi.org/10.1007/BF00268328
Holford P, Croft JH, Newbury HJ (1991) Differences between, and possible origins of, the cytoplasms found in fertile and male-sterile onions (Allium cepa L.). Theor Appl Genet 82:737–744. https://doi.org/10.1007/BF00227319
Kim S, Yoon M (2010) Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr Genet 56:177–188. https://doi.org/10.1007/s00294-010-0290-6
Pike LM (1986) Onion Breeding. In: Publishing AVI (ed) Breeding Vegetable Crops (Basset MJ. Co., Inc., Westport, Connecticut, USA, pp 357–394
Lyngkhoi F, Saini N, Gaikwad AB, Thirunavukkarasu N, Verma P, Silvar C, Yadav S, Khar A (2021) Genetic diversity and population structure in onion (Allium cepa L.) accessions based on morphological and molecular approaches. Physiol Mol Biol Plants 27:2517–2532. https://doi.org/10.1007/s12298-021-01101-3
Funding
This work was financially supported by a research grant awarded under the Core Research Grant-Science and Engineering Research Board (CRG-SERB) Scheme, Government of India (CRG/2019/006525) awarded to the first author.
Author information
Authors and Affiliations
Contributions
AK conceptualized, established and supervised the research study. MCG and AJ provided the facilities for shuttle breeding, bulb development, and field facilities. Bulb planting, DNA extraction, and molecular analysis were performed by MZ, PV and HS. AK and MM performed the data analysis. AK and MZ wrote the paper and organized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare that there are no conflicts of interest among them.
Ethical approval
Ethical approval was not required because the experiments utilized normal lab conditions only.
Informed consent
The study did not use human subjects.
Consent for Publication
All authors read the manuscript and showed their willingness to publish this study.
Research involving human and animal participants
This work does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khar, A., Zimik, M., Verma, P. et al. Molecular marker-based characterization of cytoplasm and restorer of male sterility (Ms) locus in commercially grown onions in India. Mol Biol Rep 49, 5535–5545 (2022). https://doi.org/10.1007/s11033-022-07451-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07451-9