Abstract
With the objective of identifying SSR markers that can distinguish parental lines of rice hybrids, we characterized 10 each of cytoplasmic male sterile (CMS) and restorer (R) lines along with 10 popular Indian rice varieties using a set of 48 hyperpolymorphic SSRs distributed uniformly across the rice genome. All the SSR markers were polymorphic, amplifying a total of 163 alleles, with an average of 3.36 ± 1.3 allelic variants per locus. Twenty-seven SSR markers showed amplification of an allele, which was very specific and unique to a particular parental line and not amplified in any other rice genotype tested. Through multiplex PCR, SSR marker combinations that were unique to a particular parental line or hybrid were also identified. With a set of 10 SSR markers, all the public bred Indian rice hybrids along with their parental lines could be clearly distinguished. To utilize these SSR markers effectively for detection of impurities in parental lines, a two dimensional bulked DNA sampling strategy involving a 20 × 20 grow-out matrix has been designed and used for detection of contaminants in a seed-lot of the popular CMS line IR58025A. We have also designed a multiplex PCR strategy involving single tube analysis using 2–3 markers for hybrid seed purity assessments and demonstrate its superiority over single marker analysis in accurate detection of impurities in hybrids. Implications of parental and hybrid specific SSR markers and strategies to utilize the informative SSR markers for detection of contaminants in a cost effective manner are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tropical rice growing countries including India need to step up their rice production because of increasing population and decreasing land and water resources. Hybrid rice cultivation offers an opportunity to increase rice yields and thereby ensure a steady supply of rice (Virmani and Kumar 2004). Since the development of the first CMS line in China (Yuan 1977), many hybrids have been released in China, India, Vietnam, Philippines, Indonesia, and Bangladesh. These hybrids have recorded a yield advantage of 15–20% over semi dwarf high-yielding varieties (HVYs) in farmers’ fields (Rangaswamy and Jayamani 1996; Mishra et al. 2003). The success of hybrid rice technology beside other factors, depends on the production and timely supply of genetically homogenous seeds to farmers. This ensures that the gains of heterosis can be harnessed through enhanced yields by growing a genetically pure hybrid crop. It has been estimated that 1% impurity in the hybrid seed brings down the potential yield of hybrid by about 100 kg ha−1 (Mao et al. 1996). In a country like India, where contract farming is practiced at many places for hybrid seed production, with the active participation of private sector (Mishra et al. 2003), monitoring genetic purity at each stage of seed production becomes necessary.
Assessment of seed purity is one of the most important quality control components in hybrid seed production. Traditionally, it has been the practice to carry out a grow-out test (GOT), based on morphological traits, for assessment of purity of seeds. GOT is time consuming (takes one full growing season for completion), space demanding and often does not allow the unequivocal identification of genotypes. Earlier, we reported the use of Simple Sequence Repeat (SSR) and Sequence Tagged Site (STS) markers for rapid assessment of hybrid and parental line seed purity, as an alternative to GOT (Yashitola et al. 2002; Yashitola et al. 2004). Subsequently, Nandkumar et al. (2004) also showed the utility of SSR markers for fingerprinting rice hybrids. But these studies involved a limited set of markers (<12) and the assessment of genetic purity was based on a single marker and single seed/seedling based analysis. For accurate detection of impurities in seed lots, it is essential to identify a set of informative SSR markers which can clearly distinguish the parental lines and amplify specific or unique allele combinations in the hybrids, not present in any other rice line. Moreover, the assay should preferably be based on analysis of bulked samples rather than single seed assays so as to bring down the cost of the assay. With this objective, in the present study, we have (i) characterized 10 lines each of popular public bred CMS lines and restorer (R) lines used for hybrid rice production along with 10 inbred rice varieties using a set of 48 uniformly distributed hyperpolymorphic rice SSR markers, (ii) identified parental line and hybrid specific markers/marker combinations and (iii) utilized them in parental line and hybrid seed purity assessments.
Materials and methods
Rice genotypes
Thirty genotypes which include 10 CMS lines and 10 R lines used for production of rice hybrids in India and 10 elite rice varieties adapted to different rice growing regions of India were analyzed (Table 1). In addition, three commercially released rice hybrids developed using popular CMS line IR58025A, viz.; DRRH1, KRH2 and Sahyadri (Table 1) were also analyzed.
DNA isolation and polymerase chain reaction (PCR) analysis to identify informative SSR markers
For parental line characterization, total DNA was extracted from freshly germinated young seedlings following the protocol of Dellaporta et al. (1983). A total of 48 rice SSR primer pairs were used for PCR amplification. The SSR markers were selected from the rice SSR linkage map (Temynkh et al. 2000, 2001; available online at http://www.gramene.org) based on the following criteria: (i) uniformity in distribution across the genome and (ii) High polymorphic information content (PIC). The details of selected microsatellite markers and their positions on respective chromosomes are given in Table 2. DNA samples (40 ng) were amplified in 10-μl reaction volumes containing 1X PCR buffer [10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% (v/v) gelatin] (Bangalore Genei, India), 0.2 mM of each dNTPs (Bangalore Genei, India), 10 pmol of each primer and 1 U of Taq polymerase (Bangalore Genei, India). PCR was carried out in a Thermal cycler (Perkin–Elmer-Gene Amp PCR System 9700, USA). The basic PCR profile was 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 55°C, 2 min at 72°C and 7 min at 72°C for final extension. In the preliminary parental line characterization studies, the amplification products were size fractioned in a 15% native polyacrylamide gel (Sigma, USA) in a Protean II gel casting and electrophoresis apparatus (BioRad, USA), stained in 0.5 μg/ml ethidium bromide, visualized under ultraviolet light as per the procedure described in Sambrook and Russell (2001) and documented in a gel documentation system (Alpha Innotech, USA). The sizes of the amplified fragments were estimated with the help of Alphaease software utility of the gel documentation system using 50 and 100 bp DNA ladders (MBI Fermentas, Lithuania) as the size standards. If a certain allele with respect to a particular SSR marker was observed uniquely in just one of the rice genotypes under study and absent in all the other rice genotypes, it was considered to be specific for that genotype and such SSR markers were categorized as informative SSR markers. SSR marker combinations (i.e., 2–3 markers), which did not show significant primer sequence matches and with amplicon sizes that were well apart from each other were tested for multiplex PCR in a single tube reaction. The markers combinations exhibiting clear amplification of specific allele combinations in hybrids and parental lines were also considered as ‘informative SSR markers’. In seed purity assays of parental lines and hybrids, the amplicons were resolved on 3% agarose gels, stained with ethidium bromide and visualized under UV in a gel documentation system (Alpha Innotech, USA) and impurities were identified based on deviations in expected amplification pattern.
Purity assessment of CMS line IR58025A
A two dimensional DNA sampling strategy suggested by Nas et al. (2002) was adopted for this purpose. Four hundred seeds of the CMS line IR58025A collected from a commercial seed-lot were planted in a 20-row × 20-column grow-out matrix (Fig. 2B) during Wet season 2004 in the experimental farm of Directorate of Rice Research, Hyderabad, India. When the seedlings were 20-d-old, ∼2 cm leaf bits were collected from each of the seedling on the rows of the matrix, bulked and used for isolation of total genomic DNA by following the protocol of Zheng et al. (1995). Similarly, DNA was isolated from bulked leaf bits from each column of the matrix. Thus a total of 40 DNA bulks representing 20 rows and 20 columns were prepared and these were amplified through multiplex PCR using two informative SSR markers RM202 and RM276, which amplified alleles unique to IR58025A. The rows and columns, which showed amplification pattern different from that of IR58025A, were considered as impure rows or columns. Plants located on hills where the impure row(s) and column(s) intersected were considered as suspected admixture(s). The seedlings in the 20 × 20 grow out matrix was grown till maturity and the genotype as deduced from marker profiles was verified with the phenotype as deduced from GOT. The experiment was repeated once again in Dry season 2005 to reconfirm the results.
Purity assessment of the rice hybrid-KRH-2
Four hundred seedlings of the rice hybrid KRH-2 were planted in a grow-out plot in the experimental farm of Directorate of Rice Research during wet season 2004 and dry season 2005. DNA was isolated from 20 days old seedlings of the 400 coded plants, individually as per the procedure of Zheng et al. (1995). Genotyping of the 400 seedlings was done through multiplex PCR involving two informative SSR markers RM164 and RM206, which exhibited amplification of unique alleles in the hybrid. Genotyping of the seedlings was also done using these markers individually to compare the efficiency of single marker assays with that of multiplex assay. The genotype inferred from the marker profile was compared with the phenotype at maturity to verify the results derived from marker analysis with GOT.
Results and discussions
DNA fingerprinting approaches based on polymerase chain reaction have become methods of choice for germplasm characterization, diversity studies and seed purity assays. A variety of DNA markers are now available for fingerprinting cultivars and for marker assisted selection. Of these, SSRs are the preferred ones for rice due to their abundance, co-dominant nature and their distribution throughout the genome and user-friendly nature (Mc Couch et al. 2002). Through the present study, we have assessed the potential of SSR markers in distinguishing rice hybrids and their parental lines and utilized ‘informative’ SSR markers for testing purity of seeds of a parental line and rice hybrid. The results obtained are discussed below.
Microsatellite polymorphism in the parental lines
All the 48 microsatellite markers used in the present study detected polymorphism among the 30 rice lines studied and amplified a total of 163 alleles. These include 2 alleles in 15 markers, 3 in 12 markers, 4 in 13 markers, 5 in 5 markers, 6 in 1 marker and 7 alleles in 2 markers giving an average 3.39 ± 1.3 allelic variants per marker. The number of alleles amplified by each SSR marker is shown in Table 2. Figure 1 shows the amplification pattern generated by the marker RM70 among the 30 rice lines studied.
Molecular profiles of 30 rice lines obtained with microsatellite marker RM70. (a) lanes 1—IR58025A, 2—IR62829A, 3—IR68886A, 4—IR68888A, 5—IR68897A, 6—IR62928A, 7—DRR2A, 8—PMS10A, 9—CRMS31A, 10—CRMS32A, 11—KMR3R, 12—BR827-35R, 13—IR40750R, 14—NDR3026R, 15—Ajaya, 16—C20R, 17—IR10198R, 18—UPRI192-33R, 19—IR66R, 20—IR9761R, 21—BPT5204, 22—Swarna, 23—Jaya, M—100 bp DNA ladder. (b) Lanes 24—Vijetha, 25—Krishnahamsa, 26—Pusa Basmati-1, 27—PR106, 28—Vibhava, 29—Mandya Vijaya, 30-IR64, M—100 bp DNA ladder
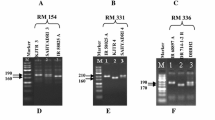
Among the ten CMS lines studied, 98 different alleles with molecular weights ranging from 85 to 400 bp were amplified. Twelve primer pairs generated alleles, which were ‘specific’ for six CMS lines with molecular weights ranging from 90 to 240 bp (Table 3). One hundred and fifteen different alleles with molecular weights ranging from 85 to 280 bp were amplified in the R-lines analyzed. Ten microsatellite primer pairs generated ‘specific alleles’ for nine R-lines (Table 3). The preliminary analysis using 48 evenly distributed hyperpolymorphic SSR markers allowed us to identify several markers, which exhibited amplification of alleles ‘specific’ or ‘unique’ to a particular parental line. Using a few SSR markers, which were highly ‘informative’, all the CMS and R lines used in the present study could be easily distinguished. These ‘informative SSR markers’ were validated in a set of 10 rice varieties to check for cross amplification of the ‘specific’ alleles seen in a parental line (Table 3). No cross amplification of the parental line specific alleles was noticed in these rice inbred lines which included varieties like Swarna, BPT5204, IR64, Jaya etc, which are normally grown in locations where hybrid and parental seed production is taken up in India. By this way it was insured that the specific alleles amplified in a particular parental line are indeed ‘unique’ and not amplified in potential contaminants grown in its vicinity. We have also identified SSR marker combinations amenable for multiplex PCR and capable of producing amplification pattern which is very unique to particular parental line. For eg., when the markers RM202 and RM276 were multiplexed, they gave amplification of alleles of size 165 and 90 bp respectively in the CMS line IR58025A, which was not seen in any other rice line tested.
Use of informative SSR markers in assessment of impurities in a commercial seed lot of the CMS line IR58025A
Genetic purity within a crop variety or hybrid is imperative for ensuring its agronomic performance and protection of intellectual property rights through Plant Variety Protection (PVP) or Plant Breeder’s Rights (PBRs). To meet the standard specifications of purity, the parental lines used in hybrid seed production should have a very high (>99%) level of purity (Yashitola et al. 2002). Among the parental lines (i.e., CMS and R lines), purity of CMS seeds is critical, as CMS lines can only be perpetuated by open pollination with its cognate isonuclear maintainer line. Therefore, pollen contamination in seed multiplication plots of CMS lines is not uncommon. We had earlier reported (Yashitola et al. 2004) the utility of a CMS mitochondria specific PCR marker for distinguishing the CMS lines of wild abortive (WA) cytoplasm background from their cognate maintainer lines. This marker, though highly useful in detecting the contamination of maintainer line seeds in CMS seed stocks, cannot distinguish the contaminants wherein the CMS line is pollinated by pollen shedders other than the cognate maintainer line. To address this problem, we utilized the two-dimensional DNA sampling strategy involving a 20 × 20 grow-out matrix (suggested by Nas et al. 2002). Two SSR markers, RM202 and RM276 amplified allelic fragments of size 165 and 90 bp respectively in IR58025A, which are very ‘specific’ and ‘unique’ to this genotype. Bulked DNA samples prepared from leaf bits collected from seedlings grown in the 20 × 20 matrix during wet season 2005, from each row and column of the matrix were analyzed through multiplex PCR involving the two markers. It was observed that at two matrix intersections (R6C18 and R8C3; Fig. 2a & b), heterozygous amplifications were observed with respect to both the SSR markers and the seedlings located on these two hills R6C18 (6th row, 18th column) and R8C3 (8th row, 3rd column) could be considered as contaminants. To further investigate the nature of these contaminants, the DNA isolated from these two plants was amplified with the CMS mitochondria specific PCR marker (Yashitola et al. 2004). The analysis revealed that both these plants amplified the CMS mitochondria specific ∼330 bp fragment indicating that the two contaminants were products of out crossing of IR58025A by a pollen shedder. Subsequently, the two plants were confirmed to be contaminants based on morphological features assayed through GOT at maturity. Similar results were obtained when the assay was repeated in dry season 2005 and the contaminants were detected accurately by the marker based assay.
A Two-dimension assay involving a 20 × 20 grow-out matrix for assessment of purity of IR58025A with the help of SSR markers RM202 and RM276. (a) Row-wise lanes 6 & 8 and Column-wise lanes 3 & 18 (indicated by arrows) represent contaminants. (b) Schematic representation of the 20 × 20 matrix based method for rapid identification of contaminants in IR58025A. Plants at intersections of 6th row 18th column and 8th row and 3rd column (indicated by arrow) were identified as contaminants
In general, two major sources of contaminants are observed in commercial CMS seed multiplication plots: (i) the maintainer line getting admixed with the seeds of CMS line and, (ii) out-pollination of CMS line by pollen shedders other than the maintainer line. The former can be easily detected using the CMS mitochondria specific PCR marker. Through multiplex PCR, we have demonstrated that even the contamination due to cross-pollination by non-maintainer pollen shedders, (that cannot be detected by the CMS specific PCR based marker) could also be detected easily and reliably. In GOT, usually a sample of ∼400 seeds is collected from a seed-lot and assayed. The same sample size can be considered for the DNA marker based assay. If the assay is done at seedling stage by adopting a 20 × 20 matrix strategy involving DNA bulks, analysis of all the 400 seedlings is not required and the total number of PCRs can be restricted to just 20 reactions as demonstrated in the present study. Nas et al. (2002) have also demonstrated utility of a similar two-dimensional DNA sampling strategy for assessing hybridity.
Identification of microsatellite markers and marker combinations suitable for hybrid purity testing
A list of markers which gave amplification of ‘specific’ and ‘unique’ alleles among the parental line constituting nine public bred Indian rice hybrids was prepared (Table 4). With a set of 10 SSR markers (RM70, RM334, RM475, RM219, RM206, RM336, RM547, RM164, RM335, and RM276), all the nine hybrids could be clearly distinguished easily. Therefore, these markers could be considered ‘highly informative’. In addition to single markers, we also tried marker combinations, which were amenable for multiplex PCR and capable of distinguishing the hybrids. A list of such markers is given in Table 4. For all the three hybrids tested, the multiplex combination of the SSR markers RM164, RM206 and RM276 generated very specific and unique amplification patterns for each hybrid, which is not shared by others (Fig. 3). We propose that these three SSR can be used for purity analysis of most of all the public bred Indian rice hybrids through multiplex PCR involving two or three SSR markers.
Purity testing of a commercial hybrid, KRH2 through multiplex PCR
Two informative microsatellite markers, RM164 and RM206 were used for assessment of purity of a sample of KRH2 consisting of 400 seeds planted in a grow-out plot during wet season of 2005. Based on single marker and multiplex PCR analysis, RM164 identified eight contaminants (sample numbers 1, 23, 28, 222, 225, 282, 311 and 392) and RM206 identified seven (sample numbers 1, 23, 28, 225, 231, 282 and 311) as contaminants. Though both the markers individually detected six contaminants, they differed in respect of the seedling numbers detected as contaminant, showing that screening using a single SSR marker may not be always accurate. When all the 400 seedlings were analyzed through multiplex PCR using the two markers, nine seedlings were found to be contaminant (Fig. 4; sample numbers 1, 23, 28, 222, 225, 231, 282, 311 and 392). Similar results were obtained when the experiment was repeated during dry season of 2005, with the multiplex assay detecting contaminants more accurately as compared to single marker assays.
Detection of impurities in the Indian rice hybrid-KRH2 through multiplex PCR using the microsatellite markers RM164 and RM206. M—50 bp ladder, A—CMS line (IR58025A), H—Hybrid (KRH2), R—Restorer line (KMR3), 221 to 240—Samples of hybrid KRH2 collected from a commercial seed-lot. Arrow indicates contaminants
Earlier, it was proposed that a single polymorphic microsatellite marker might be sufficient for routine analysis of purity of commercial hybrid seed samples (Yashitola et al. 2002). However, the additional information generated through the present study warrants us to propose that, in certain cases, the use of single microsatellite marker for hybrid purity testing may not be sufficient for accurate detection of impurities. Multiplex PCR is a cost saving strategy, wherein analysis can be carried out simultaneously using 2–3 markers through a single tube PCR, with negligible addition to the total cost of assay and with enhanced accuracy. Therefore, we propose that analysis using single markers (as proposed by Yashitola et al. 2002 and Nandkumar et al. 2004) may not help in accurate estimation of seed impurities in certain cases, and wherever possible, it is better to deploy more than one marker through multiplex PCR.
A set of morphological descriptors are currently used for varietal identification, description and seed purity assessment. Though widely adopted and practiced, purity assessments based on morphology is often affected by environment, beside the on time and resources. Furthermore, many of the modern high yielding varieties and hybrids are phenotypically less distinct making morphological evaluation more difficult. Microsatellite markers have been used for genetic characterization of cultivars in wheat, maize, sunflower and tomato (Karkousis et al. 2003; Wang et al. 2002; Zhang et al. 2005; Smith and Register 1998). The Biochemical and Molecular Techniques Group of the International Union for the Protection of New Varieties of Plants (UPOV) is evaluating different DNA marker parameters prior to its routine use in establishing distinctness, uniformity and stability (DUS) of plant varieties (Bredemeijer et al. 2002; UPOV-BMT 2002). Through the present study we have demonstrated that the hyperpolymorphic microsatellite markers, if carefully chosen, are highly useful in distinguishing all the major A and R-lines used in hybrid rice breeding in India. Thus a set of 10 hyperpolymorphic SSR markers (RM70, RM334, RM475, RM219, RM206, RM336, RM547, RM164, RM335, and RM276) are informative enough to distinguish all the parental lines and hybrids considered in the present study. In addition, we have also identified ‘cultivar specific microsatellite profile’ for eight varieties (which includes popular varieties like Pusa Basmati-1, BPT5204 and Jaya) involving the above-mentioned markers, which could be used for their seed purity estimation at different stages of seed multiplication (Table 3).
With increasing number of public as well as private bred rice hybrids under commercial cultivation, quality control in terms of monitoring seed genetic purity at both parental and hybrid seed production stages is vital for the success of hybrid rice technology. Considering the innate disadvantages of GOT for seed purity analysis, marker based seed purity assay which could be an alternative, is receiving the attention. Replacement of GOT with a marker-based assay demands characterization of the parental lines with a large set of hyperpolymorphic markers to identify ‘informative’ markers. The present study is unique due to the fact that a comprehensive set of microsatellite markers (48 Nos.) were utilized for distinguishing the popular CMS and R lines to develop a microsatellite database consisting of ‘informative’ SSR markers. The utility of these markers in detection of impurities is also clearly demonstrated through cost saving strategies of 20 × 20 grow out matrix based bulked sample analysis and multiplex PCR. Further, our study has brought out that analysis using single marker might not be sufficient for detection of impurities in hybrids and multiplex PCR assays involving more than one marker can aid in accurate detection of contaminants. The SSR marker information developed through this study will be of immense help for hybrid rice seed industry to select appropriate marker combinations and assess purity at each stage of seed multiplication.
References
Bredemeijer GMM, Cook RJ, Ganal MW, Peeters R, Issac P, Noordijk Y, Rendell S, Jackson J, Roder MS, Wendehake K, Dijcks M, Amelaine M, Wickaert V, Bertrand L, Vosman B (2002) Construction and testing of microsatellite database containing more than 500 tomato varieties. Theor Appl Genet 105:1019–1026
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation. Plant Mol Biol Rep 1:19–21
Karkousis A, Barr AR, Chalmers KJ, Ablett GA, Holton TA, Henry RJ, Lim P, Langridge P (2003) Potential of SSR markers for plant breeding and variety identification in Australian Barley germplasm. Aust J Agric Res 54:1197–1210
Mao CX, Virmani SS, Kumar I (1996) Technological innovations to lower the costs of hybrid rice seed production. In: Virmani SS et al (eds) Advances in hybrid rice technology. Proceedings of Third International Symposium on Hybrid Rice, Directorate of Rice Research, Hyderabad, India
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono Z, Yano M, Fjellstrom R, Declerck G, Schneider D, Cartinhour S, D.Ware D, Stein L (2002) Development and Mapping of 2240 New SSR Markers for Rice (Oryza sativa L.). DNA Res 9:199–207
Mishra B, Viraktamath BC, Ilyas Ahmed M, Ramesha MS, Vijayakumar CHM (2003) Hybrid rice research and development in India. In: Virmani SS, Mao CX, Hardy B (eds) Hybrid rice for food security, poverty alleviation, and environmental protection. Proceedings of the 4th International Symposium on Hybrid Rice, 14–17 May 2002, Hanoi, Vietnam. Los Baños (Philippines), International Rice Research Institute, pp 265–283
Nandakumar N, Singh AK, Sharma RK, Mohapatra T, Prabhu KV, Zaman FU (2004) Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers. Euphytica 136:257–264
Nas TM, Toledo RS, Li Z, Virmani SS (2002) Two-dimensional DNA sampling: a new strategy for assessing hybridity using molecular markers. Paper presented at the 4th International Symposium on Hybrid Rice, International Rice Research Institute, Hanoi, 14–17 May 2002
Rangaswamy M, Jayamani P (1996) Hybrid rice research in Tamil Nadu. In: Rangaswamy M (ed) Hybrid rice technology. School of Genetics, Tamil Nadu Agricultural University, Coimbatore, India
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor, NY, USA, pp 5.40–5.46
Smith JSC, Register III JC (1998) Genetic purity and testing technologies for seed quality: a company perspective. Seed Sci Res 8:285–293
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipvich L, Cho YG, Ishi T, McCouch SR (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch SR (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
UPOV-BMT (2002) BMT/36/10 Progress Report of the 36th Session of the Technical Committee, the technical working parties and working group on biochemical and molecular techniques and DNA-profiling in particular, Geneva
Virmani SS, Ish Kumar (2004) Development and use of Hybrid rice technology to increase rice productivity in the tropics. Intl Rice Res Newsl 29:10–20
Wang J, Zhong GY, Chin ECL, J.C. Register III JC, Riley RD, Niebur WS, Smith JSC (2002) Identification of parents of F1 hybrids through SSR profiling of maternal and hybrid tissue. Euphytica 124:29–34
Yashitola J, Thirumurugan T, Sundaram RM, Naseerullah MK, Ramesha MS, Sarma NP, Sonti RV (2002) Assessment of purity of rice hybrids using microsatellite and STS markers. Crop Science 42:1369–1373
Yashitola J, Sundaram RM, Biradar SK, Thirumurugan T, Vishnupriya MR, Rajeshwari R, Viraktamath BC, Sarma NP, Sonti RV (2004) A sequence specific PCR marker for distinguishing rice lines on the basis of wild abortive cytoplasm from their cognate maintainer lines. Crop Sci 44:920–924
Yuan LP (1977) The execution and theory of developing hybrid rice. Zhonggue Nongye Kexue (Chin Agric Sci) 1:27–31
Zhang LS, Clerc VL, Li S, Zhang D (2005) Establishment of an effective set of simple sequence repeat markers for sunflower variety identification and diversity assessment. Euphytica 83:66–72
Zheng K, Huang N, Bennett J, Khush GS (1995) PCR-based marker assisted selection in rice breeding. IRRI Discussion Paper Series No. 12. International Rice Research Institute, Manila, The Philippines
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sundaram, R.M., Naveenkumar, B., Biradar, S.K. et al. Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163, 215–224 (2008). https://doi.org/10.1007/s10681-007-9630-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9630-0