Abstract
Bacterial wilt (BW) caused by Ralstonia solanacearum is an important disease of many plant species especially Solanaceae. To compensate for lack of BW resistance in cultivated potato, we fused UV-treated protoplasts of a resistant eggplant variety with protoplasts of a susceptible potato clone to obtain 32 somatic hybrids. Although asymmetric protoplast fusion has the potential to transfer traits from distant species, introgression frequency and preference of alien fragments remain obscure, as well as the genetic basis for control of a trait. In the present research, the genome components of 32 somatic hybrids were determined by parent-specific SSRs. Each hybrid had integrated from one to eight alien chromosome fragments, providing a foundation for selection of BW resistance transmitted from eggplant. When the selected eggplant sequences were aligned with potato genome sequence it showed a similarity of 46.7 %, suggesting a large genetic distance between these two species. The results also revealed that introgression of eggplant fragments is non-selective, which may allow any part of alien chromosomes to be integrated. Distribution of eggplant loci in individual hybrids suggested a possible relationship between markers emk03O04, emi04P17 and emd13E02a and BW resistance, which are potential loci that control target traits and therefore deserve further investigation. With genome-wide selection of parent-specific molecular markers and sequence alignment, the present research substantiated interspecific introgression of a trait lacking in potato. Moreover, an efficient strategy was established to estimate genomic components of the somatic hybrids, and to explore the candidate loci that associate with target traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ralstonia solanacearum, one of the most devastating pathogens of potato transmitted by soil, causes bacterial wilt (BW) or brown rot disease in hundreds of plant species, including many important crops such as potato, tomato, tobacco, pepper, banana, ginger, peanut, etc. Wilt caused by this pathogen is an enormous risk for crops in both tropical and temperate climates (Miao et al. 2009). R. s olanacearum persists in the soil, plant debris, weed rhizospheres and alternate hosts, and it is spread in infected planting material and in irrigation water (Olsson 1976; Elphinstone 1996; Laferriere et al 1999). The pathogen invades plant vascular tissues from wounded roots or natural openings (Sagar et al. 2013). With a vigorous ability to overwinter and wide distribution around the world, R. s olanacearum has been demonstrated as a great threat to plants. However, because there is no efficient chemical control of this disease, crop rotation is the only means of control but may not be practiced in regions with limited land resources.
As the pathogen infecting more than 200 plant species belonging to 53 botanical families (Álvarez et al. 2010), R. s olanacearum is historically subdivided into five races based on host range and five biovars based on the ability to acidify 5–8 carbohydrates (Champoiseau et al. 2009). According to the understanding of the species complex, R. s olanacearum is geographically classified into phylotypes I–IV, which are found in Asia, America, Africa, and Oceania. The majority of host plants belongs to the Solanaceae and Musaceae families. Wilting occurs at high bacterial population in the xylem and is partially due to vascular dysfunction in which water cannot reach the leaves sufficiently. Also the degradation of occluded xylem vessels and the destruction of surrounding tissues lead to the collapse and death of the plant (Álvarez et al. 2010). EPS (exopolysaccharide), as the single most important virulence factor of R. s olanacearum, plays an essential role in blocking and breaking down the vascular tissue (Genin and Denny 2012). In susceptible hosts, accumulation of EPS is largely responsible for the vascular dysfunction that causes wilt symptoms. The expression of the ethylene and salicylic acid defense response pathways will be enhanced by EPS triggers in the wilt resistant tomato breeding line H7996 but not in a susceptible cultivar (Milling et al. 2011). In addition, extracellular protein derived from a type II secretion system (T2SS) helps to degrade the cell wall, which then leads to further invasion to the plant. The type III secretion system (T3SS) is not unique to R. s olanacearum, but it is a major determinant controlling virulence (Angot et al. 2006). Single effector protein cannot alter pathogenicity of R. s olanacearum, but it can cause disruption of certain subsets and strongly effect virulence of the pathogen, which indicates that T3E diversity (the various effectors secreted by T3SS) may play a role in determining the broad host range of the R. s olanacearum species complex (Vasse et al. 2000). Up to the time of this research, resistance to R. s olanacearum is still unclear. This is due to the different conditions of inoculation including resistant materials, strains of R. s olanacearum and different evaluation methods used. Resistance genes have been defined as recessive, incompletely dominant or dominant, the number of genes described as monogenic, oligogenic, polygenic; also additive and non-additive effects between genes have been mentioned (Miao et al. 2009).
Potato (Solanum tuberosum) regarded as the fourth most important food crop and one of the most significant solanaceous crops, is widely cultivated across the world. Potato is known to be affected by two races of R. s olanacearum, and the soil-borne nature of the pathogen makes breeding of resistant cultivars the favored strategy for BW control (Deslandes et al. 2002). Modern potato cultivars have a narrow range of disease resistance, especially lack of high level resistance to R. s olanacearum (Fock et al. 2001; Kim-Lee et al. 2005; Chen et al. 2013). Although BW resistance has been reported in potato species S. v ernei (Gao et al. 2000), S. c ommersonii (Laferriere et al. 1999; Kim-Lee et al. 2005) and some other plants like S. lycopersicum (Nazeem et al. 2001), S. m elongena (Lebeau et al. 2011; Rotino et al. 2014) and Capsicum annuum L (Lebeau et al. 2011), interspecific sexual-incompatibility makes conventional breeding fail to transfer alien genes from distantly related species. Efforts at interspecific somatic hybridization in potato have realized somatic hybrids with introgressed resistance to potato virus Y and late blight (Thieme et al. 2008; Chandel et al. 2015), potato leafroll virus (Valkonen et al. 1994), nematode resistance (Austin et al. 1993), and resistance to R. s olanacearum (Fock et al. 2001; Iovene et al. 2012; Chen et al. 2013; Yu et al. 2013). Protoplast fusion also realized alien interspecific gene-transfer in other plants, e.g., salt-tolerance from Thinopyrum ponticum to wheat (Chen et al. 2004) and from T. i ntermedium to wheat (Li et al. 2014), to obtain fertile somatic hybrids from S. a ethiopicum to S. m elongena (Daunay et al. 1993), and Fusarium oxysporum f. sp. melongenae resistance from Solanum integrifolium to eggplant (Rotino et al. 2001). However, few new varieties have been bred from the somatic hybrids. Possible explanation could be a long circle backcross which is needed to get rid of the genetic drag especially when wild species is involved, elimination of the alien chromosome occurred in meiosis during gametogenesis, or loss of the target trait owing to difficulties in following pedigree selections.
To reduce the genetic drag of somatic hybrids, asymmetric fusion has been adopted in many plants, particularly when distant parents are fused. In wheat, donor parent pre-irradiated with UV, maize nuclear and mitochondrial DNA were integrated into the wheat genome (Xu et al. 2003), and fertile somatic hybrids were produced with genetic material transferred from T. i ntermedium into a wheat background Li et al. (2014). Xu et al. (2007) obtained asymmetric somatic hybrids between UV-irradiated Citrus unshiu and C. s inensis for transfer of limited amount of favorable traits. Asymmetric hybridization was employed to intensively study the detection of asymmetric hybrids and genome elimination (Puite and Schaart 1993; Oberwalder et al. 1997), effect of recipients and genome stability (Fehér et al. 1992; Oberwalder et al. 1998; Rakosy-Tican et al. 2015), and possessed the potentials of alternative approach for potato cultivar improvement. In our study, we took the advantage of asymmetric fusion to create hybrids with target resistance to BW disease and aimed to explain the resistance-related alien sequence.
Eggplant (S. m elongena) variety 508.1, with BW resistance was used as donor parent in asymmetric protoplast fusion with potato (S. t uberosum) clone 8# that is BW susceptible. In addition to phenotyping the derived somatic hybrids in plant morphology for general evaluation of their application prospects, we focused the present research on: (1) genome variation of the hybrids to elucidate possible preference of eggplant chromosome fragments likely to introgress, and (2) potential eggplant loci associated with BW resistance to provide possible information for further use of the hybrids.
Materials and methods
Plant material
Seeds of S. m elongena cv. 508.1 (2n = 2x = 24), highly resistant to BW disease, were provided by Professor Lian Yong of the Chinese Academy of Agricultural Sciences. S. t uberosum cv. 8# (2n = 4x = 48) was generated by spontaneous doubling of a potato dihaploid clone (2n = 2x = 24), which was generated from a Chinese potato cultivar Zhongshu 2 by female parthenogenesis. The fusion parents were maintained in tissue culture on MS medium (Murashige and Skoog 1962) supplemented with 4 % sucrose and 8 % agar at 22 ± 1 °C with a photoperiod of 16 h/day under a light intensity of 60 µmol m−2 s−1.
Asymmetric protoplast fusion and plant regeneration
Protoplasts of eggplant (variety 508.1) irradiated with ultraviolet light (UV) at an intensity of 630 μW/cm2 for 20 s were fused by electrofusion with the protoplasts of potato clone 8#. Isolated protoplasts of both fusion parents were diluted to a density of 2 × 105 ml−1 and mixed in equal proportions. The cell suspension was placed into a helix fusion chamber (Eppendorf Multiporator 4308, Germany) for cell fusion. The methods for electrofusion and protoplast culture were both described previously (Yu 2013; Yu et al. 2013). The regenerated somatic hybrids were propagated and grown in vitro on MS or 1/2 MS salt medium (when rooting was difficult) for root development. The regenerated plants were numerically named by the order of callus and shoot formation. For example, 4-2 represents the second shoot regenerated from the forth callus formed.
Ploidy level determination and SSR analysis
The ploidy level was determined by flow cytometry. Approx. 1 mm2 of young leaves were collected from in vitro plantlets and analyzed by Partec Ploidy Analyzer flow cytometry (CYFLOW Space, Partec, Germany). The leaves of each regenerant and the fusion parents were mixed and sliced in a plastic dish containing nuclear extraction buffer (Extraction buffer of CYSTAIN DNA 2 Kit, Partec, Germany). After filtration, the samples were stained for 5 min with DAPI (4,6-diamine-2-phenylindol) before running the nuclei through the flow cytometry. Each histogram was generated by FloMax software yielding peak position, coefficient of variation and the relative ploidy index.
Total genomic DNA was isolated from young leaves of in vitro plantlets according to the CTAB method (Dellaporta et al. 1983). The fusion parents and regenerants were analyzed with 206 SSR markers located on the genome map of eggplant (Nunome et al. 2009). The PCR reaction which consisted of 20 μl volume with 30 ng genomic DNA, 0.5 U of Taq polymerase, 0.2 μM each primer, 200 μM dNTPs, 1.5 mM MgCl2 and 1× Taq buffer, was conducted as below: one cycle of 94 °C for 3 min; ten cycles of 94 °C for 0.5 min, 65–55 °C decreasing by 1 °C per cycle for 1 min, and 72 °C for 1 min; 30 cycles of 94 °C for 0.5 min, 55 °C for 1 min, and 72 °C for 1 min; and a final cycle of 72 °C for 5 min (Nunome et al. 2009) by C1000 Thermal Cycler (Bio-Rad Inc, Hercules, CA, USA). Amplified products were separated by 6 % denatured polyacrylamide gel electrophoresis and stained with silver.
Measurement of plant morphology
Four-week-old plantlets of the fusion parents and hybrids were transplanted into plastic pots (35 cm diam) in a greenhouse. Six plants of each tested material were grown under normal conditions favorable for potatoes. Plant height, stem diameter, leaf and flower morphology were measured about 7 weeks after transplanting when first flowers appeared. Stolon length, tuber morphology and tuber dormancy were recorded after harvest or during storage at room temperature.
Evaluation of resistance to R. s olanacearum
R. s olanacearum strain HA4-1 (phylotype 1, sequevar 14) was used to inoculate the fusion parents and the somatic hybrids. The strain was isolated from peanut in Hongan (northeast Hubei of China) which infected potato plants with serious wilt symptoms in the pre-test. All the tested materials were cultured on MS medium at 22 ± 1 °C with 16 h/day for 3 weeks before inoculation of root cuttings with a sterile knife and inoculated with 5 ml bacterial solution (1 × 108 colony forming units per ml) on the medium of each chamber. Each material was cultured in four boxes with six plants each (one plant for eggplant parent 508.1). The test was repeated three times. The control consisted of inoculation with sterile water. Disease score (DS) was according to Kim-Lee et al. (2005) for severity of wilting (0, no wilted leaves; 1, <25 % leaves wilted; 2, 26–50 % leaves wilted; 3, 51–75 % wilted; and 4, 75–100 % wilted). The disease index (DI) was calculated by DI = ∑ (number of plants with a specific DS × DS)/(total number of inoculated plants × 4) (Winstead and Kelman 1952). For comparison among tested materials, we calculated the relative disease index (RDI) using the following formula: RDI = DI of the tested plant material × correction coefficient K, where K = the DI expectation of susceptible control/DI of the tested susceptible control (Wu et al. 2004; Fan et al. 2014). For each plant material, the degree of resistance to R. s olanacearum was classified as: highly resistant (HR), 0 ≤ RDI < 20; resistant (R), 20 < RDI < 40; medium resistant (MR), 40 < RDI < 60; medium susceptible (MS), 60 < RDI < 80; susceptible (S), 80 < RDI < 90; highly susceptible (HS), 90 < RDI ≤ 100.
Results
Plant regeneration and ploidy level
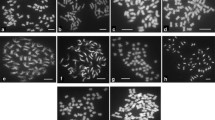
Cell division first occurred on CM-I medium 3–4 days after protoplast fusion, and cell colonies of 1–2 mm diam formed in 3–4 weeks. Calli were then transferred to the two-layer culture medium for 2–3 weeks, and then to the proliferation medium (CM-II) for 3–4 weeks. After 3 weeks the calli that were approximately 5 mm in size were transferred onto the regeneration medium. In total, 1080 calli were obtained, whereas 50 vigorous plants regenerated from 20 calli (Fig. 1).
The recipient potato parent was tetraploid, and donor eggplant was diploid. The flow cytometry revealed the 47 regenerated plants to vary in ploidy level, including tetraploid, octoploid, mixoploid and aneuploid (Table 1, Supplement Fig. 4). For breeding consideration, we focused on the tetraploid plantlets for further analysis of the chromosome composition and alien trait transmission for bacterial wilt resistance.
Chromosome composition of hybrids via SSR analysis
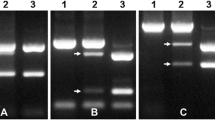
The SSR markers were arrayed to distinguish differences in genome components of the somatic hybrids. In the present study, 26 of 177 eggplant parent-specific SSR markers yielded a total of 38 clear and repeatable polymorphic bands in 32 somatic hybrids, whereas 48 of 52 potato parent-specific SSR markers could be detected in all the hybrids. Selected primers are shown in Fig. 2a and the results summarized in Table 2. Each band was taken as a genetic locus, we detected about 8.3 % eggplant loci in the hybrids, indicating that the eggplant parent genome was partially introgressed into the recipient potato genome. The introgression frequency of each hybrid was 1-8. Uneven distribution of the eggplant loci among the hybrids implied a randomized integration of eggplant chromosome fragments. Novel bands (Hu et al. 2002; Liu et al. 2002; Yang et al. 2007) which were present in hybrids and absent in the fusion parents were observed on five occasions (Fig. 3). The UV radiation protocol used for producing asymmetric somatic hybrids employed in this study appears to have been effective for this interspecific genome introgression.
The genome components of somatic hybrids and assessment of resistance to R. s olanacearum. a The genome component of somatic hybrids in SSR analysis. Scatterplot presents the SSR locus of eggplant genome detected in somatic hybrids. All the SSR loci are listed as the order from chromosome I–XII of eggplant, which was published in prior research (Nunome et al. 2009). Twenty-six SSRs generated bands in somatic hybrids containing 38 loci in 32 somatic hybrids. Resistant and susceptible hybrids are separated by a vertical blue line. The potato parent 8# stands for susceptible one, eggplant parent for resistant one. The three markers emk03O04, emi04P17, emd13E02a in red circle appear in MR hybrids but absent in MS or more susceptible ones and b the resistance level of the somatic hybrids (tetraploid hybrids) to R. s olanacearum. RDIs (relative disease index) are shown in bar chart and the difference of RDIs are indicated by the letters below (different letter represents the difference at p < 0.05 by Duncan). (Color figure online)
SSR analysis of the fusion parents and hybrids. a Amplification of primer emi04P17. The downward pointing arrows indicate somatic hybrids with emi04P17 insertion and b amplification of primer emf01O01. This primer indicates the identical bands with potato parent, and has no insertion of eggplant genome fragment, c amplification of primer of emg11M09. In lane 9, the somatic hybrids have the emg11M29 insertion, and most hybrids have the new band which is absent in both fusion parents and d amplification of primer emf11F24. Another new band is amplified by emf11F24, and no eggplant introgression is observed. Lane E, the eggplant fusion parent 508.1; P, the potato fusion parent 8#; M, marker φX174 DNA-HaeIII; 1-33, hybrids. Arrow in c and d indicates novel bands, these bands are presented on a l ine, which are absent in the aera of both fusion parents
There were about 88.5 % potato parent-specific loci that could be identified in the hybrids, reflecting the fact that asymmetric somatic hybrids retained most of the genome of the recipient parent and more resembled the potato phenotype (Supplement Fig. 4). The frequency of potato loci remaining varied from 69.0 to 85.1 % among the hybrids, which may reflect a loss of chromosome fragments during the process from protoplast fusion to plant regeneration.
Clarification of introgression bias
Since potato and eggplant are related species in Solanaceae, a question arises as to whether sequence similarity is related to uneven insertion of eggplant fragments in the somatic hybrids. For instance, there were eight eggplant loci detected in hybrid 12-1 but only one in hybrids 22-1, 16-1 and 10-5 (Fig. 2a). Also, eggplant specific band emk03O04 locus 2 of chromosome IV was integrated in 14 hybrids whereas the several loci (emk04N11, emi05P17, emf01D24, emh01F12 or eme36B08) were present in only a single hybrid (Fig. 2a). We first compared the DNA sequence similarity between the two species by aligning all 177 eggplant specific SSRs of the fusion parent 508.1, as well as 1000 other eggplant sequences of 500–1000 bp randomly selected from the Eggplant Genome Database (http://eggplant.kazusa.or.jp/blast.html) to the potato PGSC database (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml). The results showed that, with a threshold value of 1e−20, 44.6 % of them have corresponding hits (Table 3), suggesting a rather low similarity between potato and eggplant.
Of 26 eggplant specific SSRs that produced bands in the hybrids, 18 could be found in the Eggplant Genome Database. Further alignment of these matched sequences to the PGSC potato genome resulted in only four hits (22.2 %) which were identical. No relationship was established between sequence similarity and insertion frequency of eggplant fragments. For instance, the four sequences homologous to potato (i.e., the eggplant markers that can be matched to potato genome) did not have the most insertions (Fig. 2a) and, as to the 131 eggplant specific markers absent in the somatic hybrids, 42 were homologous with potato. The results indicate that eggplant homologous sequence does not preferentially integrate into potato genome, and introgression of asymmetric fusion appears to be random, which provides a possibility of transmitting any trait at an unbiased frequency from donor to recipient.
Identification of BW resistance-related introgressions
The resistance of all 32 somatic hybrids and the fusion parents was assessed and the resistance level was assigned to each material according to their RDI (Fig. 2b, Supplement Fig. 4). The eggplant parent 508.1 was resistant to R. s olanacearum (R with RDI 29.6 %). There were six hybrids (3-1.52, 3-1.30, 3-1.37, 22-1, 16-1, 18-3) that showed middle level resistance (MR) with RDI varying 41.7–59.7 %. The potato fusion parent 8# was BW susceptible (S) and its RDI was calculated as 70.5 %. In the present research, the tested somatic hybrids varied in BW resistance with 18.8 % MR, 53.1 % MS and 28.1 % S or HS. Four somatic hybrids (3-1.52, 3-1.30, 3-1.37 and 22-1) were not significantly different in resistance level from the donor parent 508.1. The results implies that the eggplant BW resistance could be controlled by the loci that have been successfully introgressed into the potato genome.
Considering together the data of eggplant loci distribution of the hybrids (Fig. 2a) and data of their BW resistance (Fig. 2b), an association between the loci and the resistance was established. Dots appearing exclusively in R plants and absent in S plants may be possibly associated with resistance. As a result, three loci (emk03O04 locus 1 located on chromosome IV, emi04P17 and emd13E02a locus 1 located on chromosome VI of eggplant), present in one or two MR hybrids but absent in MS and more susceptible hybrids, may be positively correlated with the resistance to R. s olanacearum (Fig. 2a in red circles). These loci could be considered as markers of the fragments carrying BW resistance of the eggplant parent.
In addition, three bands were amplified with emd13E02a but only the band emd13E02a locus 1 was likely associated with BW resistance. We speculate that there could be at least three homologous sequences matched by primer emd13E02a and the R locus may be closer to emd13E02a locus 1 than the other two.
Morphology of somatic hybrids
Variation in plant morphology of the somatic hybrids was investigated twice. Plant growth, leaf shape and size, flower color, stolon length and tuber shape were recorded. Most hybrids displayed a similar morphology to the potato parent 8# and grew vigorously (Supplement Fig. 4). There was variation in leaf shape (data not shown) and number of leaflets per compound leave. Changes in tuber shape, in terms of length/width ratio, were also observed. Most of 26 hybrids that formed tubers showed oval tuber shape similar to the potato parent 8#, while several hybrids appeared to have fusiform-shaped tubers with greater length/width ratio. A large difference was found in tuber dormancy among hybrids. When stored at room temperature for 65 days, more than half of hybrids had a germination rate over 60 %, similar to or higher than the potato parent, while the rest had germination rates from 0 to about 40 %.
Nine hybrids formed flower buds and two flowered with normal floral organs under greenhouse (Supplement Fig. 4). Although no pollen was produced by flowering hybrids, the potato parent 8# likewise did not form fertile pollen, although it has been known to produce viable pollen under other conditions (data not published). Hybrids that are capable of developing flowers, particularly hybrid 18-3 that possesses BW resistance derived from eggplant, have great potential in potato breeding.
Discussion
In the study, we estimated the genomic composition of asymmetric somatic hybrids with specific parental SSR markers. Among 32 tetraploid somatic hybrids, single or multiple fragments of the eggplant genome were detected, indicating transfer of the alien fragments by interspecific protoplast fusion. The somatic hybrids derived by asymmetric fusion exhibited plant morphology similar to the potato parent with some variation in leaf shape, number of leaflets of compound leaves, tuber shape and tuber dormancy. The particular alien insertion and its location in potato chromosomes are likely to be major reasons for the variation (Chen et al. 2004; Li et al. 2014; Maćkowska et al. 2014).
It is well-known that R. s olanacearum infects hundreds of plant species belonging to 53 botanical families (Álvarez et al. 2010). Within a broad range of infected plants, the symptoms and resistant systems of Solanum species like potato, tomato, eggplant and pepper may share certain similarities. A conclusion drawn by Lebeau and colleagues pointed out that a common resistance mechanism of different plant hosts may be reflected by successful infection of the same pathogen race (Lebeau et al. 2011). This lays the foundation of transferring resistant loci from one species to another. In the present study, we observed that the hybrids which acquired resistance to R. s olanacearum shared some loci of the eggplant parent in common, by arraying the alien loci of each hybrid against their resistance level (Fig. 2). The loci present in the hybrids with resistance level similar to the eggplant parent but absent in the hybrids with significantly lower resistance level than the eggplant parent may be considered as potential loci associated with bacterial wilt resistance. By this standard, three resistance-related loci, Emk03O04 locus 1 (chromosome IV of eggplant), emi04P17 and emd13E02a locus 1 (chromosome VI of eggplant), may reflect the eggplant chromosome fragments containing the resistance genes against R. s olanacearum.
Confronted with RSSC (R. solanacearum species complex), studies on the resistance to R. s olanacearum are sophisticated and results are varied, for different materials involved and inoculations performed. Meanwhile, with high and consistent genetic variation, virulence related gene sequences are more diverse, which indicates R. s olanacearum is a highly diverse bacterial species. In previous studies, Nishi et al. (2003) found one QTL for bacterial wilt resistance among 125 doubled haploid lines of tobacco (derived from F1 hybrids between resistant and wilt-susceptible varieties), which spanned 32 cM on linkage group 5 and explained 43.8 % of the variation for bacterial wilt resistance. In tomato, two AFLP markers were linked to BW resistance through an F2 population derived from pair-cross between BW resistant and susceptible tomato cultivars (Miao et al. 2009). The cDNA-AFLP patterns were found different between resistant and susceptible genotypes in tomato for newly expressed fragments, which occurred on the 2nd day but absent in susceptible ones on the 5th day after inoculation (Nazeem et al. 2001). Recently, in hot pepper near isogenic lines (NILs) differing for resistance to bacterial wilt, the resistance was estimated to be governed by homozygous recessive (rr) gene. AFLP products of primer combination, EcoACT + MseCAC, yielded three polymorphic bands (103, 117, and 161 bp) which were linked to the recessive resistant allele and three polymorphic bands (183, 296, 319 bp) linked to the dominant susceptible allele of the bacterial wilt resistance gene (Thakur et al. 2014). All the above results suggest that the markers may partially possess or play important roles in the resistance against bacterial wilt. Other works in eggplant (Lebeau et al. 2013) and potato (Zhi et al. 2014) showed that the resistance to bacterial wilt may be controlled by one or more quantitative trait loci. In the present research, three candidate loci from eggplant were identified to be associated with bacterial wilt resistance. Interestedly, the resistance phenomenon was also observed in the symmetric hybrids which contain intact chromosome set of eggplant, a sister line of the present eggplant parent (Yu et al. 2013), demonstrating that the chromosome fragments possessing the resistant loci were introgressed from eggplant to potato. Our results may indicate that asymmetric protoplast fusion can not only transfer the alien fragments controlling resistance as symmetric hybridization, but also introgress less genetic background of the eggplant genome.
We aligned the marker sequences, assumed to be associated with bacterial wilt resistance, to the Eggplant Genome Database and found that there were two genes flanking the marker emk03O04 in each of two accessions (Sme2.5_00491.1_g00010.1, Sme2.5_00491.1_g00011.1; Sme2.5_00949.1_g00023.1, Sme2.5_00949.1_g00024.1), and one gene (Sme2.5_12673.1_g00001.1) contains the sequence of the marker emd13E02a. In peptide alignment, four proteins had identity ranging from 43 to 67 % with gag-pol polyprotein and retrotransposon protein of Solanum demissum. In published studies, gag-pol polyproteins were encoded by gag-pol genes of LTR (long terminal repeats)-retrotransposons (Feschotte et al. 2002; Sha et al. 2005), and methylation status of the gag-pol genes may be involved in APR (adult plant resistance) response upon bacterial blight pathogen attack (Sha et al. 2005). It is widely accepted that retrotransposons in plants seem to be tightly linked to molecular pathways activated by stress (Grandbastien 1998). As a stress-induced generator, retrotransposon activation is under control of cis-regulatory sequences which are strikingly similar to those of plant defense genes, and it displays finely tuned responses to external stress (Wessler 1996; Grandbastien 1998, 2014). In addition, abiotic and biotic stresses were the major factors in transcriptional and transpositional activation of retrotransposon (Grandbastien 1998; Alzohairy et al. 2014). In the present research, our results may demonstrate that speculated genes encoding gag-pol polyproteins or/and retrotransposon proteins may function under R. s olanacearum invasion. The inserted sequences linked with the markers will provide a starting point for exploring new insight into the mechanism of BW resistance as well as for expanding broad resistant resource for potato.
In the present study, we obtained somatic hybrids with bacterial wilt resistance by interspecific asymmetric protoplast fusion between S. t uberosum and S. melongena, and the resistance-related loci derived from the eggplant parent had been preliminarily identified by using the parent-specific SSRs. Several hybrids with elevated resistance shared similar growth habit and blooming characteristics with potato, which widens the potential resources for resistance breeding against R. s olanacearum in potato.
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- APR:
-
Adult plant resistance
- BW:
-
Bacterial wilt
- DI:
-
Disease index
- DS:
-
Disease score
- EPS:
-
Exopolysaccharide
- LTR:
-
Long terminal repeats
- NIL:
-
Near isogenic lines
- QTL:
-
Quantitative trait locus
- RDI:
-
Relative disease index
- RSSC:
-
Ralstonia solanacearum species complex
- SSR:
-
Simple sequence repeat
- T2SS:
-
Type II secretion system
- T3SS:
-
Type III secretion system
References
Álvarez B, Biosca EG, López MM (2010) On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. In: Méndez-Vilas A (ed) Current research, technology and education topics in applied microbiology and microbial biotechnology, vol 1. Formatex, Badajoz, pp 267–279
Alzohairy AM, Sabir JSM, Gyulai G, Younis RAA, Jansen RK, Bahieldin A (2014) Environmental stress activation of plant long-terminal repeat retrotransposons. Funct Plant Biol 41:557–567
Angot A, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, Sartorel E, Genschik P, Boucher C, Genin S (2006) Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc Natl Acad Sci USA 103:14620–14625
Austin S, Pohlman JD, Brown CR, Mojtahedi H, Santo GS, Douches DS, Helgeson JP (1993) Interspecific somatic hybridization between Solanum tuberosum L. And S. b ulbocastanum dun. as a means of transferring nematode resistance. Am Potato J 70:485–495
Champoiseau PG, Jones JB, Allen C (2009) Ralstonia solanacearum race 3 biovar 2 causes tropical losses and temperate znxieties. Plant Health Prog. doi:10.1094/PHP-2009-0313-01-RV
Chandel P, Tiwari JK, Ali N, Devi S, Sharma S, Sharma S, Luthra SK, Singh BP (2015) Interspecific potato somatic hybrids between Solanum tuberosum and S. c ardiophyllum, potential sources of late blight resistance breeding. Plant Cell Tissue Organ Cult 123:579–589
Chen L, Guo X, Xie C, He L, Cai X, Tian L, Song B, Liu J (2013) Nuclear and cytoplasmic genome components of Solanum tuberosum + S. c hacoense somatic hybrids and three SSR alleles related to bacterial wilt resistance. Theor Appl Genet 126:1861–1872
Chen S, Xia G, Quan T, Xiang F, Jin Y, Chen H (2004) Introgression of salt-tolerance from somatic hybrids between common wheat and Thinopyrum ponticum. Plant Sci 167:773–779
Daunay MC, Chaput MH, Sihachakr D, Allot M, Vedel F, Ducreux G (1993) Production and characterization of fertile somatic hybrids of eggplant (Solanum melongena L.) with Solanum aethiopicum L. Theor Appl Genet 85:841–850
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Deslandes L, Olivier J, Theulières F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA 99:2404–2409
Elphinstone JG (1996) Survival and possibilities for extinction of Pseudomonas solanacearum (Smith) Smith in cool climates. Potato Res 39:403–410
Fan J, Liu Y, Li Y, Zhou J (2014) Evaluation of tobacco seedling resistance to bacterial wilt and comparison of resistance evaluation system. J Yunnan Agric Univ 29:487–493 (in Chinese)
Fehér A, Preiszner Z, Litkey Z, Csanadi G, Dudits D (1992) Characterization of chromosome instability interspecific somatic hybrids obtained by X-ray fusion between potato (Solanum tuberosum L.) and S. b revidens Phil. Theor Appl Genet 84:880–890
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Fock I, Collonniera C, Luisettib J, Purwitoc A, Souvannavongd V, Vedele F et al (2001) Use of Solanum stenotomum for introduction of resistance to bacterial wilt in somatic hybrids of potato. Plant Physiol Biochem 39:899–908
Gao G, Qu D, Lian Y, Jin L, Feng L (2000) Identification molecular markers linked with resistance to bacterial wilt (Ralstonia solanacearum) in diploid potato. Acta Hortic Sin 27(1):37–41
Genin S, Denny TP (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89
Grandbastien MA (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3(5):181–187
Grandbastien MA (2014) LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochem Biophys Acta 1849:1–14
Hu Q, Andersen S, Dixelius C, Hansen L (2002) Production of fertile intergeneric somatic hybrids between Brassica napus and Sinapis arvensis for the enrichment of the rapeseed gene pool. Plant Cell Rep 21:147–152
Iovene M, Aversano R, Savarese S, Caruso I, Di Matteo A, Cardi T et al (2012) Interspecific somatic hybrids between Solanum bulbocastanum and S. t uberosum and their haploidization for potato breeding. Biol Plant 56:1–8
Kim-Lee H, Moon JS, Hong YJ, Kim MS, Cho HM (2005) Bacterial wilt resistance in the progenies of the fusion hybrids between haploid of potato and Solanum commersonii. Am Potato J 82:129–137
Laferriere LT, Helgeson JP, Allen C (1999) Fertile Solanum tuberosum + S. c ommersonii somatic hybrids as sources of resistance to bacterial wilt caused by Ralstonia solanacearum. Theor Appl Genet 98:1272–1278
Lebeau A, Daunay MC, Frary A et al (2011) Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101:154–165
Lebeau A, Gouy M, Daunay MC et al (2013) Genetic mapping of a major dominant gene for resistance to Ralstonia solanacearum in eggplant. Theor Appl Genet 126:143–158
Li C, Cheng A, Wang M, Xia G (2014) Fertile introgression products generated via somatic hybridization between wheat and Thinopyrum intermedium. Plant Cell Rep 33:633–641
Liu JH, Pang XM, Cheng YJ, Meng HJ, Deng X (2002) Molecular characterization of the nuclear and cytoplasmic genomes of intergeneric diploid plants from cell fusion between Microcitrus papuana and rough lemon. Plant Cell Rep 21:327–332
Maćkowska K, Jarosz A, Grzebelus E (2014) Plant regeneration from leaf-derived protoplasts within the Daucus genus: effect of different conditions in alginate embedding and phytosulfokine application. Plant Cell Tissue Organ Cult 117:241–252
Miao L, Shou S, Cai J, Jiang F, Zhu Z, Li H (2009) Identification of two AFLP markers linked to bacterial wilt resistance in tomato and conversion to SCAR markers. Mol Biol Rep 36:479–486
Milling A, Babujee L, Allen C (2011) Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS One 6:e15853
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nazeem PA, Jose S, Sheeba NK, Madhavan S, Babya A, Kumarb PGS, Devib N (2001) Differential gene expression for bacterial wilt incidence in tomato (Solanum lycopersicum L.) revealed by cDNA-AFLP analysis. Physiol Mol Plant Pathol 76:197–203
Nishi T, Tajima T, Noguchi S, Ajisaka H, Negishi H (2003) Identification of DNA markers of tobacco linked to bacterial wilt resistance. Theor Appl Genet 106:765–770
Nunome T, Negoro S, Kono I, Kanamori H, Miyatake K, Yamaguchi H, Ohyama A, Fukuoka H (2009) Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theor Appl Genet 119:1143–1153
Oberwalder B, Ruoß B, Schilde-Rentschler L, Hemleben V, Ninnemann H (1997) Asymmetric protoplast fusion between wild and cultivated species of potato (Solanum ssp.)—detection asymmetric hybrids and genome elimination. Theor Appl Genet 94:1104–1112
Oberwalder B, Schilde-Rentschler L, Ruoß B, Wittemann S, Ninnemann H (1998) Asymmetric protoplast fusions between wild species and breeding lines of potato: effect of recipients and genome stability. Theor Appl Genet 97:1347–1354
Olsson K (1976) Experience of brown rot caused by Pseudomonas solanacearum (Smith) in Sweden. EPPO Bull 6:199–207
Puite K, Schaart J (1993) Nuclear genomic composition of asymmetric fusion products between irradiated transgenic Solanum brevidens and S. tuberosum: limited elimination of donor chromosomes and polyploidization of recipient genome. Theor Appl Genet 86:237–244
Rakosy-Tican E, Thieme R, Nachtigall M, Molnar I, Denes T (2015) The recipient potato cultivar influences the genetic makeup of the somatic hybrids between five potato cultivars and one cloned accession of sexually incompatible species Solanum bulbocastanum Dun. Plant Cell Tissue Organ Cult 122:395–407
Rotino GL, Mennella G, Fusari F, Vitelli G, Tacconi MG, D’Alessandro A, Acciarri N (2001) Towards introgression of resistance to Fusarium oxysporum f. sp. melongenae from Solanum integrifolium into eggplant. Antalya Turk XI:303–307
Rotino GL, Sala T, Toppino L (2014) Alien gene transfer in crop plants, vol 2. Springer, New York, pp 381–409
Sagar V, Jeevalatha A, Mian S, Chakrabarti SK, Gurjar MS et al (2013) Potato bacterial wilt in India caused by strains of phylotype I, II and IV of Ralstonia solanacearum. Eur J Plant Pathol 138:51–65
Sha AH, Lin XH, Huang JB, Zhang DP (2005) Analysis of DNA methylation related to rice adult plant resistance to bacterial blight based on methylation-sensitive AFLP (MSAP) analysis. Mol Genet Genom 273:484–490
Thakur PP, Mathew D, Nazeem PA, Abida PS, Indira P, Girija D, Shylaja MR, Valsala PA (2014) Identification of allele specific AFLP markers linked with bacterial wilt [Ralstonia solanacearum (Smith) Yabuuchi et al.] resistance in hot peppers (Capsicum annuum L.). Physiol Mol Plant Pathol 87:19–24
Thieme R, Rakosy-Tican E, Gavrilenko T, Antonova O, Schubert J, Nachtigall M, Heimbach U, Thieme T (2008) Novel somatic hybrids (Solanum tuberosum L. + Solanum tarnii) and their fertile BC1 progenies express extreme resistance to potato virus Y and late blight. Theor Appl Genet 116:691–700
Valkonen JPT, Xu Y-S, Pulli S, Pehu E, Rokka V-M (1994) Transfer of resistance to potato leafroll virus, potato virus Y and potato virus X from Solarium brevidens to S. t uberosum through symmetric and designed asymmetric somatic hybridisation. Ann Appl Biol 124:351–362
Vasse J, Genin S, Frey P, Boucher C, Brito B (2000) The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol Plant Microbe Interact 13:259–267
Wessler SR (1996) Plant retrotransposons: turned on by stress. Curr Biol 6:959–961
Winstead NN, Kelman A (1952) Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42:628–634
Wu S, Fang S, Pan J, Lin H, Chen S, Chen Y, Gu G (2004) Screening and evaluation of tobacco germplasm resistant to Ralstonia solanacearum. Acta Tabacaria Sinica 10:22–24 (English abstract)
Xu C, Xia G, Zhi D, Xiang F, Chen H (2003) Integration of maize nuclear and mitochondrial DNA into the wheat genome through somatic hybridization. Plant Sci 165:1001–1008
Xu X, Hu Z, Li J, Liu J, Deng X (2007) Asymmetric somatic hybridization between UV-irradiated Citrus unshiu and C. s inensis: regeneration and characterization of hybrid shoots. Plant Cell Rep 26:1263–1273
Yang XY, Zhang XL, Jin SX, Fu LL, Wang LG (2007) Production and characterization of asymmetric hybrids between upland cotton Coker 201 (Gossypium hirsutum) and wild cotton (G. k lozschianum Anderss). Plant Cell Tissue Organ Cult 89:225–235
Yu Y (2013) Germplasm creation via protoplast fusion between potato and eggplant. Ph.D. dissertation, Huazhong Agricultural University, China
Yu Y, Ye W, He L, Cai X, Liu T, Liu J (2013) Introgression of bacterial wilt resistance from eggplant to potato via protoplast fusion and genome components of the hybrids. Plant Cell Rep 32:1687–1701
Zhi Y, Li H, Zhang H, Gang G (2014) Identification and utility of sequence related amplified polymorphism (SRAP) markers linked to bacterial wilt resistance genes in potato. Afr J Biotechnol 13:1314–1322
Acknowledgments
This research was partially supported by the China Agriculture Research System [CARS-10-P06], The Ministry of Education of the People’s Republic of China (CN) (IRT13065) and Special Fund for Agro-scientific Research in the Public Interest (201303007).
Author contributions
J L, C X and X C designed and supervised the study; T L, Y Y and W T performed the experiments; T L and C X wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11240_2016_958_MOESM1_ESM.doc
The growth habit, flower characteristic of the fusion parents and somatic hybrids a, d, g, j, l, m Plant architecture of the fusion parents and somatic hybrids; b, e, h, k Flower morphology of the fusion parents and somatic hybrids; c, f, i Tuber shape of the potato parent and somatic hybrids. The octoploid and mixoploid did not form flowers and tubers. a, b, c Potato parent 8# (tetraploid); d, e, f Hybrid 10-5 (tetraploid); g, h, i Hybrid 18-3 (tetraploid); j, k Eggplant parent 508.1 (diploid); l Hybrid 24-3 (octoploid); m Hybrid 6-1 (mixoploid) (doc 9605 kb)
Rights and permissions
About this article
Cite this article
Liu, T., Yu, Y., Cai, X. et al. Introgression of bacterial wilt resistance from Solanum melongena to S . t uberosum through asymmetric protoplast fusion. Plant Cell Tiss Organ Cult 125, 433–443 (2016). https://doi.org/10.1007/s11240-016-0958-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-0958-9