Abstract
Asymmetric somatic hybrids were obtained between Gossypium hirsutum Coker 201 and wild cotton G. klozschianum Anderss. An investigation on the effect of ultraviolet (UV) irradiation on donor protoplasts was carried out, and the lethal dose was determined to be 38.7 J cm−2. We firstly screened the putative hybrids by the color of the calli produced, followed by morphological, cytological, and molecular analysis of putative hybrid plants. Most regenerated plants derived from fused protoplasts displayed a recipient-like morphology, while some showed an intermediate phenotype between Coker 201 and G. klozschianum. Chromosome numbers in these somatic hybrids ranged from 54 to 74. The hybrids were verified by random amplified polymorphic DNA (RAPD) and simple sequence repeat (SSR). Absence or co-existence of parents’ genome DNA fragments was identified through molecular analysis. The heredity of cytoplasm was investigated by cleaved amplified polymorphic sequence (CAPS) analysis using mitochondrial and chloroplast universal primer pairs. The results indicated that recombination and rearrangements might have occurred in some regions of mitochondria (mt) and chloroplast (cp) DNA. To our knowledge, this is the first report about asymmetric protoplast fusion in cotton, and the hybrids obtained would be useful for breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main resources of natural fiber, cotton production have attracted more governmental attention as the demands increased. But the unit yield increase has been limited by numerous diseases and pests that cause serious yield reduction. The transfer of polygenic traits such as pathogen resistance is of great importance in cotton (Kumria et al. 2003; Sun et al. 2004, 2006). However, conventional breeding using sexual crossing is often restricted due to sexual incompatibility, especially if the gene of interest is only present in the wild species (Liang 1999). An available way to bypass sexual-crossing barriers is via protoplast fusion through which we can transfer desirable agronomical relevant traits from wild cotton to cultivars. Symmetric protoplast fusion between wild and cultivated species has been achieved successfully in cotton (Sun et al. 2004, 2005, 2006). However, besides desirable traits, some undesirable traits linked to fertility or yield in wild species were also present. This disadvantage might be avoided by asymmetric protoplast fusion, which is also called donor–recipient fusion. In this manner, only a part of the donor genome is transferred to a receptor protoplast (Ramulu et al. 1996), reducing the number of undesirable traits incorporated into the receptor genome.
For transferring partial genome or cytoplasm, the donor protoplast was usually irradiated with X- or gamma-rays (ionizing irradiation) prior to fusion. X- or gamma-rays were the two widely used irradiation methods in most fusion combinations. However ultraviolet (UV) irradiation can be regarded as a substitute or alternative to ionizing irradiation in asymmetric protoplast fusion experiments (Menczel et al. 1982; Hall et al. 1992a, b; Vlahova et al. 1997; Zhou et al. 2005). While both UV and ionizing radiations induce a broad spectrum of physical and chemical modifications in plant and animal DNA, the physiological consequences of UV treatment are much more immediate than those of gamma irradiation. Moreover, the degree of DNA damage observed following UV irradiation was clearly extensive while gamma irradiation at the same biological doses resulted in considerably less DNA damage (Hall et al. 1992b). Although it has been well documented that irradiation leads to a preferential loss of donor DNA (Negrutiu et al. 1989), the genetic components of asymmetric hybrids were very variable. In some fusion combinations most donors DNA is retained (Famelaer et al. 1989; Wolters et al. 1991; McCabe et al. 1993) while in others chromosomes were eliminated more thoroughly leading to highly asymmetric lines (Dudits et al. 1987; Vlahova et al. 1997; Xia et al. 2003).
Putative hybrids analysis is an essential step in confirming hybrid status. Symmetric hybrids carrying both parents genome can be confirmed easily via morphological, biochemical, cytological, and molecular markers (Kovtun et al. 1993; Cabasson et al. 2001; Binsfeld and Schnabl 2002; Zhou et al. 2005). However, asymmetric hybrids were more difficult to identify if only a few chromosomes, chromosome fragments or little DNA contents were transferred from the donor genome to the recipient. Methods have been developed to analyze the inheritance of cytoplasm in asymmetric somatic hybrids. A common technique for studying the inheritance of mitochondria (mt) DNA and chloroplast (cp) DNA in asymmetric somatic hybrids is Southern analysis of organelle or total DNA using mitochondrial and plastidial probes (Cabasson et al. 2001; Kanno et al. 1997). But this method is comparatively expensive and time-consuming, moreover, a considerable amount of DNA is required, and in addition, to our knowledge, mitochondrial and plastidial probes were not available in cotton. Cleaved amplified polymorphic sequence (CAPS) analysis using mitochondrial or chloroplast universal primer pairs have proved to be an efficient and reliable method for characterizing the cytoplasmic genome (Zheng et al. 1999; Cheng et al. 2003).
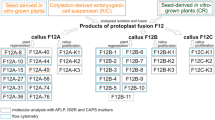
In the present paper, we report the recovery of asymmetric somatic hybrids following fusion of upland cotton protoplasts with UV irradiated wild-type protoplasts and analyze the regenerated plants in order to obtain useful information with respect to their use in breeding programs as a way to overcome sexual barriers and transfer desirable traits from related wild species to the cultivated. The hybrids were firstly screened by distinguishing the color of callus derived from fused protoplasts mixture, and then analyzed at morphological, cytological, and molecular levels.
Materials and methods
Plant materials and protoplast isolation
Embryogenic calli of G. klozschianum Anderss, which have been maintained in our laboratory for more than 3 years, were green in color. The newly induced embryogenic calli of Coker 201 produced according to Wu et al. (2004), were light yellow in color. The establishment of suspension cultures was according to Sun et al. (2004). About 1 g fresh suspensions were mixed with 3 ml filter-sterilized enzyme solution in a 60 mm diameter plate. The enzyme solution was 1.5% (m/v) cellulase Onozuka R-10 (Yakult Inc., Tokyo, Japan), 1% (m/v) macerozyme R-10 (Yakult Inc.), 1.5% (m/v) hemicellulose (Sigma, St. Louis, Mo, USA) dissolved in CPW9M solution [CaCl2 · 2H2O 10 mM, KH2PO4 · 0.2 mM, KNO3 1.0 mM, MgSO4 · 7H2O 1.0 mM, CuSO4 · 5H2O 0.1 μM, KI 10 μM, 2, (N-morpholino) ethane sulfonic acid (MES) 15.37 μM, 9% (m/v) mannitol, and pH 5.8]. The mixture was incubated on a shaker (40 rpm) at 28–30°C in the dark for about 14–16 h, and then the protoplasts–enzyme mixtures were firstly passed through double layered stainless steel sieves (100 and 38.5 μm) and followed by centrifugation in CPW9M at 80×g for 5 min. The protoplasts were resuspended in 1.5 ml CPW9M after removing the supernatant, and then gently added to the top of 3 ml CPW25S (replace 9% mannitol by 25% sucrose in CPW9M) in another tube. The floating protoplasts were collected from the solution interface after centrifugating at 80×g for 7 min, and then resuspended at 106 ml−1 in electro-fusion buffer [10% (w/v) mannitol, 0.25 mM CaCl2] for fusion.
UV irradiation, protoplast fusion and culture
Before protoplast fusion, the effect of UV irradiation on the donor protoplast (G. klozschianum) was investigated. The protoplasts resuspended in electrofusion buffer were put into 6 mm Petri dishes in a thin layer and irradiated with an UV lamp (30 W) at an intensity of 1,290 μW cm−2 for 0, 30, 60, and 90 s, respectively. UV doses under these conditions were 0 (K 0), 38.7 J cm−2 (K 1), 77.4 J cm−2 (K 2), 116.1 J cm−2 (K 3), respectively. After UV irradiation, agarose gel electrophoresis in combination with a fast DNA preparation technique (Hall et al. 1992b) was used to determine the degree of DNA damage caused by UV irradiation treatment. Moreover, the viability and division percentage of the irradiated protoplast and control was assessed after a 20-day culture period. The viability was determined by staining with fluorescein diacetate (FDA) under fluorescent microscope (Leica DM2500, Leica, Bensheim, Germany) (Sun et al. 2004) (Fig. 2a). Viability was assessed as the mean percentage of fluorescent protoplasts from the total protoplasts at the same visual field of microscope of ten visual fields. Cell division and plating efficiency was calculated on the basis of the percentage of protoplasts that initiated division and continued to form cell groups and calli.
For protoplast fusion, equal amounts of donor (K) and recipient protoplasts (C) were mixed and resuspended at a density of 1.0 × 106 ml−1 for fusion. An SSH-2 somatic hybridizer (Shimadzu, Toyota, Japan) was used to mediate protoplast fusion. Approximately 1.6 ml of the mixed protoplasts was pipetted into the FTC-4 fusion chamber. The fusion proceeded as described by Sun et al. (2004). The mixed protoplasts were aligned in an alternate current field of 100 V cm−1 at a frequency of 1 MHz for 60 s, then fusion was facilitated by application of direct current to cause reversible breakdown of the aligned protoplasts for five times at 0.5 ms intervals at a field strength of 1,250 V cm−1 and a duration of 10 s to induce protoplast fusion followed by 20 min resting period in order that the fusion products could regain normal shape. Products were then centrifuged at 80×g for 5 min.
Protoplasts were resuspended and cultured in double-layer culture in KM8P (Kao and Michayluk 1975) medium at the density of 5 × 105 ml−1. About 5 ml melted solid MS medium (Murashige and Skoog 1962) plus B5 (Gamborg et al. 1968) vitamins (MSB) medium were put into the plate, after solidification, 2 ml protoplasts–KM8P medium mixtures at the density of 5 × 105 ml−1 were added to the plate. Parental protoplast cultures (C and K 1) were used as controls. Two plates were used for each treatment and the experiment was repeated five times. All plates were sealed with parafilm and incubated at 28 ± 1°C in darkness. Fifteen days later, 2 ml fresh liquid medium with half amount of mannitol was gradually added to the cultures to accelerate cell division and micro-calli formation. When micro-calli (1–5 mm in size) were formed, all single calli were transferred to solid MSB medium supplemented with 2.460 μM indole-3-butyric acid (IBA), 0.698 μM kinetin, 6.8 mΜ glutamine, 3.8 mM asparagine, 0.25% (w/v) Phytagel, and 3% (w/v) glucose for proliferation. Subculture was made at 15-day intervals for somatic embryo induction. The vigorous embryo were transferred to half strength MS basal medium containing 2% glucose and solidified with 0.27% Phytagel. Regenerated plants were grafted and transferred to the soil as described by our previous report (Jin et al. 2005).
Cytology
Metaphase plates were prepared according to the method of Sun et al. (2004). The proliferated single calli, globular embryos and heart embryos 7–10-day post-subculture and 1-cm long young root tips collected from fusion hybrids and their parents were put into saturated pdichlorobenzene for 3.5–4 h at room temperature and subsequently fixed in Carnoy’s solution of ethanol–acetic acid mixture (3:1) for at least 24 h at 4°C. They were then rinsed in water and hydrolyzed in 5 N HCl for 25 min before transfer to a slide and staining with Carbol Fuchsin solution. The materials were examined and photographed under a light microscope. Approximately 5–10 metaphase plates were analyzed per sample.
Genomic DNA extraction and molecular analysis
Total genomic DNA was isolated from fresh single-cell derived calli and young leafs of hybrids and their parents according to the cetyltrimethyl ammoniumbromide (CTAB) procedure of Paterson et al. (1993). Random amplified polymorphic DNA (RAPD) analysis was according to Sun et al. (2004). Forty-eight 10-mer primers (He et al. 2007) were used for amplification of the template DNA.
Additional verification for somatic hybrid status was provided by simple sequence repeat (SSR) analysis using 104 pairs of primers (He et al. 2007). SSR amplifications were carried out in a PTC-100 (MJ Research Inc., Watertown, MA, USA) thermocycler in 10-μl reaction volumes each containing 25 ng DNA, 0.4 μM primer, 3.6 mM MgCl2, 0.25 mM dNTP, 1× reaction buffer and 0.5 U Taq polymerase (MBI. Jingmei Biotech Co. Ltd., Shenzhen, China). Amplification was programmed for a predenaturation of 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, 55°C for 45 s, 72°C for 60 s, and a final extension of 72°C for 10 min. About 2 μl Amplification products were surveyed by polyacrylamide gel electrophoresis (PAGE)/silver staining as described by Zhang et al. (2003).
The organelle DNA amplification of the regenerants and their parents was performed using three mitochondria (nad4 exon 1: 5′-CAGTGGGTTGGTCTGGTATG-3′/nad4 exon 2: 5′-TCATATGGGCTACTGAGGAG-3′, nad1 exonB: 5′-GCATTACGATCTGCAGCTCA-3′/nad1 exonC: 5′-GGAGCTCGATTAGTTTCTGC-3′, 18S rRNA: 5′-GTGTTGCTGAGACATGCGCC-3′/5S rRNA: 5′-ATATGGCGCAAGACGATTCC-3′) and one chloroplast (TrnK 5′-AACCCGGAACTAGTCGGATG-3′/TrnK 5′-TCAAT-GGTAGAGTACTCGGC-3′) universal primer pairs (Cheng et al. 2003) in a PTC-100 thermocycler. The PCR reaction mixture (50 μl) contained 1× reaction buffer, 3.6 mM MgCl2, 0.2 mM dNTP, 1.2 U Taq DNA polymerase, 0.2 μM of each primer pairs and 100 ng of genome DNA. The amplification parameters were the same as SSR. About 8 μl PCR products was digested with 5 U of restriction endonucleases (TaqI, EcoRI, HindIII, and HaeI, respectively), in a 0.5 ml volume at 37°C for 4 h. The digested DNA samples were electrophoresed on a 2.0% agarose gel with 0.5× TBE and 0.5 μg ml−1 ethidium bromide at 2 V cm−1 for 2 h, and then the samples were photographed under UV light.
Results
Effect of UV irradiation on protoplast culture
We conducted four doses (0, 38.7, 77.4, and 116.1 J cm−2) of UV irradiation on G. klozschianum protoplasts. The results of agarose gel electrophoresis indicated that considerable fragmentations of the DNA happened when irradiated with the dose of 77.4 and 116.1 J cm−2, but less fragmentation with the dose of 38.7 J cm−2 compared with control (Fig. 1). The viability and division percentage detected during the 7-day culture period are shown in Table 1. The viability and first division percentage of the irradiated protoplasts decreased along with the increase of irradiation dose, and the viability of irradiated protoplasts decreased along with the increase of culture time, while the protoplasts without irradiation showed no signification difference during the 5-day culture period. The first division time of protoplasts without irradiation took place at day 3, but 4 days later for the protoplasts irradiated with the dose of 38.7 J cm−2 (Table 2). The protoplast irradiated with the dose of 77.4 J cm−2 could divide at low percentage and displayed a normal shape at early period of culture, but gradually broke after 10 days. The protoplasts irradiated with the dose of 116.1 J cm−2 broke up along with increasing of culture time. Generally, the protoplasts irradiated with the dose of 77.4 J cm−2 stopped growth after the first division, but a few cell-groups could be observed from the protoplasts irradiated with a dose of 38.7 J cm−2. None of the protoplasts treated with UV could form mass callus although plantlets developed from untreated protoplasts. As a result, we choose 38.7 J cm−2 UV treatment as the lethal dose for the following asymmetric cell fusion experiments.
Protoplast culture and plant regeneration of fused protoplasts via somatic embryogenesis. (a) Protoplast viability as monitored by FDA. (b) Calli formation from protoplast of Coker 201. (c) Calli formation from fused protoplast. (d) Single calli proliferation after transfer to solid medium individually. (e) Fascicular plants regenerated from abnormal embryo. (f) Vigorous stem and bigger stipules of hybrid plant. (g) Normal plant development after grafting. (h) Coker 201. (i) G. klotzschianum. (j–k) Asymmetric somatic hybrids
Morphology of the regenerated plants
The first post-fusion cell division took place after about 5 days, later than Coker 201 but earlier than protoplasts irradiated with the dose of 38.7 J cm−2 (Table 2), and this was followed by the formation of small calli visible to the naked eye in about 35–40 days (Fig. 2c; Table 2). In total, 47 single green calli were selected from the products (Fig. 2d; Table 2). The selected calli grew vigorously and were able to differentiate rapidly. After 2–4 months, most of the selected calli developed green plantlets. Most regenerants displayed a recipient-like morphology (Fig. 2k), whereas some plantlets were intermediate to the two parents, in which their first three or four leafs showed full round edge, and the upper leaves had three nicks. The stipule of one hybrid was bigger than both parents, and the stem was thicker (Fig. 2f), showing stronger growth to the parents. And some fascicular plants were regenerated from abnormal embryos (Fig. 2e). When the hybrids were grafted and transferred to a greenhouse, their growth was less vigorous than the two parents, and some nearly stopped growing (Fig. 2j), which resulted in a smaller size of the somatic hybrids. These regenerated plants were considered for further analysis.
True hybrid confirmation by chromosome counting and molecular tools
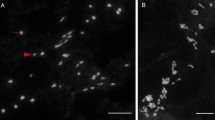
Several hybrids were examined cytologically. Among the cultures derived from the fused protoplasts, the mini calli, globular embryos, heart-shaped embryos and roots of regenerated plants were picked out for chromosome counting. The chromosome numbers ranged from 2n = 54 to 74 (Fig. 3), showing a complex chromosome variation in number.
To determine the origin of the putative hybrid, we conducted RAPD and SSR analysis. The eight morphologically and cytologically determined hybrids were analyzed. Among the 48 RAPD primers used, eight (S68, S118, S124, S127, S176, S353, S429, and S439) showed polymorphisms between the two parents. S127 (5′-CCGATATCCC-3′) and S353 (5′-CCACACTACC-3′) clearly detected polymorphisms between the hybrids and their two parents. The presence of one or more distinct parental or novel bands in the individual hybrids confirmed their hybrid status. Primer S127 generated multiple banding profiles in hybrid plants, two specific bands of Coker 201 (∼650 and 750 bp) and three specific bands (∼600, 1,000, and 1,300 bp) of G. klozschianum (Fig. 4a). Four hybrids (Lanes 1, 4, 5, 6) showed full Coker 201 bands and parts of G. klozschianum bands. Rearrangements might happen because a novel band appeared in one hybrid (Lane 3) compared to the two parents. One hybrid (Lane 2) had only one band of Coker 201, meaning the loss of recipient DNA fragments might occur. Primer S353 produced two specific bands of Coker 201 (550 and 650 bp) and one specific band (800 bp) of G. klozschianum (Fig. 4b). It was easy to distinguish the asymmetric hybrids from parents.
Among the 104 SSR primer pairs used, 46 primer pairs showed polymorphisms between the two parents. The SSR patterns of two primer pairs BNL3030 (f: 5′-TTGCCCAACACTTCATCAAA-3′, r: 5′-CGTAGAAAGAGACCCAACGG-3′), BNL3232 (f: 5′-AGCTCACCAACCCCATATTG-3′, r: 5′-TCTATTGTATGTTATTGCTGCCC-3′) were distinctly different between the hybrids and their parents (Fig. 5). For the eight putative hybrids analyzed, the variation or absence of G. klozschianum specific bands in asymmetric hybrids confirmed the hybridity of the putative hybrid plantlets.
SSR analysis of somatic hybrids and their parents. (a) SSR band patterns of the parental species and regenerated plantlet by the primer pair BNL3232 (f: 5′-AGCTCACCAACCCCATATTG-3′, r: 5′-TCTATTGTATGTTATTGCTGCCC-3′). (b) SSR band patterns of the parental species and regenerated plantlet by the primer pair BNL3030 (f: 5′-TTGCCCAACACTTCATCAAA-3′, r: 5′-CGTAGAAAGAGACCCAACGG-3′). Lane C, Coker 201; k, G. klotzschianum; 1–8, Asymmetric somatic hybrids
In order to confirm the cytoplasmic genome fusion, CAPS analysis was employed to determine the cytoplasmic constitutions of the somatic hybrids. When the PCR products derived from amplification by universal primer pairs were digested with the restriction endonucleases, some polymorphic loci were found between the fusion parents. The results showed that cpDNA primer pair/enzyme combinations of trnK–trnK/EcoRI and trnK–trnK/TaqI were effective in distinguishing both parents (Fig. 6). The cpDNA of three asymmetric somatic hybrids plant were from Coker 201 (Fig. 6; Lanes 1, 2, 4), and one asymmetric somatic hybrids displayed the specific bands of both parents (Fig. 6a; Lane 5). For mtDNA, primer pair/enzyme combination of nad4exon1-nad4exon2/EcoRI was able to distinguish both parents of Coker 201 and G. klotzschianum, and a novel band was detected in the mtDNA of the somatic hybrid (Fig. 7; Lane 1), indicating that rearrangements or recombination might have occurred in some regions of the mtDNA in the somatic hybrid. The profiles of most hybrids were the some as Coker 201.
Discussion
The effect of UV irradiation as a pretreatment for the donor protoplast in asymmetric protoplast fusion has been assessed and some detailed results presented. A correlation between irradiation dose and the rate of DNA loss was observed in our study, and similar results have been reported by Hall et al. (1992a, b) and Melzer and O’Connell (1992). However, DNA loss in asymmetric somatic hybrids is a complex process. The exact mechanisms underlying chromosome elimination are unknown. Factors such as culture conditions and genetic distance may play a significant role in chromosome elimination (Oberwalder et al. 1998). Several alternatives for directing chromosome elimination, such as the use of cytoplast–protoplast (Dudits et al. 1987) and micro-protoplasts (Ramulu et al. 1993) have also been proposed. It is clear that further investigations on the determinants responsible for nuclear elimination in somatic cells are needed to fully exploit asymmetric protoplast fusion technology to produce morphologically normal and fertile hybrids for breeding program.
The early selection of regenerated hybrids in asymmetric protoplast fusion is complex and necessary. There are many cell groups with different genetic backgrounds in the post-fusion mixture, such as fused cells, unfused cells, and poly fused cells. Nutrition-sensitive, chlorophyll-absence or resistance genes combined with molecular selection has proved to be a means for hybrid verification. Some scientists utilize the regenerative capability of hybrids to select (Xia and Chen 1996). Marker genes, such as green fluorescent protein (GFP) were used for hybrids selection in some cases (Masako et al. 2001), while others use morphological markers (Derks et al. 1992). In this paper, we used light yellow calli as recipient and green calli as donor, and select green calli post-fusion as putative asymmetric hybrids, molecular analyses indicated that almost all putative hybrids were green in color.
This seems to be the first report about analysis of cytoplasm inheritance in somatic hybrids of cotton. We selected some mitochondrial and chloroplast universal primer pairs used in other plants to analyze cytoplasm of cotton asymmetric hybrids. The result showed that chloroplast might be recombinant in somatic hybrids (Fig. 6; Lane 5). A strange band profile was present in one hybrid (Fig. 6; Lane 6), the profile of one fragment amplification by trnK–trnK showed different origins fragmented by two restriction endonucleases. However, in most cases, the regenerated hybrid plants have only one chloroplast type (Fig. 6; Lanes 1, 2, 4). In contrast to cpDNA, when the PCR products amplified by universal primer pairs were digested with the restriction endonucleases, fewer polymorphic loci were found in mtDNA between the fusion parents. The PCR products amplified by three mtDNA primer pairs were digested by four restriction endonucleases (TaqI, EcoRI, HindIII, and HaeI), respectively, only one polymorphic locus was found in the combination of nad 4 exon 1–2/EcoRI. Nevertheless, as shown in Fig. 7, a novel band was detected in the mtDNA of one somatic hybrid, indicating that rearrangements or recombination might have occurred in some regions of the mtDNA in the somatic hybrid.
Asymmetric somatic hybrids were obtained using UV irradiation of the donor protoplasts prior to fusion in cotton. To our knowledge, this is the first report about production of asymmetric somatic hybrids in cotton, and this progress might be a useful tool for producing novel germplasm for breeding programs. Protoplast culture in upland cotton and wild species and symmetric protoplast fusion in cotton had been achieved successfully and reported in detail in our laboratory (Sun et al. 2004, 2005, 2006). Until now, we have done experiments on protoplast culture and protoplast fusion, but the advantages or limitations of asymmetric and symmetric somatic hybrids in breeding programs have not yet been systematically investigated and require more information for further analysis. Asymmetric hybrids as well as symmetric hybrids can be used in different breeding programs. Symmetric protoplast fusion appeared to produce a wide range of variability in genomic controlled traits, while the aim of asymmetric protoplast fusion was to minimize the disadvantages of donor genome as well as transfer desired traits, chromosomes, or chromosome fragments (Binsfeld and Schnabl 2002). Unfortunately, in most cases the hybrids gained much more donor DNA than desired, resulting in an aberrant morphology and partially sterilility (Kovtun et al. 1993; McCabe et al. 1993), although a stable genomic transfer through asymmetric protoplast fusion has also been reported (Binsfeld et al. 2000). Furthermore, asymmetric protoplast fusion has been successfully used for transfer of one or few chromosomes, the production of specific addition lines and the introgression of genes between sexually incompatible species (Ramulu et al. 1996; Binsfeld et al. 2000; Wardrop et al. 2004; Chen et al. 2004). Highly asymmetric fertile hybrids have only occasionally been described (Vlahova et al. 1997; Xia et al. 2003). In our present study, no highly asymmetric hybrids were obtained though the molecular analysis indicated that some might be asymmetric hybrids, but with more than 52 chromosomes. We also conducted asymmetric protoplast fusion with iodoacetamide (IOA) lethal dose pretreated Coker 201 and UV lethal dose irradiated G. klotzschianum and observed cell groups, but no calli were generated. For further improving the fusion process for asymmetric protoplast fusion, many factors should be concerned, such as culture density, fusion parameters, and other conditions.
Abbreviations
- CAPS:
-
Cleaved amplified polymorphic sequence
- cpDNA:
-
Chloroplast DNA
- CPW:
-
Cell and protoplast washing solution (Frearson et al. 1973)
- CTAB:
-
Cetyltrimethyl ammoniumbromide
- FDA:
-
Fluorescein diacetate
- GFP:
-
Green fluorescent protein
- IBA:
-
Indole-3-butyric acid
- IOA:
-
Iodoacetamide
- MES:
-
2, (N-morpholino) ethane sulfonic acid
- MSB:
-
MS medium (Murashige and Skoog 1962) and B5 (Gamborg et al. 1968) vitamins
- mtDNA:
-
Mitochondrial DNA
- PAGE:
-
Polyacrylamide gel electrophoresis
- RAPD:
-
Random amplified polymorphic DNA
- SSR:
-
Simple sequence repeat
- UV:
-
Ultraviolet
References
Binsfeld PC, Wingender R, Schnabl H (2000) Characterization and molecular analysis of transgenic plants obtained by microprotoplast fusion in sunflower. Theor Appl Genet 101:1250–1258
Binsfeld PC, Schnabl H (2002) Molecular and cytogenetic constitution of plants obtained via two different somatic hybridization methods. Plant Cell Rep 21:58–62
Cabasson CM, Luro F, Ollitrault P, Grosser W (2001) Non-random inheritance of mitochondrial genomes in Citrus hybrids produced by protoplast fusion. Plant Cell Rep 20:604–609
Chen JF, Luo XD, Qian CT, Jahn MM, Staub JE, Zhuang FY, Lou QF, Ren G (2004) Cucumis monosomic alien addition lines: morphological, cytological and genotypic analyses. Theor Appl Genet 108:1343–1348
Cheng YJ, Guo WW, Deng XX (2003) Molecular characterization of cytoplasmic and nuclear genomes in phenotypically abnormal Valencia orange (Citrus sinensis) + Meiwa kumquat (Fortunella crassifolia) intergeneric somatic hybrids. Plant Cell Rep 21:445–451
Derks FHM, Hakkert JC, Verbeek WHJ, colijn-Hooymans CM (1992) Genome composition of asymmetric hybrids in relation to the phylogenetic distance between the parents: nucleus-chloroplast interaction. Theor Appl Genet 84:930–940
Dudits D, Maroy E, Praznovszky T, Olah Z, Gyorgyey J, Cella R (1987) Transfer of resistance traits from carrot into tobacco by asymmetric somatic hybridization: regeneration of fertile plants. Proc Natl Acad Sci USA 84:8434–8438
Famelaer I, Gleba YY, Sidorov VA, Kaleda VA, Parokonny AS, Boryshuk NV, Cherep NN, Negrutiu I, Jacobs M (1989) Intrageneric asymmetric hybrids between Nicotiana plumbaginifolia and Nicotiana sylvestris obtained by “gamma-fusion”. Plant Sci 61:105–117
Frearson EM, Power JB, Cocking EC (1973) The isolation, culture and regeneration of petunia leaf protoplasts. Dev Biol 33:130–137
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension culture of soybean roots cells. Exp Cell Res 50:150–158
Hall RD, Rouwendal GJA, Krens FA (1992a) Asymmetric somatic cell hybridization in plants. I. The early effects of (sub) lethal doses of UV and gamma radiation on the cell physiology and DNA integrity of cultured sugarbeet (Beta vulgaris L.) protoplasts. Mol Gen Genet 234:306–314
Hall RD, Rouwendal GJA, Krens FA (1992b) Asymmetric somatic cell hybridization in plants. II. Electrophoretic analysis of radiation-induced DNA damage and repair following exposure of sugarbeet (Beta vulgaris L.) protoplasts to UV and gamma rays. Mol Gen Genet 234:315–324
He DH, Lin ZX, Zhang XL, Nie YC, Guo XP, Zhang YX (2007) QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum × G. barbadense. Euphytica 153:181–197
Jin SX, Zhang XL, Liang SG, Nie YC, Guo XP, Huang C (2005) Factors affecting transformation efficiency of embryogenic callus of upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 81:229–237
Kao KN, Michayluk MR (1975) Nutrient requirements for growth of Vicia hajastana cells and protoplasts at a very low population density in liquid media. Planta 126:105–110
Kanno A, Kanzaki H, Kameya T (1997) Detailed analyses of chloroplast and mitochondrial DNAs from the hybrid plant generated by asymmetric protoplast fusion between radish and cabbage. Plant Cell Rep 16:479–484
Kovtun YV, Korostash MA, Butsko YV, Gleba YY (1993) Amplification of repetitive DNA from Nicotiana plumbaginifolia in asymmetric hybrids between Nicotiana sylvestris and Nicotiana plumbaginifolia. Theor Appl Genet 86:221–228
Kumria R, Sunnichan VG, Das DK, Gupta SK, Reddy VS, Bhatnagar RK, Leelavathi S (2003) High-frequency somatic embryo production and maturation into normal plants in cotton (Gossypium hirsutum) through metabolic stress. Plant Cell Rep 21:635–639
Liang ZL (1999) Genetics and breeding of distant hybridization in cotton, 2nd edn. Science press, Beijing, pp 1–5
Masako T, Yousuke T, Kuniya A, Norio N, Takashi T (2001) Supplementary material nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol 2:1553–1558
McCabe PF, Dunbar LJ, Guri A, Sink KC (1993) T-DNA-tagged chromosome 12 in donor Lycopersicon esculentum x L. Pennellii is retained in asymmetric somatic hybrids with recipient Solanum lycopersicoides. Theor Appl Genet 86:377–382
Melzer JM, O’Connell MA (1992) Effect of radiation dose on the production of, and the extent of, asymmetry in tomato asymmetric somatic hybrids. Theor Appl Genet 83:337–344
Menczel L, Galiba G, Nagy F, Maliga P (1982) Effect of radiation dosage on efficiency of chloroplast transfer by protoplast fusion in Nicotiana. Genchc 100:457–495
Murashige T, Skoog F (1962) A revised medium for rapid grouth and bio assay with tobacco tissue cultures. Physiol Plant 15:473–479
Negrutiu I, Hinnisdaels S, Mouras A, Gill BS, Gharti-Chhetri GC, Davey MR, Gleba YY, Sidorov V, Jacobs M (1989) Somatic versus sexual hybridization: features, facts and future. Acta Bot Neerl 38:253–272
Oberwalder B, Schilde-Rentschler L, Ruoû B, Wittemann S, Ninnemann H (1998) Asymmetric protoplast fusions between wild species and breeding lines of potato—effect of recipients and genome stability. Theor Appl Genet 97:1347–1354
Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP and PCR analysis. Plant Mol Biol Rep 11:112–127
Ramulu KS, Dijkhuis P, Famelaer I, Cardi T, Verhoeven HA (1993) Isolation of sub-diploid microprotoplasts for partial genome transfer in plants—enhancement of micronucleation and enrichment of microprotoplasts with one or a few chromosomes. Planta 190:190–198
Ramulu KS, Dijkhuis P, Rutgers E, Blass J, Krens FA, Verbeek WHJ, Colijn-Hooymans CM, Verhoeven HA (1996) Intergeneric transfer of a partial genome and direct production of monosomic addition plants by microprotoplast fusion. Theor Appl Genet 92:316–325
Sun YQ, Zhang XL, Nie YC, Guo XP, Jin SX, Liang SG (2004) Production and characterization of somatic hybrids between upland cotton (Gossypium hirsutum) and wild cotton (G. klotzschianum Anderss) via electrofusion. Theor Appl Genet 109:472–479
Sun YQ, Zhang XL, Nie YC, Guo XP (2005) Production of fertile somatic hybrids of Gossypium hirsutum + G. bickii and G. hirsutum + G. stockii via protoplast fusion. Plant Cell Tissue Organ Cult 83:303–310
Sun YQ, Nie YC, Guo XP, Huang C, Zhang XL (2006) Somatic hybrids between Gossypium hirsutum L. (4×) and G. davidsonii Kellog (2×) produced by protoplast fusion. Euphytica 151:393–400
Vlahova M, Hinnisdaels S, Frulleux F, Claeys M, Atanassov A, Jacobs M (1997) UV irradiation as a tool for obtaining asymmetric somatic hybrids between Nicotiana plumbaginifolia and Lycopersicon esculentum. Theor Appl Genet 94:184–191
Wardrop J, Fuller J, Powell W, Machray GC (2004) Exploiting plant somatic radiation hybrids for physical mapping of expressed sequence tags. Theor Appl Genet 108:343–348
Wolters AMA, Schoenmakers HCH, van der Meulen-Muisers JJM, van der Knaap E, Derks FHM, Koorneef M, Zelcer A (1991) Limited DNA elimination from the irradiated potato parent in fusion products of albino Lycopersicon esculentum and Solanum tuberosum. Theor Appl Genet 83:225–232
Wu JH, Zhang XL, Nie YC, Jin SX, Liang SG (2004) Factors affecting somatic embryogenesis and plant regeneration from a range of recalcitrant genotypes of Chinese Cottons (Gossypium hirsutum L.). In Vitro Cell Dev Biol Plant 40:371–375
Xia GM, Chen HM (1996) Plant regeneration from intergeneric somatic hybridization between Triticum aestivum L and Leymus chinesis (Trin) Tzvel. Plant Sci 120:197–203
Xia GM, Xiang FN, Zhou AF, Wang H, Chen HM (2003) Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Neviski. Theor Appl Genet 107:299–305
Zhang TZ, Yuan YL, Yu J, Guo WZ, Kohel RJ (2003) Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theor Appl Genet 106:262–268
Zheng XY, Wolff DW, Baudracco-Arnas S, Pitrat M (1999) Development and utility of cleaved amplified polymorphic sequences (CAPS) and restriction fragment length polymorphisms (RFLPs) linked to the Fom-2 fusarium wilt resistance gene in melon (Cucumis melo L.). Theor Appl Genet 99:453–463
Zhou CE, Xia GM, Zhi DY, Chen Y (2005) Genetic characterization of asymmetric somatic hybrids between Bupleurum scorzonerifolium Willd and Triticum aestivum L.: potential application to the study of the wheat genome. Planta 223:714–724
Acknowledgments
The author X. Y. Yang thanks Y. X. Zhang and L. L. Tu for technical assistance with the molecular analysis. This research was supported by program for New Century Excellent Talents in University (NCET-04-0739) and National Basic Research program of China (2004CB117301).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, Xy., Zhang, Xl., Jin, Sx. et al. Production and characterization of asymmetric hybrids between upland cotton Coker 201 (Gossypium hirsutum) and wild cotton (G. klozschianum Anderss). Plant Cell Tiss Organ Cult 89, 225–235 (2007). https://doi.org/10.1007/s11240-007-9245-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-007-9245-0