Abstract
Brain tumors pose a significant health concern globally, with their detection and diagnosis being crucial for timely intervention and treatment planning. These abnormal growths can develop within the brain or originate from other parts of the body, spreading to the brain. They can be benign or malignant, and their impact on cognitive and physical function can vary widely depending on their location, size, and type. Detecting brain tumors involves a combination of imaging techniques and clinical assessment. Magnetic Resonance Imaging (MRI) is one of the primary imaging modalities used for this purpose due to its ability to provide detailed images of the brain's anatomy and pathology. Advanced image processing techniques and machine learning algorithms play an increasingly important role in enhancing the accuracy and efficiency of brain tumor detection. This study presents a comprehensive approach for enhancing brain tumor detection in MRI images using a combination of advanced techniques. Firstly, an improved K-means clustering algorithm is introduced to segment tumor regions effectively. Following this, Median Filtering is applied to refine image quality and reduce noise interference. Subsequently, a deep neural network architecture is employed for precise tumor classification utilizing the BRATS dataset. The proposed methodology aims to improve the accuracy and efficiency of brain tumor detection, offering potential advancements in medical image analysis and diagnosis. Experimental results demonstrate promising outcomes, indicating the effectiveness of the proposed approach in accurately identifying tumor regions in MRI scans.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The brain is the most complex part of the Central Nervous System (CNS). The doctors' First task is to find the tumors, which would be a time-consuming process that they are overburdened with. A brain tumor is a solid intracranial neoplasm. The only optimal approach for this problem is to use image segmentation [1]. The location and size of the tumor determine the symptoms. The right place and grade level are detected with improved accuracy using a deep learning approach. Gliomas make up 80% of all malignant tumors in the glial cells of the central nervous system. The World Health Organization divides gliomas into three categories: high (Grade 4), low (Grade 1), and intermediate (Grade 2 and 3) [2].
MRI is currently used to provide a medical examination, and doctors can use the data to predict the position of the tumor [3]. However, because an MRI scan may contain distortion, edge contour, and produce an image that is too imprecise for doctors to detect efficiently, the accuracy is just 80.6 percent. A biopsy is then conducted on the area discovered by the specialists to establish the tumor's grade. Histopathological classification, which would be predicated on the stereotactic biopsy technique, is the gold standard for identifying tumor grade [4]. The neurosurgeon drills a tiny hole in the skull and collects tissue with specialized equipment. Biopsy tests include a multitude of hazards, including malignancy and brain bleeding. This can cause a serious stroke, coma, or even death. Further dangers include infection and convulsions. Patients die when the location is misled by making a hole indicated using MRI (which has an accuracy of 85 percent). The convolutional networks technique is used with the tensor-flow backend to find the correct position.
Figure 1 shows the MRI scans which represent the tumor region. The MRI images project the shape of the tumor without segmentation. Figure 1a illustrates the presence of the tumor region with an undetermined shape. Figure 1b represents the presence of the tumor with a small size and less intensity. Figure 1c represents the presence of the tumor, the intensity of the tumor and the size of the tumor. Figure 1d represents the tumor's presence and size, and Fig. 1d represents the partial presence of the tumor with less intensity and small size. An incorrect mitotic mechanism affects the function of morphological interstitial cells in the brain. Throughout the operation, cancerous cells were reconstructed with morphological qualities, including size, shape, and intensity. Low-grade and high-grade brain tumors are the two types. The bulk of cancerous cells shows low contrast when compared to adjacent cells. As a result, precisely diagnosing a brain tumor is a difficult task.
The most often used MRI modality for brain tumor segmentation is a painless procedure that allows for tumor investigation from a variety of angles. As a result, MRI image analysis is the most effective method for detecting brain tumors, though it is costlier and has a higher accuracy rate. The existing systems are all about the classification of brain tumors by using conventional algorithms like Support Vector Machine (SVM) [5], Random Forest [6], K-means algorithm [7], Fuzzy algorithm [8], K-Nearest Neighbor algorithm [9] and Convolution Neural Network [10]. These learning approaches, particularly CNN, have proven reliable and are widely used for medical image analysis. Moreover, they have drawbacks over conventional methods, including high temporal complexity, inefficiency in application with a small dataset, and the need for powerful GPUs, all of which increase user expenses. Choosing the appropriate deep learning tools is challenging because of the various segmentation factors, training techniques, and topology complexity.
In this paper, we introduce a pioneering approach to brain tumor segmentation and classification leveraging deep neural networks on MRI-FLAIR images. Our model represents a significant advancement in the field, offering a comprehensive solution that addresses the challenges of accurate tumor delineation and subtype classification. By harnessing the power of deep learning, our proposed method achieves superior segmentation accuracy and enables precise classification of tumor subtypes, thus facilitating personalized treatment strategies. Unlike existing techniques, our model integrates multi-scale features and contextual information, resulting in robust performance across diverse datasets. Additionally, we introduce a novel modified K-means algorithm that enhances the model's ability to focus on relevant tumor regions, further improving segmentation accuracy. Through extensive experiments and comparative analysis, we demonstrate the efficacy and innovation of our approach, establishing new benchmarks in brain tumor segmentation and classification research.
The significant contributions of the proposed approach are as follows:
-
1.
A feature-based constraint approach for detecting brain tumors using the deep neural network is proposed.
-
2.
A deep feature extraction model is developed using a median filtering approach and the improved K-means algorithm.
-
3.
An efficient segmentation method using 2D images from the BRATS dataset using FLAIR modalities is developed.
The remaining sections of the research paper are as follows: Sect. 2 describes the literature survey explored for the research problem. Section 3 defines the proposed research methodology, including the improved k-means algorithm and DNN model. Section 4 provides the details about implementation tools, datasets incurred, parameter tuning, and the experimental results. Finally, Sect. 5 illustrates the conclusion and future work of the proposed research.
2 Literature review
Nowadays, tumors are the main cause of death in many people. Tumors are a group of unwanted cells formed together and cause cancer. Tumors are primarily seen in organs like the brain, lungs, skin, blood, etc. Among all those tumors, the brain tumor is the most dangerous and can also lead to death in its early stages. The tumors are detected by using MRI scans and CT scans. MRI scans are a little bit expensive but produce more accurate results when compared to CT scans.
Many studies have been conducted to understand better how to spot brain tumors, and some of the most recent discoveries are discussed here. Medical image analysis uses a fuzzy rough set with statistical characteristics [11]. To detect breast anomalies, researchers have turned to a probabilistic fuzzy-c-mean method that uses textural characteristics [12]. In addition, U-network [13] is also used for meningioma categorization. For deep feature extraction on skin cancer, brain tumor, breast cancer, and colon cancer individuals, the fine-tuned ResNet-50 model achieves 95% classification accuracy [14]. For an accurate diagnosis, image fusion is essential. A sparsely convolutional decomposition model is utilized to combine MRI and CT slices. Edges are identified by contrast enhancement and the steepest descent spatial approach. A dictionary approach is built for brain image segmentation through texture decomposition and enhanced convolutional coding. Their results are measured against six high-quality benchmark datasets [15].
For the purpose of classifying brain tumors, a capsule neural model was developed with an impressive 86.56% accuracy [16]. Pretrained models have been used for tumor classification, with ResNet-50 achieving competitive results compared to inceptionv3 and VGG-16 [17]. Researchers have created a hybrid model for tumor cell categorization that combines the VGG-net, ResNet, and LSTM models, with an accuracy of 71% on Alexnet and 84% accuracy on the ResNet model [18]. In order to classify tumors more precisely, researchers have suggested a novel model based on discrete wavelet transform (DWT) [19]. To improve the image quality, a histogram equalization technique was used. The principal component analysis is used to identify informative features, which are then fed into the feed-forward network, which can classify MRI images as normal or suspicious with a 96% success rate [20]. Using the SVM model, the classification of tumors achieved 95% accuracy, 95% sensitivity and 95% specificity on the BRATS dataset [21].
Tumor categorization in [22] utilized three machine learning models. In terms of accuracy, the model scored an impressive 88% accuracy. To classify cancers, capsule networks are based on a modified version of a conventional CNN model (CapsNet). The proposed CapsNet uses the spatial interaction between the tumor and its surrounding tissues [23]. The proposed CapsNet achieves 90% accuracy, an improvement over SVM's 88% accuracy. Tumor classification for multiclass brain image classification is realized using an adversarial generative model [24]. The proposed paradigm has a whopping six levels of complexity. This was paired with other methods for improving data quality. Both defined and random splits achieved an accuracy of 96.25%.
The graph convolutional neural network has been selected for tumor classification by many authors [25]. There have been several proposed methods for categorizing brain tumors; however, these methodologies suffer from various limitations, some of which have been discussed in [26,27,28]. Until recently, several tumor classification methods required human intervention to define tumor areas [29,30,31,32]. Algorithms based on Convolutional Neural Networks (CNNs) and their offshoots could not significantly improve processing time. Therefore, non-accuracy-based performance measures gain significance. In addition, CNN models struggle when presented with sparse data.
Nassar et al. [33] focused on developing a robust MRI-based brain tumor classification system using a hybrid deep learning technique. The proposed approach leverages a majority voting strategy based on the predictions of five fine-tuned pre-trained models (GoogleNet, AlexNet, ShuffleNet, SqueezeNet, and NASNet-Mobile) to classify three types of brain tumors (meningioma, glioma, and pituitary) with high accuracy and reliability. Utilizing a public brain tumor image dataset and minimal pre-processing steps during training and testing phases, the method demonstrates improved performance metrics such as accuracy, recall, precision, specificity, and F1-score compared to existing techniques. The study showcases the effectiveness of ensemble learning and the potential of deep learning models in enhancing the efficiency of brain tumor classification, offering valuable support to radiologists in making accurate diagnostic decisions.
The research in [34] presented a novel approach for multigrade brain tumor classification in MRI images using a fine-tuned EfficientNet model. The research introduces a fine-tuned lightweight EfficientNet V2S model, showcasing state-of-the-art performance in classifying a diverse range of brain tumor classes across three datasets. The model achieved high accuracy rates ranging from 97 to 98% for all datasets through extensive performance comparisons and multigrade classification analysis. The study emphasizes the importance of explainable models using GradCam visualization to provide insights into the classification process. Overall, the research highlights the potential of the Fine-tuned EfficientNet model in accurately and efficiently classifying brain tumors in MRI images, offering valuable implications for medical imaging and diagnosis.
The proposed work in [35] focuses on utilizing deep learning methods. Specifically, CNNs and transfer learning are used to classify brain tumors in MRI images. The study compares the performance of different models, including InceptionV3, EfficientNetB4, VGG16, VGG19, and a Multi-Layer CNN, in classifying brain tumors. The VGG16 model achieved the best accuracy result (97%). The study emphasizes the importance of AI-based applications, especially in image processing, for supporting health decision-makers in early diagnosis and treatment processes. Overall, the study demonstrates the effectiveness of deep learning models, particularly VGG16, in accurately classifying brain tumors from medical images, highlighting the potential of AI in improving healthcare decision-making processes.
Kumar et al. [36] developed a multi-classification system for accurately classifying brain tumors in medical images using a combination of deep learning techniques and fuzzy-based logic algorithms. The system integrates transfer learning with texture-based classification to enhance accuracy in tumor classification. By leveraging pre-trained models and advanced neural networks, the study achieved a significant improvement in accuracy compared to existing methods, with a 2% increase in accuracy over an improved ResNet-50 model. The research highlights the potential of utilizing advanced techniques in medical imaging for solving complex problems and lays a foundation for future research in the field.
The proposed work in the study [37] focuses on enhancing the accuracy of multi-class brain tumor classification using deep transfer learning models. Five popular deep learning architectures, including Xception, DenseNet201, DenseNet121, ResNet152V2, and InceptionResNetV2, were employed and modified with a deep dense block and softmax layer to improve classification accuracy. Performance metrics such as accuracy, sensitivity, precision, specificity, and F1-score were used to evaluate the models. The results showed that the proposed Xception + DDB model achieved the highest classification accuracy of 95.87% on a 4-class brain MRI dataset, outperforming the baseline models and demonstrating the effectiveness of the deep transfer learning approach in enhancing brain tumor classification accuracy.
Rajput et al. [38] focused on utilizing transfer learning with pre-trained Convolutional Neural Network (CNN) models for the classification of brain tumors in MRI images. By leveraging pre-trained Inception-V3 and DenseNet201 models for feature extraction and classification, the study achieved a high classification accuracy on a brain tumor MRI dataset containing glioma, meningioma, pituitary tumors, and non-tumor cases. The use of transfer learning not only improved the efficiency of the model but also outperformed other deep learning architectures such as ResNet-50, Inception-V3, and VGG19. Through rigorous evaluation metrics and comparative analysis, the proposed framework demonstrated superior performance in accurately diagnosing brain tumors, showcasing the potential of transfer learning with pre-trained CNN models in enhancing medical image analysis for improved patient outcomes.
The proposed work [39] utilizes a CNN-based approach for automatic brain tumor detection, employing the AlexNet architecture and transfer learning to enhance efficiency and accuracy. The model is trained and tested on MRI image datasets by extracting 32 intensity and grain textual features from segmented tumor regions, including statistical features and matrices. The study achieves maximum accuracy, validated by medical experts, and compares favorably with existing methods in terms of system accuracy. Using popular pre-trained CNN models like AlexNet, GoogLeNet, and VGG, the research demonstrates the effectiveness of CNN architectures in extracting essential features from input images, leading to improved brain tumor identification. Performance analysis reveals the significance of selected features through partitioning and statistical feature comparison, highlighting the impact of hyperparameter tuning and training functions like Adam and RMSprop on model optimization and overall system accuracy.
Much research has gone into identifying brain tumors. However, due to the complex pattern of the lesion's sites, this field still has limitations. Due to their size, microscopic lesions are notoriously difficult to see. In addition, classification accuracy suffers when extracting and choosing informative features. While convolutional neural networks can be useful for extracting useful information, the computing demands of these models might be prohibitive. To this day, compact modeling for the diagnosis of brain tumors is demanded. To address these issues, a novel improved k-means algorithm based DNN model is proposed for the detection of tumor lesions.
The DNN models offer solutions to challenges in identifying brain tumors by automatically learning relevant features from raw image data, including microscopic lesions, without manual feature engineering. Transfer learning enables leveraging pre-trained models to improve performance even with limited labeled data, while data augmentation increases dataset diversity. Model optimization techniques reduce DNN size and computational demands, making them more feasible for diagnosis. Ensemble learning further enhances accuracy and robustness. Overall, DNNs provide a comprehensive solution for efficient and accurate brain tumor diagnosis, addressing feature extraction and computing demands complexities.
3 Proposed research methodology
The suggested work is a deep neural network-based automatic brain tumor segmentation technique. Due to several factors, the proposed DNN model with Conv2D and Maxpooling2D layers can outperform more complex architectures like residual networks. Firstly, our model is less prone to overfitting, mainly when dealing with minor or less complex datasets, as they have fewer parameters and are less likely to capture noise in the data. Secondly, our model requires fewer computational resources, making them more practical and efficient, especially in resource-constrained environments. Additionally, the proposed architecture is easier to interpret and debug, which can be crucial for understanding model behavior and diagnosing issues during training. Also, combining median filtering and an improved k-means algorithm with a simple DNN model offers several advantages over more complex architectures. Median filtering algorithm is effective in removing noise from the input data. By replacing each pixel's intensity value with the median value of its neighboring pixels, median filtering effectively reduces the impact of outliers and random noise. By improving the standard k-means algorithm, i.e., by incorporating strategies to handle outliers or optimizing the initialization of centroids, we can enhance its effectiveness in capturing the underlying structure of the data. The combination of median filtering and an improved k-means algorithm with a simple DNN model offers a compelling solution that achieves competitive performance while maintaining simplicity, interpretability, and efficiency.
The input images are from BRATS 2022 [40], which includes three MRI scans of the human brain. The key advantage of MRI over CT scan is that it is less hazardous and has a higher accuracy rate. All earlier work on the dataset was based on tumor categorization, such as malignant tumor, glioma, and pituitary tumor, and was not meant for tumor segmentation and extraction accuracy. The proposed work's key contribution is image partitioning based on the spatial of acquired MR images. To improve segmentation outcomes, multiple networks are trained individually. Pre-processing is done with median filtering, and the image is then segmented using the K-means clustering technique and presented as histogram equalization. The extracted features are transformed into a grayscale image using the Gray-level Co-occurrence Matrix (GLCM) for feature extraction, which includes border extraction and region growth, then deep neural networks are used to classify the image.
Tensor Flow is a tool for simplifying difficult numerical computations, and it was employed in the classification section. It enables users to represent any computation as a data flow graph. Keras, a Python-based open-source neural network toolkit that operates on the upper edge of Theano or Tensor Flow, is chosen to implement the code because it is modular, quick, and easy to debug. Our data collection contains a total of 253 brain MRI pictures, 155 of which are malignant tumors and 98 of which are not. Using the classical classifiers, a tumor classification and segmentation method is proposed. Because of the brain's complex anatomy, detecting and classifying tumors using MRI images is difficult. The processes include pre-processing of MRI scans, chunk segmentation, feature extraction, and classification. In our project, we used MATLAB R2017A to write the programming code and represent the output of a brain tumor. Pre-processing of MRI images, section segmentation, feature extraction, and classification are the four major phases of image processing that we have accomplished here.
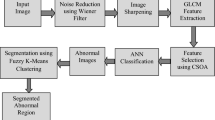
Figure 2 represents the process of the proposed system. The MRI images are taken as the input and pre-processed to extract the features of the tumor that are present in the MRI images. The feature is selected to reduce the dimensions so that the image can be segmented. The tumor part gets segmented in the segmentation process, and the remaining part is left the same. Finally, the segmented area is calculated, and the accuracy is determined.
3.1 Pre-processing
In most cases, the gathered data is jumbled, comes from many sources, and contains impurities. For noise removal, pictures are pre-processed to use the median filtering approach [41].
Median filtering is a widely used technique in image processing for noise reduction, particularly in scenarios where images are corrupted by salt-and-pepper noise or impulse noise. Unlike linear filters such as mean filters, which replace each pixel value with the average of its neighboring pixel values, median filtering replaces each pixel value with the median value of its neighboring pixels within a defined window. The chosen window size for our tumor detection is \(5\times 5\). By selecting a K value corresponding to a 5 × 5 grid, we partition the image into smaller, localized regions, each represented by a cluster centroid. This finer granularity allows the algorithm to capture more detailed information about local structures and textures, leading to more discriminative feature representations. Moreover, the chosen window size adapts the number of clusters based on the complexity of the underlying image content. This leads to a balance between capturing sufficient detail and maintaining computational efficiency.
The median filter operates by sliding a window (often a square or rectangular shape) over the image, and for each position of the window, the pixel value at the center of the window is replaced by the median value of all the pixel values within the window. This process effectively removes outliers and preserves edges in the image. Mathematically, the operation of median filtering can be represented as follows:
Let \(f\left(x,y\right)\) be the input image, where \(\left(x,y\right)\) are the spatial coordinates and \({W}_{xy}\) denote the window centered at the position \(\left(x,y\right)\). The output of the median filter is denoted as \(g\left(x,y\right)\) is computed as \(g\left(x,y\right)=median\left({W}_{xy}\right)\), where \(median\left({W}_{xy}\right)\) represents the median value of all pixels values within the window \({W}_{xy}\). The 'window' is a pattern of neighbors that glides pixel by pixel over the image sequence. The median is computed by sorting all the pixel values in the frame into numerical order, then substituting the pixel in question with the median pixel value.
Figure 3 depicts two MRI images taken from the BRATS data set. In Fig. 3a, the image represents the presence of the tumor part. It has a small part of the tumor with more intensity, but the second figure, as shown in Fig. 3b, has no tumor.
The two test images in Fig. 3 are used to illustrate the normalization process. Figure 4 lists the normalized results of the MRI scans, as shown in Fig. 3. The normalized images will then be used to predict the tumor part and segmentation.
3.2 Segmentation
A watershed method was used in the previous research for segmentation. Over-segmentation and the detection of erroneous edges were the results of the problem. The suggested study employs the K-means clustering [42, 43] technique for image segmentation. White Matter (WM), Grey Matter (GM), Cerebrospinal Fluid (CSF), and the background are typically separated in a brain MRI image using the conventional K-means algorithm. However, the K-means algorithm takes the grey pixels in the brain image for processing, ignoring the association between pixels. This results in poor brain segmentation accuracy, particularly for the reduced signal-to-noise ratio information.
This research takes advantage of the fact that neighboring pixels in a brain MRI image are highly likely to belong to the same class. Hence, a new pixel point value is generated for clustering by using the average value of the neighborhood of each image pixel. This workflow reduces the effect of distortion on the segmentation accuracy. The working of the improved K-means algorithm is explained in algorithm 1. Table 1 provides a detailed comparison between traditional and improved K-means algorithm in terms of computational complexities.

Algorithm-1. Improved K-means Algorithm
Figure 5 represents the segmentation process and levels of the segmentation process. The principle of the K-means technique is to cluster similar data points by first grouping the K cluster points in the space. Updates are periodically made to the cluster centroid value until optimal clustering results are achieved. The data point's distance from the prototype is the optimization aim. Finding function extremes is a useful technique for deriving iterative function adjusting rules. The K-means algorithm uses Euclidean distance as the similarity measure to discover the best possible categorization of an initial cluster center vector. This ensures that the evaluation index is minimized. Clustering is performed using the neighborhood criteria function. Though the K-means algorithm is effective, it requires an input of a fixed value for K, and choosing the optimal K value is difficult. In our case, 5 × 5 proved to be the optimal K value after repeated experimental trials.
3.3 Feature extraction
An image's structure, shape, size, perspective, and substance are all features. One of the most critical processes in medical image processing is feature extraction. The grey-level co-occurrence matrix [43,44,45,46,47] is one of the most qualitative-change feature extraction methods.
Figure 6 represents the process feature extracting the MRI images that undergo the filtering of the image, clustering of the image, complementing the image where the tumor part is determined, and the unwanted part is omitted, and extraction of the tumor from the original MRI scans.
3.4 DNN classification model
Figure 7 represents the realized DNN model for brain tumor prediction. An input data, an output layer, and a few layers between them make up deep neural networks. These networks are capable of handling not only unstructured and unlabeled input but also non-linearity. Comparable to the human brain, they feature a hierarchical architecture of neurons. The neurons transmit the message to other neurons based on the information received. The result will be passed if the signal value is larger than that of the threshold value. Otherwise, it will be ignored. As you can see, data is transmitted to the input layer, which produces output for the next layer, until it hits the output layer, which delivers a yes or no forecast based on likelihood. A layer is made up of numerous neurons, each with its own function termed the Activation Function. They serve as a conduit for signal transmission to the next linked neuron. The weight influences the input to the following neuron's output and, finally, the very last output layer. The weights are originally random, but when the network is trained repeatedly, the weights are tuned to ensure that the network delivers accurate predictions. Algorithm 2 explains the DNN classification workflow, and algorithm 3 explains the maximization of accuracy obtained in brain tumor classification.

Algorithm-2. DNN Classification

Algorithm-3. Maximization of Obtained Accuracy
Figure 8 depicts the block diagram explaining the workflow of the proposed DNN algorithm for classification and maximization of obtained accuracy. The terms x and y are the inputs that are provided to get the process to be done by the algorithm. x represents the list of directions and y represents the image size. Consider the human brain, which can recognize various persons despite having only two eyes, one nose, and two ears. The neurons can learn these variances and deviations and combine all these characteristics to identify persons. All of this happens in a fraction of a second.
A mathematical approach is used to apply the same rationale to deep neural networks. The information from one neuron is transmitted to another neuron based on a basic rule comparable to the brain's learning process. When the neuron's output is high, its corresponding dimensions are also high in importance. Similarly, all the signs of one layer are obtained in the form of deviation, which is then combined and sent to the following layer. As a result, the system naturally learns the process. Table 2 describes the semantic structure of the proposed DNN model. The layers in the model are comprised of one sequential layer, six 2D convolution layers, six 2D maximum pooling layers, a single flatten layer, and two dense layers. The total number of trainable parameters generated for the DNN-based tumor detection is 1,83,747.
4 Implementation and analysis
4.1 Dataset and tools
The BRATS 2022 (Multimodal Brain Tumor Segmentation Benchmark) [43,44,45,46] dataset stands as a cornerstone in the realm of medical image analysis, particularly in the context of brain tumor research. It comprises a diverse collection of multi-modal MRI scans, including T1-weighted, T1-weighted with contrast enhancement (T1ce), T2-weighted, and fluid-attenuated inversion recovery (FLAIR) images. Each scan is annotated with expert-defined tumor regions, encompassing various tumor subtypes such as glioblastoma (GBM), astrocytoma, and oligodendroglioma. One of the defining characteristics of the BRATS dataset is its complexity and variability. Brain tumors exhibit heterogeneous characteristics in terms of size, shape, location, and intensity across different imaging modalities. This variability poses significant challenges for segmentation algorithms, necessitating robust and adaptable methodologies for accurate tumor delineation. Table 3 summarizes the unique features of BRATS dataset.
Three tumor sub-regions are annotated: enhancing tumor, peritumoral edoema, and necrotic and non-enhancing tumor core. The Whole Tumor (WT), Tumor Core (TC), and Enhancing Tumor (ET) were created from the annotations. The images were gathered from 19 different institutions utilizing various MRI scanners. The most recent BRATS dataset 2022 includes FLAIR axial pictures which are tumorous and normal. A workstation with an i7 GPU from the 11th generation and 16 GB of RAM was used to test the proposed methods. The implementation was written in Python 3.10.5 using MATLAB2017a libraries.
For any neural networks and CNNs, hyperparameters like batch size, learning rate, and others play a crucial role in determining the model's performance [47,48,49]. For our implementation, we realized the batch size to be 32, the number of epochs to be 50, the Rectified Linear Unit (ReLU) to be the optimizer, and a learning rate of 0.0001. The implementation was started with a logarithmic scale 0.001 and observed the effect on training dynamics and model performance. A learning rate that is too high may lead to unstable training, while a learning rate that is too low may result in slow convergence. Larger batch sizes can lead to faster convergence but may suffer from generalization issues. Smaller batch sizes might offer better generalization but slower convergence. Keeping in mind of these limitations, 32 batch size was chosen since larger batch sizes require more memory and computational power.
Additionally, 5-fold cross-validation is employed to the dataset, to avoid overfitting during the training process. In the cross-validation process, the dataset is initially shuffled randomly. In the next step, the dataset is divided into 5-folds. For each fold, 80% of the dataset is used for training, whereas 20% of the dataset is used for testing. The proposed DNN model is used for training and evaluated on the testing set. The final evaluation score is calculated by summarizing all the model evaluation scores.
Several tests are carried out in this research to assess the functionality of the suggested system. The positive impact of the proposed strategies on the improved K-means segmentation models is further demonstrated via preliminary experiments conducted on the BRATS validation set. To gauge the effectiveness of the proposed methodology, we evaluated it using seven different parameters (Eqs. 1 to 7): accuracy, precision, sensitivity, specificity, Matthew’s Correlation Coefficient (MCC), dice score, and Jaccard index.
4.2 Results and comparison
The proposed methodology is compared with existing methods such as SVM, CNN, U-Net, VGG-19, YOLO, GAN, CapsNet, Alexnet, DenseNet, EfficientNet, MobileNet and ResNet-50 to measure the performance efficacy.
Figures 9 and 10 depict the loss and accuracy comparison of the proposed model during the training and validation phase, respectively. The statistical performance of the suggested model is assessed by contrasting the loss and accuracy of the training and validation sets. During the learning phase, a total of 50 epochs are analyzed to validate accuracy and loss of precision. The loss value obtained during the validation phase is 0.3324, which aligns efficiently with the loss value of 0.3162 obtained during the training phase. The accuracy of the proposed model in the validation phase is 0.9795, which is comparatively better than the 0.8987 training accuracy.
Figures 11 and 12 illustrate the dice coefficient and Jaccard index comparison of the proposed model during the training and testing phase, respectively. \({P}_{r}\) indicates the predicted value and \({T}_{g}\) denotes the ground truth information for brain tumor segmentation. The dice value of 0.9485 is obtained during the validation phase and is 3% better than in the training phase. During the validation phase, a 2% improvement is achieved with a final score of 0.8572.
Figure 13 depicts the overall performance of the proposed frameworks with existing methods such as SVM, CNN, U-Net, VGG-19, YOLO, GAN, CapsNet, Alexnet, DenseNet, EfficientNet, MobileNet and ResNet-50 in terms of Accuracy, Precision, Sensitivity, Specificity, and MCC. The proposed method has been compared with existing approaches based on the BRATS dataset. Table 4 includes various state-of-the-art methodologies or models, each evaluated based on the provided metrics. The proposed DNN model stands out with the highest accuracy of 0.9795, indicating superior overall performance compared to other models. EfficientNetV2 and DenseNet121 also demonstrate high accuracy scores of 0.9758 and 0.9728, respectively. Sensitivity and specificity scores vary among models, reflecting differences in their ability to classify positive and negative instances correctly. Precision measures the proportion of true positive predictions out of all positive predictions made by the model. Higher precision indicates fewer false positives. The Matthews Correlation Coefficient (MCC) provides a single value that balances true and false positives and negatives, offering a comprehensive assessment of model performance. Compared with the existing methods, the proposed technique achieved 0.9795 accuracy, 0.9754 specificity, 0.9799 precision and 0.9596 MCC scores on the BRATS database.
4.3 DNN segmentation model analysis
4.3.1 BRATS dataset analysis
In most published works, the BRATS dataset is augmented to strengthen the modality of the research. On its whole, the BRATS collection includes thousands of MRI images. However, there are numerous drawbacks to using segmentation techniques to analyze the dataset, such as (1) the ease with which anatomically inaccurate samples can be generated, (2) the fact that these methods are not easily implemented, and (3) the mode collapse problem. However, the effects of using unrealistic samples in training sets are currently unknown. For this reason, the DNN classifier fused with an improved K-means algorithm is proposed for brain image segmentation in this research.
The visual representation of the compared segmentation results is presented in Figs. 14 and 15. It is evident that the proposed approach can segment whole brain tumors; nevertheless, the differences between our methods and the ground truth are minimal. For each set of image data, we determine the median tumor volume. After arithmetically averaging all the slices, the volumes are determined by multiplying the number of tumor voxels by the voxel volume [56]. The resulting units are cubic millimeters. The volumes of the tumors that have been appropriately segmented are then computed, and the results are compared to the true values to assess the degree of agreement. Calculations show that the BRATS dataset's similarity between the tumor volume retrieved from the ground-truth pictures and the suggested approach is 94.63%. Table 5 displays the tumor volume analysis as mean values compared with the ground truth results in tabular form.
4.3.2 Figshare dataset analysis
The robustness of the proposed improved K-means-median filtering segmentation process is analyzed using the open-source figshare brain tumor dataset [57]. The figshare brain tumor collection is a compilation of medical imaging data focused on brain tumors. It encompasses a total of 3,064 T1-weighted Contrast-Enhanced Magnetic Resonance Images (CEMRI) showcasing three primary categories of brain tumors: glioma, meningioma, and pituitary tumor. Each image entry within the collection contains crucial information essential for medical analysis, including the class name (indicating the type of tumor), patient ID, raw image data, tumor borders defined by x and y coordinates outlining various points on the tumor's boundary, and a binary tumor mask representing the segmented tumor area.
Table 6 provides an overview of the initial number of tumor instances for each tumor type, aiding researchers and medical professionals in understanding the distribution of tumor types within the dataset. This collection serves as a valuable resource for medical research, particularly in developing and evaluating algorithms for brain tumor detection, segmentation, and classification.
The segmentation performance is analyzed using the performance metrics, namely the Jaccard coefficient and Dice coefficient, which are represented in Eqs. (6) and (7).
Table 7 shows the comparison of the results between the proposed DNN model and the existing frameworks in terms of accuracy, dice coefficient, and Jaccard coefficient. The Jaccard coefficient score exceeded 0.8724, and the accuracy score is 0.9891, surpassing the existing approaches.
4.4 Br35H dataset analysis
The dataset was created in 2020 by Mikhael Adriel and Pratama Gana [61]. It served as a testing platform for efficient brain tumor detection. The dataset includes 3000 brain images, of which 1500 images are classified as YES category (Tumor present) and 1500 images are classified as NO (Tumor absent) category. Figure 15 displays the sample images available in the Br35H dataset.
Table 8 shows the performance comparison of the proposed modified DNN model aided with the K-means algorithm and median filtering with convolutional architectures. From the result, it is evident that our model performed well compared to existing models.
4.5 Time complexity analysis
Analyzing the time complexity of a proposed DNN model involves considering various factors, including the number of layers, the number of neurons in each layer, the size of the input data, and the computational operations performed during training and inference. In each convolution layer, a set of filters (also known as kernels) is applied to the input feature map to produce output feature maps. There are N convolution layer with M filters each in our proposed model. Therefore, the total time of complexity of the convolution layers is calculated as \({\rm O}\left(N\times M\right)\). Max-pooling layers downsample the feature maps obtained from convolutional layers by selecting the maximum value within each pooling window. The time complexity of applying max-pooling to a single feature map is approximately \({\rm O}\left({K}^{2}\right)\), where K is the size of the pooling window. There are L max-pooling layers in our model and hence the total time complexity for max-pooling layers is \({\rm O}\left(L\times {K}^{2}\right)\). The overall time complexity of the DNN with convolutional and max-pooling layers is the sum of the time complexities of all convolution and max-pooling layers. It is approximated as \(\{{\rm O}\left({K}^{2}\right)+{\rm O}\left(L\times {K}^{2}\right)\}\).
Additionally, the computation process of median filtering and improve k means algorithm is also included in calculating the overall time complexity of the proposed DNN model. The time complexity of median filtering depends on the size of the input image and the size of the kernel used for filtering. The size of the kernel determines the number of elements considered for computing the median value. The kernel size for our model is define as \(K\times K\) and the number of comparisons required to find the median value is proportional to the number of elements in the kernel. For a kernel size of \(K\times K\), the number of comparisons is \({\rm O}\left({K}^{2}\right)\). The dimensions of the input image are denoted as \(\left(N\times M\right)\), where M is the number of rows and N is the number of columns. For each pixel in the input image, the median filtering operation is applied. The time complexity of median filtering can be expressed as \({\rm O}\left(N\times M\right)\).
Finally, the time complexity of the improved K-means algorithm is primarily determined by the number of data points (N), the number of clusters (K), the dimensionality of the data (d), and the convergence criterion. The algorithm iteratively assigns data points to clusters and updates cluster centroids until convergence, with a time complexity that scales linearly with the number of iterations and the size of the dataset. Thus, the time complexity of improved K means algorithm is represented as \({\rm O}\left(N\bullet K\bullet d\right)\). Eventually the overall time complexity of the proposed DNN model with median filtering and improved K means algorithm is the sum of all individual operational complexity. It is represented in Eq. (8). By applying the upper bound rule, the final time complexity of the proposed model is shown in Eq. (9).
Table 9 represents the comparative analysis of the proposed model and other existing approaches with respect to time complexity. It is clear that our model has the optimal time execution even though there are complex segmentation and simplified deep training processes.
4.6 Discussions
Tumors in MRI scans are often difficult to distinguish. Thus, it's essential to boost those areas for better segmentation. However, accurate localization is essential for tumor enhancement since this will ensure that the segmentation is only applied where needed. To improve the localization of previously unclear tumors, we first used a median filtering approach to pinpoint their locations based on the most salient features of the brain images. Segmentation errors for individual tumors in high-contrast images are reduced with the use of the suggested filtering approach by making the tumorous region's pixel values match those of the background. As a result, fuzzy pixels caused by tumors are made clearer, while the backdrop is kept intact, all at a modest computational cost (0.0025 s).
Also, the DNN design benefited from the addition of an improved K-means technique, allowing it to extract features more effectively and carry out segmentation with greater precision. As a result of the filtering and feature selection method, the proposed DNN model is now occupied mostly with enhanced and aesthetically better tumorous areas. This improved the DNN segmentation by a factor of 0.04 for the BRATS dataset. The findings demonstrated that modest image pre-processing considerably improves learning models' segmentation capabilities without requiring any changes to the models' core architecture. When compared to the earlier research [23, 46], the training time for the DNN architecture is reduced and datasets are trained without using any data augmentation approach. The proposed filtering and improved K-means techniques improved the segmentation competence of the DNN architecture (dice score on BRATS set = 0.9485) by 3% while decreasing training time and computational cost. From the above discussions, the proposed DNN model can greatly improve brain tumor segmentation outcomes and reduce the computational costs mentioned above for future research.
Multiple pixel-fuzzy tumorous patches in images serve as a benchmark for the proposed system's limitations. The proposed system predicted tumor because the tumor is a singular area. However, this restriction might be addressed in the future by adapting the algorithms to identify numerous areas. The study also has several gaps, such as the suggested methods for tumor segmentation not being tested using additional modalities to segment tumors as multi-class and FLAIR images alone being used for comprehensive tumor segmentation.
The primary drawback of the proposed algorithm lies in its prolonged processing time attributed to additional optimization steps. Furthermore, its efficacy diminishes notably in identifying minute lesions, particularly in instances where the dataset lacks representative instances. Augmenting the dataset with synthetic images featuring small lesions, guided by medical experts' insights, holds promise in diversifying the dataset and exposing the model to uncommon scenarios. Integration of predictions from multiple detection models trained on distinct data subsets can bolster overall detection accuracy, particularly in scenarios with varying lesion dimensions. Active engagement of medical professionals in iteratively annotating and selecting training samples can prioritize the inclusion of challenging cases with small lesions, progressively refining the model's proficiency in their detection.
5 Conclusions
This research has demonstrated a comprehensive approach for identifying brain tumors in BRATS dataset through segmentation utilizing an improved K-means algorithm, complemented by median filtering, and subsequent classification employing a deep neural network. Through the integration of these techniques, we have achieved promising results in accurately delineating tumor boundaries and classifying tumor types. The refined K-means algorithm coupled with median filtering has enhanced the precision of segmentation, effectively isolating tumor regions from healthy brain tissue. Furthermore, the utilization of a deep neural network for classification has enabled robust categorization of tumor types, contributing to clinical decision-making processes. On the benchmark BRATS datasets, the proposed method achieves an accuracy of 0.9795, 0.9485 dice score, 0.8572 Jaccard index value, and 0.9896 MCC score, respectively, for segmenting entire brain tumors, demonstrating its exceptional segmentation ability. Overall, this integrated methodology presents a promising avenue for improving the efficiency and accuracy of brain tumor identification, potentially enhancing patient outcomes and facilitating personalized treatment strategies in clinical settings. Further validation and refinement of this approach hold considerable potential for advancing the field of medical image analysis and improving patient care in neuro-oncology.
Data availability
The brain MRI image data used to support the findings of this study is publicly available in the repositories (https://www.kaggle.com/datasets/awsaf49/brats2020-training-data).
References
Zhang W, Yong Wu, Yang Bo, Shunbo Hu, Liang Wu, Dhelim S (2021) Overview of multi-modal brain tumor mr image segmentation. Healthcare 9(8):1051
Prakash Tunga P, Singh V (2021) Compression of MRI brain images based on automatic extraction of tumor region. Int J Electrical Comput Engineering 11(5):3964–3976
Gutmann DH, Kettenmann H (2019) Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron 104(3):442–449
Srikanth B, Venkata Suryanarayana S. Multi-class classification of brain tumor images using data augmentation with deep neural network. Materials Today: Proceedings. 2021. https://doi.org/10.1016/j.matpr.2021.01.601
Hussain, Ashfaq, and Ajay Khunteta. "Semantic segmentation of brain tumor from MRI images and SVM classification using GLCM features." In 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), pp. 38–43. IEEE, 2020.
Sankaran KS, Poyyamozhi AS, Ali SS, Jennifer Y. A Conceptual and Effective Scheme for Brain Tumor Identification Using Robust Random Forest Classifier. In Proceedings of International Conference on Information Technology and Applications. Springer. Singapore. 2022. pp. 109-118.
Chaudhary A, Bhattacharjee V (2020) An efficient method for brain tumor detection and categorization using MRI images by K-means clustering & DWT. Int J Inf Technol 12(1):141–148
Zotin A, Simonov K, Kurako M, Hamad Y, Kirillova S (2018) Edge detection in MRI brain tumor images based on fuzzy C-means clustering. Proc Comput Sci 126:1261–1270
Maruthi KD, Satyanarayana D, Prasad MN (2021) MRI brain tumor detection using optimal possibilistic fuzzy C-means clustering algorithm and adaptive k-nearest neighbor classifier. J Ambient Intell Human Comput 12(2):2867–2880
Deepak S, Ameer PM (2021) Automated categorization of brain tumor from mri using cnn features and svm. J Ambient Intell Humaniz Comput 12(8):8357–8369
Chaki J (2022) Two-fold brain tumor segmentation using fuzzy image enhancement and DeepBrainet2. 0. Multimedia Tools Appl 81:30705–30731
Devi RS, Perumal B, Rajasekaran MP (2022) A hybrid deep learning based brain tumor classification and segmentation by stationary wavelet packet transform and adaptive kernel fuzzy c means clustering. Advances in Engineering Software 170:103146
Fang L, Wang X (2023) Multi-input Unet model based on the integrated block and the aggregation connection for MRI brain tumor segmentation. Biomed Signal Process Control 79:104027
Bodavarapu PNR, Srinivas PVVS, Mishra P, Mandhala VN, Kim H-J. Optimized deep neural model for cancer detection and classification over ResNet. In Smart Technologies in Data Science and Communication. Springer. Singapore. 2021. pp. 267–280.
Gu X, Shen Z, Xue J, Fan Y, Ni T (2021) Brain tumor MR image classification using convolutional dictionary learning with local constraint. Front Neurosci 15:679847
Adu K, Yongbin Yu, Cai J, Asare I, Quahin J (2022) The influence of the activation function in a capsule network for brain tumor type classification. Int J Imaging Syst Technol 32(1):123–143
Akinyelu AA, Zaccagna F, Grist JT, Castelli M, Rundo L (2022) Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. J Imaging 8(8):205
Sedik A, El-Hag NA, El-Hoseny HM, El Banby G, Khalaf AA, Abd El-Samie FE, El-Shafai W. Retinal Disorder Diagnosis Based on Hybrid Deep Learning Models. Available at SSRN 4111795.
Toufiq DM, Sagheer AM, Veisi H (2021) Brain tumor identification with a hybrid feature extraction method based on discrete wavelet transform and principle component analysis. Bullet Electr Eng Inform 10(5):2588–2597
Ahmadi M, Sharifi A, Jafarian Fard M, Soleimani N. Detection of brain lesion location in MRI images using convolutional neural network and robust PCA. Int J Neurosci. 2021:1–12.
Rao CS, Karunakara K (2022) Efficient detection and classification of brain tumor using kernel based SVM for MRI. Multimedia Tools Appl 81(5):7393–7417
Toufiq DM, Sagheer AM, Veisi H (2021) A Review on Brain Tumor Classification in MRI Images. Turkish J Comput Math Educ 12(14):1958–1969
Aminian M, Khotanlou H (2022) CapsNet-based brain tumor segmentation in multimodal MRI images using inhomogeneous voxels in Del vector domain. Multimedia Tools Appl 81(13):17793–17815
Ahmad B, Sun J, You Qi, Palade V, Mao Z (2022) Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 10(2):223
Mishra L, Verma S (2022) Graph attention autoencoder inspired CNN based brain tumor classification using MRI. Neurocomputing 503:236–247
Díaz-Pernas FJ, Martínez-Zarzuela M, Antón-Rodríguez M, González-Ortega D (2021) A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare 9(2):153
Kumar A, Manikandan R, Kose U, Gupta D, Satapathy SC. Doctor's Dilemma: Evaluating an Explainable Subtractive Spatial Lightweight Convolutional Neural Network for Brain Tumor Diagnosis. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM). 2021;17(3s): 1–26.
Nayak DR, Padhy N, Mallick PK, Singh A (2022) A deep autoencoder approach for detection of brain tumor images. Comput Electr Eng 102:108238
Abd El Kader I, Xu G, Shuai Z, Saminu S, Javaid I, Salim Ahmad I (2021) Differential deep convolutional neural network model for brain tumor classification. Brain Sci 11(3):352
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: Deep convolutional neural networks. Fut Gen Comput Syst 87:290–297
Vankdothu R, Hameed MA (2022) Brain tumor MRI images identification and classification based on the recurrent convolutional neural network. Measurement Sensors 24:100412
Shah HA, Saeed F, Yun S, Park JH, Paul A, Kang JM (2022) A Robust Approach for Brain Tumor Detection in Magnetic Resonance Images Using Finetuned EfficientNet. IEEE Access 10:65426–65438
Nassar SE, Yasser I, Amer HM, Mohamed MA (2024) A robust MRI-based brain tumor classification via a hybrid deep learning technique. J Supercomput 80(2):2403–2427
Priyadarshini P, Kanungo P, Kar T. Multigrade Brain tumor classification in MRI images using Fine tuned EfficientNet. e-Prime-Advances in Electrical Engineering, Electronics and Energy. 2024:100498.
Khaliki MZ, Başarslan MS (2024) Brain tumor detection from images and comparison with transfer learning methods and 3-layer CNN. Sci Rep 14(1):2664
Kumar S, Choudhary S, Jain A, Singh K, Ahmadian A, Bajuri MY (2023) Brain tumor classification using deep neural network and transfer learning. Brain Topography 36(3):305–318
Asif S, Zhao M, Tang F, Zhu Y (2023) An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. Multimedia Tools Appl 82(20):31709–31736
Rajput IS, Gupta A, Jain V, Tyagi S. A transfer learning-based brain tumor classification using magnetic resonance images. Multimedia Tools Appl. 2023:1–20.
Bairagi VK, Gumaste PP, Rajput SH, Chethan KS (2023) Automatic brain tumor detection using CNN transfer learning approach. Med Biol Eng Comput 61(7):1821–1836
Brain Tumor Segmentation, Dataset from BraTS2020 Competition, available at https://www.kaggle.com/datasets/awsaf49/brats2020-training-data.
Zhang C, Shen X, Cheng H, Qian Q. Brain tumor segmentation based on hybrid clustering and morphological operations. Int J Biomed Imaging. 2019.
Elmontaser NM, Abushaala AM (2022) Brain Tumor Detection in Real MRI Images Based on Otsu and K-means Cluster Algorithms. Brain 21:22
Kshirsagar PR, Yadav AD, Joshi KA, Chippalkatti P, Nerkar RY (2020) Classification and Detection of Brain Tumor by using GLCM Texture Feature and ANFIS. J Res Image Signal Proc 5(1):15–31
Rastogi R, Rastogi AR, Chaturvedi DK, Sagar S, Tandon N. Massive data classification of brain tumors using DNN: opportunity in medical healthcare 4.0 through sensors. In Handbook of Machine Learning for Computational Optimization, pp. 95–111. CRC Press, 2021.
Ahmed AA, Jabbar WA, Sadiq AS, Patel H. Deep learning-based classification model for botnet attack detection. J Ambient Intell Human Comput. 2020:1–10.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J et al (2015) The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS et al (2017) Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat Sci Data 4:170117. https://doi.org/10.1038/sdata.2017.117
S. Bakas, M. Reyes, A. Jakab, S. Bauer, M. Rempfler, A. Crimi, et al., "Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge." arXiv preprint arXiv:1811.02629, 2018.
S. Bakas, H. Akbari, A. Sotiras, M. Bilello, M. Rozycki, J. Kirby, et al., Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-GBM collection. Cancer Imaging Arch. 2017. https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q.
S. Bakas, H. Akbari, A. Sotiras, M. Bilello, M. Rozycki, J. Kirby, et al., Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-LGG collection. Cancer Imaging Arch. 2017. https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF.
Sethy PK, Behera SK (2021) A data constrained approach for brain tumour detection using fused deep features and SVM. Multimedia Tools Appl 80:28745–28760. https://doi.org/10.1007/s11042-021-11098-2
Sharif MI, Li JP, Amin J, Sharif A (2021) An improved framework for brain tumor analysis using MRI based on YOLOv2 and convolutional neural network. Complex Intell Syst 7:2023–2036. https://doi.org/10.1007/s40747-021-00310-3
Sunsuhi GS, Jose SA (2022) An Adaptive Eroded Deep Convolutional neural network for brain image segmentation and classification using Inception ResnetV2. Biomed Signal Proc Control 78:103863
Sachdeva J, Sharma D, Ahuja CK. Comparative Analysis of Different Deep Convolutional Neural Network Architectures for Classification of Brain Tumor on Magnetic Resonance Images. Arch Computational Methods Eng. (2024):1–20.
Vatanpour M, Haddadnia J (2024) Brain tumour segmentation of MR images based on custom attention mechanism with transfer-learning. IET Image Proc 18(4):886–896
Mayer R, Simone CB, Turkbey B, Choyke P (2021) Algorithms applied to spatially registered multi-parametric MRI for prostate tumor volume measurement. Quant Imaging Med Surg 11(1):119
Cheng, J. Brain tumor dataset. Figshare. Dataset. Accessed: Sep. 19, 2019. [Online]. Available: https://doi.org/10.6084/m9.figshare.1512427.v5.
Roy Choudhury A, Vanguri R, Jambawalikar SR, Kumar P. Segmentation of brain tumors using DeepLabv3+. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Revised Selected Papers, Part II 4, pp. 154–167. Springer International Publishing, 2019.
Zhao X, Yihong Wu, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111
Zhang Y, Zhong P, Jie D, Jiewei Wu, Zeng S, Chu J, Yilong Liu EdXWu, Tang X (2021) Brain tumor segmentation from multi-modal MR images via ensembling UNets. Frontiers in radiology 1:704888
Mikhael Adriel and Pratama Gana, Br35H – Brain Tumor dataset (2020), Available at: https://www.kaggle.com/datasets/mikhaelapg/brain-tumor-detection/data.
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X (2018) Supervised learning based multimodal MRI brain tumour segmentation using texture features from supervoxels. Comput Methods Programs Biomed 157:69–84
Salçin K (2019) Detection and classification of brain tumours from MRI images using faster R-CNN. Tehnički glasnik 13(4):337–342
Amin J, Sharif M, Yasmin M, Fernandes SL (2020) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett. 139:118–127
Kang M, Ting CM, Ting FF, Phan RC. "Bgf-yolo: Enhanced yolov8 with multiscale attentional feature fusion for brain tumor detection." arXiv preprint arXiv:2309.12585 (2023).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they do not have any conflict of interests that influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rajendirane, R., Ananth Kumar, T., Sandhya, S.G. et al. A novel brain tumor segmentation and classification model using deep neural network over MRI-flair images. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19487-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-19487-z