Abstract
The primary objective of this paper is to develop a methodology for brain tumor segmentation. Nowadays, brain tumor recognition and fragmentation is one among the pivotal procedure in surgical and medication planning arrangements. It is difficult to segment the tumor area from MRI images due to inaccessibility of edge and appropriately visible boundaries. In this paper, a combination of Artificial Neural Network and Fuzzy K-means algorithm has been presented to segment the tumor locale. It contains four phases, (1) Noise evacuation (2) Attribute extraction and selection (3) Classification and (4) Segmentation. Initially, the procured image is denoised utilizing wiener filter, and then the significant GLCM attributes are extricated from the images. Then Deep Learning based classification has been performed to classify the abnormal images from the normal images. Finally, it is processed through the Fuzzy K-Means algorithm to segment the tumor region separately. This proposed segmentation approach has been verified on BRATS dataset and produces the accuracy of 94%, sensitivity of 98% specificity of 99%, Jaccard index of 96%. The overall accuracy of this proposed technique has been improved by 8% when compared with K-Nearest Neighbor methodology.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Different types of restorative imaging systems such as Computed Tomography (CT), Single-Photon Emission Computed Tomography (SPECT), and Magnetic Resonance Imaging (MRI) are utilized to give important data about nature, dimension, vicinity and metabolism of cerebrum tumor aiding determination. These modalities are utilized in combination to give the most elevated data about the cerebrum tumor. MRI is a non-intrusive system that uses radio recurrence signals to create the internal images under the influence of an extremely amazing attractive field. Distinctive MRI modalities produce various kinds of tissue contrast images, subsequently giving important auxiliary data and empowering analysis and segmentation of tumor along with their subregions. Cerebrum tumor segmentation from MRI is a critical errand which includes different covering pathology, medical resonance materal science, radiologist’s discernment, and image analysis dependent on intensity and shape. There are numerous difficulties related to tumor fragmentation. The tumor might be of any dimension, may have an assortment of shapes, and may emerge at any location. The manual segmentation is a troublesome and tedious task, which makes an attractive computerized cerebrum tumor segmentation technique. The challenges related with programmed cerebrum tumor segmentation have offered to various methodologies.
The identification of the brain tumor from the MRI image is the challenging task due to trademark requirements. Feature extraction and determination are the significant strides for brain tumor location. There are various types of features such as GLCM, statistical, texture, region based, wavelet features, and so forth [1]. Also, there are numerous methods are available to extract the required attributes from the image. Using huge number of features for classification is an extraordinary deterrent [2]. Therefore, the significant features must be chosen. It is the process of increasing the classification accuracy and also reducing the computational intricacy. There are numerous deep learning and optimization algorithm which includes Fisher Linear Discriminant Analysis [3], K-Nearest Neighbor [4] Decision Tree, Multilayer Perceptron [5], and Support Vector Machine (SVM) [6] have been utilized for selecting features from the image [7]. It was found that, Artificial Neural Network (ANN) classification methods give an powerful approach for multi-category classification problem [8]. Similarly, even though number of segmentation methods has been used for brain tumor segmentation, still the accurate segmentation technique has not found out.
The fundamental target of this paper is to develop an efficient cerebrum tumor segmentation approach for MRI imageries using ANN classifier and Fuzzy-K Means (FKM) clustering technique. The accuracy of cerebrum tumor diagnosis has been exaggerated by the segmentation process. Recently, Deep Learning based methods have been widely used in Medical Image Segmentation. Since, the medical images have high correlation among the neighborhood pixels, it must be accounted while characterizing the segmentation method. The FKM in its original state does not take this into consideration. Therefore, the FKM method has been combined with ANN classifier. The MRI images are affected by multiplicative type of speckle noise. Wiener filter has been utilized for speckle noise reduction. After that, GLCM attributes are extricated from every image. In order to decrease the complexity and to increase the ANN classifier accuracy, significant attributes are selected by using Crow Search Optimization Algorithm (CSOA). Then, the usual and unusual images are classified using ANN classifier. After the classification process, Fuzzy K-means clustering technique has been utilized for cerebrum tumor segmentation.
Remaining part of this paper is arranged in the following manner. Section 2 presents about a summary of related works, Sect. 3 gives the detailed description about the proposed methodology for brain tumor segmentation. Section 4 presents the simulation result and findings of proposed methodology, and Sect. 5 concludes the efficiency of the proposed methodology for brain tumor segmentation.
2 Related Works
Different types of medical imaging data has been developed in clinical research centers which are hard to segment within the adequate time. Furthermore, manual audit of image is hard, tiring and tedious [9]. The significant aim of any program based technique is right recognition and classification of tumor region. The brain tumor segmentation comprises of four phases which includes standardization, tumor segmentation, attribute extraction and classification. Among this, standardization plays a significant role in tumor segmentation, because the other phases are reliant on it. A few ancient rarities exist in MRI imageries which reduces the segmentation accuracy [10]. In [11], the noise present in the MRI image has been removed by using structural information. In [12], morphological filtering technique has been used to expel the ancient rarities. In [13], neighborhood spatial information has been utilized to reduce the noise effects. In the standardization process, various filters are utilized to evacuate noise and other ancient rarities. These filters includes high pass filter, sharpening filter, histogram equalization. In [14], developed local binary pattern and histogram of gradient based features for tumor segmentation. In this paper, after the extraction of features, it is classified using Random forest classifier. In [15], presented a combination of spatial Fuzzy C- Means and K-means clustering technique for cerebrum tumor fragmentation from MRI imageries. In [16], author developed another tumor classification strategy dependent on attribute extraction and probabilistic neural network (PNN). In [17], improved binomial thresholding and multi-feature selection has been developed for tumor segmentation. Initially, the noise is removed by using Gaussian filter and the region of interest has been extracted using manual skull stripping. Then the tumor has been segmented using improved thresholding method. In [18], a novel RescueNet structure has been proposed for brain tumor segmentation. This network structure has been trained by using GAN based training approach. Additionally, Scale-invariant algorithm has been utilized to increase the segmentation accuracy. In [19], Grab cut method and deep learning method is utilized for brain tumor detection. In [20], authors proposed novel brain tumor segmentation and classification method depends on fusion of MRI sequences. The different MRI sequences such as T1, T1C, T2 and Flair are amalgamated using discrete wavelet transform. Universal thresholding method has been used for the segmentation of brain tumor and Convolutional Neural Network is used for the tumor classification. The above mentioned segmentation techniques are significance to fragmentation and attribute extraction methods for cerebrum tumor detection and classification from MRI images. In [21], Deep learning based HCNN and CRF-Recurrent regression neural network (CRF-RRNN) model has been proposed to analyze the types of tumor in MRI images. In this method, initially, the HCNN network was pretrained by using image patches and the CRF-RRNN model was pretrained by using image slices. Then voting fusion technique has been utilized to segment the tumor region on the internet of medical things platform. In [22], One-pass Multi-task Network (OM-Net) has been proposed to resolve class imbalance for cerebrum tumor segmentation. This network classifies the segmentation tasks into one deep model and the OM-Net has been optimized. The cross-task guided attention (CGA) module has also been used for tumor segmentation. CGA can adaptively recalibrate channel-wise feature responses based on the category-specific statistics. The proposed methodology has been verified on the BraTS 2015 testing set and BraTS 2017 online validation set.
In early days, simple rule-based techniques were utilized for brain tumor segmentation. Due to high robustness, these techniques are not suitable for huge variety of data. Thereafter, different adaptive algorithms were utilized based on the geometric shape and fuzzy systems. These algorithms suffer from human biases and cannot deal with the variances in real-world data. In the recent years, deep learning based techniques have been used to overcome the above mentioned drawbacks. In this regard, ANN has been used along with the FKM clustering technique for tumor segmentation of MRI images.
3 Proposed Methodology
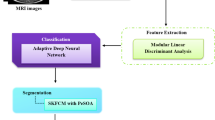
The overall workflow diagram of proposed brain tumor classification and segmentation method has been illustrated in Fig. 1. The main stage is the classification of MRI images into ordinary or unusual image. At first, the input MRI imageries are preprocessed for noise expulsion. Then, significant attributes are extricated from the preprocessed imageries. The extracted attributes are given to the ANN classifier to discover the image as a normal or abnormal image. The final stage separates the tumor area from the original image utilizing FKM algorithm. At last, this proposed strategy separates the tumor region from the input MRI image.
3.1 Noise Removal Using Wiener Filter
The MRI images are affected by multiplicative noise. Therefore, noise evacuation is a significant procedure in medical images and it is the preprocessing step in medical image processing. In this paper, Wiener filter has been used for noise removal. It limits the Mean Square Error during the process of noise smoothing. The filtering technique depends on a stochastic structure. The Wiener filter can be describes as,
where \( S_{xx} \left( {f_{1} ,f_{2} } \right), S_{\eta \eta } \left( {f_{1} ,f_{2} } \right) \) are the power spectrum of undegraded MRI image and the noise, \( H\left( {f_{1} ,f_{2} } \right) \) is the blurring filter. This filter conserves the edges through inverse filtering and removes the noise with low pass filtering.
3.2 Feature Extraction
Extrication of significant attributes of an object is an errand to increase the ANN classifier accuracy. In this work, GLCM properties of an image have been extracted from the image. The attribute set comprises of morphological and statistical information. Here, sixteen sorts of GLCM highlights from each image have been extracted. The feature includes mean, standard deviation, contrast, entropy, correlation, energy, inverse difference moment, variance, Dissimilarity, homogeneity, Sum variance, Sum entropy, Auto correlation, Difference entropy, Maximum probability and sum average.
3.3 Feature Selection Using CSOA
After the feature extraction procedure, the significant characteristics are chosen by using CSOA. Since, large number of highlights influences the execution of classification procedure and increases the computational intricacy. This algorithm relying upon the attributes and knowledge of crows. The feature selection procedure has been described in Fig. 2.
3.3.1 Initialization
Randomly select the characteristics from the feature set and the quantity of the feature is represented by N. The features are arranged in a matrix form and it is represented as,
3.3.2 Fitness Estimation
In this fitness estimation, the fitness of every crow has been determined. Each crow location is assessed by utilizing a predetermined fitness function It is used to quantify the integrity of alternate arrangements. The fitness is determined by utilizing the below Eq. (3),
where TP, FP is the true positive and false positive, and Tn, Fn is the true negative and false negative respectively.
3.3.3 Updation Based on CSOA
After the completion of fitness estimation, find the tumor location by utilizing CSOA algorithm. For finding the tumor location, there are two cases which are discussed beneath;
Case 1: The tumor region is updated by using the below equation,
where, Rm is a random integer in range [0,1], \( FL_{m}^{t} \) is the flight length of tumor m at iteration t.
Case 2: When the tumor region is wrongly identified with the non tumor region. Then the location of the tumor region is updated randomly. It is updated by using the below equation,
where \( P_{n}^{t} \) is the possibility of tumor n at iteration t.
3.3.4 Termination Criteria
This process ends, when it accomplishes the outrageous number of cycle or when the best arrangement has been achieved. In this paper, outrageous number of cycle is utilized. The highest number of cycle set for this work is 40.
3.4 ANN Classification Stage
Once the optimized feature selection process using CSOA have been completed, then the attributes are bolstered to the classification stage. The extricated GLCM properties have been utilized as the input of an ANN classifier. ANN comprises of 3 layers specifically input, concealed, and output layer. The optimized chosen attributes are given to the input layer. Then, the arbitrary weights are doled out to input-hidden (\( W_{ij}^{h} \)) and hidden-output layer (\( W_{ij}^{o} \)). The structure of ANN classifier has been illustrated in Fig. 3.
Initially, the feature value of the input layer has been multiplied with the input-hidden weights. Therefore, the concealed node j gets the function Hj, and it is given by,
Here, xj is the input bias value. The activation function is described as follows,
The output function is described as,
where xi is the output bias value. The obtained error value can be described as,
Here, n is the number of parameters, Yi is the output value, and Ti is the target value. The obtained MSE has been minimized using back-propagation algorithm. During the training, the minimum achieved MSE based weight values have been stored, and during the testing, the chosen attributes are given to ANN classifier and trained weights are given to the ANN. In this network, two classes are assigned to the output layer. Depending on the obtained value, the testing image is classified into equivalent class. The required condition can be represented as,
Here, N1 is the normal image and N2 is the abnormal tumor image.
3.5 Segmentation Using Fuzzy K-Means (FKM) Algorithm
The classified abnormal images are given to the FKM algorithm for tumor segmentation. The FKM algorithm has been used to reducing the distortion [23],
where N is the number of data points, m is the fuzzifiness parameter (m = 2), k is the number of clusters and dij is the squared Euclidean distance (ED) amid the image pixels and the cluster center.
The membership ui,j must satisfy the following constraints [24],
Since the objective function depends on the Euclidean Distance (ED), it is most essential to reduce the Euclidean Distance. If the ED is minimum, then the distortions in the objective function is also minimum. FKM begins with a set of initial cluster centers chosen randomly, and no two or more clusters have the same cluster centroid. Then update the membership function using Euclidean Distance for calculating new centroids. After having K-new centroids, a new grouping must be done between the same image pixels and the nearest new cluster center. Thus, an iterative process has been performed. The K-new cluster centers change their location for every iteration until the cluster center becomes stable. In such a way, FKM clustering minimizes the ED between images pixels and cluster center. When ED is minimum, the distortion is also minimum. Hence the distortions in the objective function have been reduced. The value of membership function lies in the range of [0, 1]. In this technique, the new membership function is determined by taking the average of previous membership function values. The ED 5 has a membership value 0.65 in cluster 2 which makes us appoint it to that cluster. In addition, it has a membership value of 0.21 in cluster 1. The FKM algorithm is as follows,
Step. 1 Choose a set of initial clusters randomly and set p = 1.
Step. 2 Calculate the squared Euclidean distance dij.
Step. 3 Update the membership function ui,j using the below equation [24],
Here, \( l \ne j \). If dij< ɳ, then set ui,j= 1, where ɳ is the small positive number.
Step. 4 Compute the new set of cluster centers for each initial cluster center by using the equation given below [24],
Step. 5 If \( \left\| {C_{j} - C_{j - 1} } \right\| < \varepsilon \) for j = 1 to k, then stop the iteration. Otherwise set \( p + 1 \to p \) and repeat step 2.
The computational intricacy of step. 3 is higher than step. 4. Here, 12 iterations have been performed. During 12th iteration, it satisfies the above condition. Therefore the iteration process has been stopped.
4 Simulation Results
The proposed method has been verified on different MRI images and it has been simulated using MATLAB 2017a software. BRATS dataset has been used for this experiment. The obtained segmented results are contrasted with the tumor region given in the dataset in terms of over segmentation and under segmentation limits and are tabulated in Table 2.
The simulation results are presented in Fig. 4. In this figure, the noise removed image is represented in Fig. 4a. Figure 4b represents the contrast sharpened image, Fig. 4c represents the optimized image using CSOA, Fig. 4d represents the Fuzzy K-Means clustered image Fig. 4e represents the clustered image at 12th iteration and the final segmented image is given in Fig. 4f.
4.1 Assessment Metrics
The performance of the proposed brain tumor segmentation method has been evaluated using various measures such as sensitivity, specificity and accuracy. These parameters are described as follows,
4.1.1 Sensitivity
The ratio of actual positives which are accurately distinguished is the measure of the sensitivity. It relates to the ability to identify positive results.
4.1.2 Specificity
The ratio of negatives which are effectively distinguished is the measure of the specificity. It relates to the ability to identify negative results.
4.1.3 Accuracy
It is nothing but the precision of the proposed method. It is related to the specificity and sensitivity. It can be determined by utilizing the below equation,
4.2 Performance Analysis on Classification Stage
The performance of classification stage must be analyzed separately in order to test the efficiency of the ANN classifier. It has been analyzed by using various above mentioned parameters. The performance of the ANN classifier has been analyzed by varying the hidden neurons. The number of neurons has been used in the increasing order such as 10, 20, 30, and 40 respectively and the values are tabulated in Table 1. From Table 1, we can observe that the ANN provides better classification when the number of Hidden Neurons (HN) = 12. When the number of HN are much lesser, ANN classifier does not produce better results. It gives the sensitivity of 42.25%, specificity of 91.98% and the accuracy of 85.61% respectively.
The graphical representations of performance analysis based on hidden neurons have been illustrated in Fig. 5. When the number of HN increases, the ANN classifier produces lesser value of sensitivity, specificity and accuracy. Therefore, choosing number of neurons in ANN classifier is very important. To choose the number of neurons, Rule-of-Thump has been used. In this work, 12 number of hidden neurons have been used for classification of abnormal and normal brain images.
4.3 Performance Analysis on Segmentation Stage
In this subsection, the efficiency of the segmentation technique has been analyzed. From the segmented region, the tumor area has been evaluated and compared with the ground truth images and the values are tabulated in Table 2. The efficiency of the Fuzzy K-means clustering technique has been evaluated using Structural Similarity Index Measure (SSIM). SSIM is one of the evaluation parameter used to find the similarity among the obtained segmented result and the ground truth images. After analyzing the segmented results, we can conclude that, this method does not affect by over segmentation. This method produces the average SSIM value of 98%.
The proposed brain tumor segmentation has been compared with other existing methods such as K-Nearest Neighbor (K-NN), Computational Neural Network (CNN), Fully Computational Neural Network (FCNN) and Skull-stripping based 2D Convolutional Network (SS-2D ConvNet). The various evaluation parameters for different methods have been calculated and listed in Table 3. The graphical illustration of this comparison has been represented in Fig. 6.
From the comparative analysis, the proposed segmentation methodology achieves the maximum accuracy of 94%, SS-2D ConvNet achieves the accuracy of 91%, FCNN segmentation method produces the accuracy of 89%, CNN based segmentation produces the accuracy of 87%, and K-NN based segmentation gives the accuracy of 86%. From the obtained comparative analysis, we can clearly identify that the proposed method gives the better performance when compared with other existing tumor segmentation methodologies.
5 Conclusion
In this paper, an automated deep learning based Fuzzy K-Means clustering segmentation approach have been developed for brain tumor segmentation. This method includes four stages. Initially, the MRI images are preprocessed using wiener filter for noise expulsion. From the filtered images, the significant features are extricated by using CSOA algorithm. Then, the normal and abnormal images are classified through ANN. Finally, the fuzzy K-means algorithm has been utilized on the abnormal images to segment the tumor region. The performance of proposed strategy has been verified by utilizing different assessment parameters such as sensitivity, specificity and accuracy measures. The simulation results clearly indicates that the proposed segmentation technique accomplishes the better segmentation results with the maximum accuracy of 94%. The segmentation accuracy of 8% has been improved when compared with K-NN, 7% has been improved when compared with CNN method, 5% has been increased when compared with FCNN, and 3% has been improved with SS-2D ConvNet method. In future, we have planned to develop various types of disease segmentation and classification using different algorithms.
Change history
04 April 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s11063-024-11601-4
References
Osareh A, Shadgar B (2011) A computer aided diagnosis system for breast cancer. IJCSI Int J Comput Sci 8(2):535–545
Sapra P, Singh R, Khurana S (2013) Brain tumor detection using neural network. Int J Sci Mod Eng (IJISME) 1(9):2319–6386
Dudoit S, Fridlyand J, Speed TP (2002) Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc 97(457):77–87
Li L, Darden TA, Weinberg CR, Levine AJ, Pedersen LG (2001) Gene, assessment and sample classification for Gene expression data using a genetic algorithm and k-nearest neighbor method. Comb Chem High Throughput Screen 4(8):727–739
Khan J, Wei JS, Ringner M et al (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 7(673):679
Jong K, Mary J, Cornuejols A, Marchiori E, Sebag M (2004) Ensemble feature ranking. In: Proceedings of European conference on machine learning and principles and practice of knowledge discovery in databases
Acır N, Ozdamar O, Guzelis C (2006) Automatic classification of auditory brainstem responses using SVM-based feature selection algorithm for threshold detection. Eng Appl Artif Intell 19:209–218
Statnikov A, Aliferis CF, Tsamardinos I, Hardin D, Levy S (2005) A comprehensive evaluation of multicategory classification methods for microarray gene expression cancer diagnosis. Bioinformatics 21(5):631–643
Popuri K, Cobzas D, Murtha A, Jägersand M (2012) 3D variational brain tumor segmentation using Dirichlet priors on a clustered feature set. Int J Comput Assis Radiol Surg 7(4):493–506
Patil S, Udupi V (2012) Preprocessing to be considered for MR and CT images containing tumors. IOSR J Electr Electron Eng 1(4):54–57
Wu W, Chen AY, Zhao L, Corso JJ (2014) Brain tumor detection and segmentation in a CRF (conditional random felds) framework with pixel-pairwise afnity and superpixel-level features. Int J Comput Assis Radiol Surg 9(2):241–253
Chaddad A (2015) Automated feature extraction in brain tumor by magnetic resonance imaging using gaussian mixture models. Int J Biomed Imaging 2015:868031. https://doi.org/10.1155/2015/868031
Damodharan S, Raghavan D (2015) Combining tissue segmentation and neural network for brain tumor detection. IAJIT 12:1
Abbasi S, Tajeripour F (2017) Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing 219:526–535
Ariyo O, Zhi-guang Q, Tian L (2017) Brain MR segmentation using a fusion of K-means and spatial fuzzy C-means. In: 2017 international conference on computer science and application engineering (CSAE 2017), pp 863–873
El Abbadi NK, Kadhim NE (2017) Brain cancer classifcation based on features and artifcial neural network. Brain 6:1
Sharif M, Tanvir U, Munir EU, Khan MA, Yasmin M (2018) Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J Ambient Intell Humanized Comput. https://doi.org/10.1007/s12652-018-1075-x
Shubhangi N, Dudhane A, Murla S, Naidu S (2020) RescueNet: an unpaired GAN for brain tumor segmentation. Biomed Signal Process Control 55:101641
Saba T, Mohamed AS, Affendi M, Amin J, Sharif M (2020) Brain tumor detection using fusion of hand crafted and deep learning features. Cognit Syst Res 59:221–230
Amin J, Sharif M, Gul N, Yasmin M, AliShad S (2020) Brain tumor classification based on DWT fusion of MRI sequences using convolutional neural network. Pattern Recogn Lett 129:115–122
Deng W, Shi Q, Wang M, Zheng B, Ning N (2020) Deep learning-based HCNN and CRF-RRNN model for brain tumor segmentation. IEEE Access 8:26665–26675
Zhou C, Ding C, Wang X, Lu Z, Tao D (2020) One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Process 29:4516–4529
Emami H, Derakhshan F (2015) Integrating fuzzy K-means, particle swarm optimization, and imperialist competitive algorithm for data clustering. Arab J Sci Eng 40(12):3545–3554
Chang C-T, Lai JZC, Jeng M-D (2011) A fuzzy K-means clustering algorithm using cluster center displacement. J Inf Sci Eng 27:995–1009
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s11063-024-11601-4
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pitchai, R., Supraja, P., Victoria, A.H. et al. RETRACTED ARTICLE: Brain Tumor Segmentation Using Deep Learning and Fuzzy K-Means Clustering for Magnetic Resonance Images. Neural Process Lett 53, 2519–2532 (2021). https://doi.org/10.1007/s11063-020-10326-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-020-10326-4