Abstract
Glioma is a type of brain tumor that is the most typical and most aggressive tumor. Magnetic resonance imaging (MRI) has a widespread utilization as an imaging method for assessing the tumor; however, a lot of information obtained from MRI would prevent manual segmentation in an acceptable timeframe. Therefore, we need reliable and automatic segmentation techniques. Nonetheless, broad structural and spatial variability amongst the brain tumors made automatic segmentation a difficult issue. Capsule Network (CapsNet) is an improved convolutional neural network, which involves responding to challenges. Consequently, CapsNet has been considered to be a perfect candidate to perform brain tumor segmentation. In this paper, a new model containing CapsNet, that operates in the Del vector domain and works with inhomogeneous voxels, is presented. Flair and T1 MR images are first transformed from the time domain to the vector domain using the Del operator. MVGC (Mean Value Guided Contour) algorithm is then applied on the Flair image to segment ROI (Region of Interest). Inhomogeneous voxels are then extracted from ROI of Flair and T1 MR images. In the final phase, a new two-path CapsNet architecture is applied to classify voxels. The introduced technique is verified according to BRATS 2015 and BRATS 2013 datasets. The outputs show that this technique achieves a competitive result with the average Dice similarity equal to 0.90 for the whole tumor in Brats2013 and 0.88 for the entire tumor in Brats2015. In addition, this technique took just ~86.2 s for the segmentation of a patient case.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Precise detection and classification of brain tumors are necessary for performing an effective treatment of tumors. The stage of the tumor when is diagnosed, the pathological type of it, and the grade of the tumor are three factors that create the type and strategy of treatment [8, 38]. Gliomas have been introduced as the most occurred primary brain tumor, apparently from the glial cells, which infiltrated the around tissues. According to the World Health Organization (WHO) [3], brain tumors are classified into four grades with enhancing aggressiveness. The grade III and IV tumors have been considered the high-grade gliomas (HGG), and the rest are low-grade gliomas (LGG) [30].
Various sequences of Magnetic Resonance Imaging (MRI) would be specifically beneficial for assessing gliomas and the treatment success in clinical practices [9, 22]. Figure 1 illustrates a patient’s tumor case in a single layer with T1, T1c, T2, T2Flair MRIs.
The brain tumor segmentation task is the partitioning of a brain tumor into several segments. In general, researchers divide the brain tumors into four sections such as edema, necrosis, the enhancing tumor, and the non-enhancing tumor [ 1, 42].
Notably, the automatic or semi-automatic tumor segmentation would be a difficult issue, for which several reasons have been presented. A problem in this area is the large variation of the structure, location, and shape of the brain tumors [45]. Besides, the mass effect of the tumor changes the surrounding normal tissue arrangement [35]. Other issues like the intensity in-homogeneity [53], or various intensity ranges amongst similar sequences and acquisition scanner [41], could affect segmentation accuracy. Moreover, similar tumorous cells could finish very distinct gray-scale values pictured in various hospitals [14].
Regarding what we said before, the main problem is to segment MR Images which have Glioma tumors. To perform this task, we present an automatic algorithm that benefits Del vector space [23] characteristics, inhomogeneous voxel vise formulation, and a two-path Capsule network model. For each of these features we have a major motivation:
-
Using vector space let us extracting ROI with no more preprocess steps which reduce complexity of model and make it more general.
-
Using voxel vise approaches reduces complexity of model and let us solve the problem with more simple patterns so final deep model can be also more simple.
-
We choose CapsNet [48] as a core deep algorithm so we can handle affine transformation in similar patterns.
-
Also CapsNet based model benefits capsule layer which reduce vanishing gradient problem. Our capsule model have two paths which take into account both local and global information of decision area.
-
Our model is a rapid system to segment an MRI taking to account more than one modality of MRI and considering voxel based local and global information.
The rest of this paper is structured as follows. In Section 2 the related works are presented. In Section 3, the proposed method is described. Section 4 demonstrates the experimental outputs and Section 5 discusses the results, and limitations of the work. Finally, Section 6 concludes the paper.
2 Related works
Modern studies on brain tumor segmentation could be classified into region level segmentation (mostly unsupervised methods), pixel level segmentation (mostly supervised methods), and a combination of both of them. However, as one of the unsupervised learning methods, clustering has a widespread utilization for brain tumor segmentation due to its abilities for grouping data with given similar criteria. In addition, researchers provided one of the well-automated frameworks using the characteristics derived from the Gabor filter banks as the similarity index [37]. Regarding the feature reduction, Kaya emphasized the clustering on T1-weighted MRI images to segment the brain tumors using the Principle Component Analysis (PCA) [19]. Authors in [17] introduced their technique for a proper segmentation that combines the fuzzy c-mean algorithm with the region growing and utilizes the T1-weighted and the T2-weighted MRI images. Authors in [50] selected a fuzzy c-means cascade algorithm and applied the multimodal MR images to segment the tumor. Even though clustering has been considered to be brief and efficient, the output accuracy has not been satisfactory [20]. In paper [51], a model based on texture and kernel sparse coding from the Flair channel is introduced. The introduced method in [12] utilizes different segmentation models for various pulse sequences, fusing texture features, and an ensemble classifier to carry out three levels of the classification.
Another approach to enhance the precision of segmentation is the fusion of different modalities of medical images into one more transparent and informative image, which can help machine learning models to diagnose problems more effectively. As an example of such approaches a method is presented in [27], which uses sparse representation along with histogram similarity to effectively perform the segmentation task. Also, in [29] the authors presented a deep learning method that contains convolutional layers in order to create decompositions of two different modalities of image (for example, MRI) and fuse them using Deep Boltzmann Machine (DBM).
In [4] authors presented their newly developed strategy through Random-Forests model, which combines the voxel-wise texture with the abnormality features on a Flair MRI and contrast-enhanced T1. Authors in [44] proposed a framework in longitudinal multi-modal MRIs, utilizing two steps; the fusion of features and joint labels. Step one fuses features from stochastic multi-resolution texture, and step two mixes segmentation labels gained from the texture features combined with a random forest ensemble algorithm.
The method in [34], which could be one of the discriminant models or generative models, is a graphical model to segment the multidimensional images. In fact, context information has been utilized for smooth segmentation via MRF. In addition, the other techniques considered voxels as the independent and identical distribution and introduced Conditional Random Field (CRF) for using the neighborhood information [3, 26, 32, 33]. Because of natural capabilities in monitoring the multiclass problems and larger feature vectors, the Random Forests (RF) based methods [11, 46, 49, 54] have been introduced in brain tumor segmentation. Nevertheless, features applied for the segmentation of the images in most of techniques like gradients, brain symmetry, and first-order texture have been manually made.
Notably, significant development is observed in the deep learning, particularly in deep convolutional neural networks (DCNN). DCNN could obtain the complicated function mapping and automatically learn the features. Such a condition helps to address the complicated images. Different types of networks, including CNN and RNN have been introduced, and most of them are utilized in machine vision problems [7]. Therefore, it has a widespread usage both in natural image processing [15, 24, 31, 47] and in biological image processing [6]. Moreover, it has an extensive utilization in segmenting brain tumors [10, 16, 21, 55,56,57].
As an instance, the DCNN adopts the multi-scale [13] or cross-modality convolution [52] for extracting more plentiful features from the MRI images for tumor segmentation. The authors in [2] proposed a patch-based automated segmentation of the brain tumors using DCNN with little convolutional kernels and the leaky rectifier linear units (LReLU) as the activation function. However, the presence of little convolutional kernels allowed greater layers for forming a deeper architecture and a smaller number of kernel weights in all layers in the course of training.
The authors of [43], offered brain tumor detection based on fuzzy C-means with the super- resolution and the convolutional neural networks (CNNs) with the extreme learning machine (ELM) algorithms (SR-FCM-CNN) strategy. In [39], authors aimed to segment and grade glioma tumors using deep learning approach. They introduced a deep convolutional method that combines CNN based on the U-net architecture to deal with brain tumor segmentation task and transfer learning method on a pre-trained Vgg16 architecture and an FCN classifier for tumor grading. They used the Flair channel as an input and reported results for segmentation and classification on 110 LGG patients separately.
Paper [45] proposed an automatic segmentation method on the basis of the CNNs, which uses the little 3 × 3 kernels. Therefore, a deeper architecture has been designed using little kernels that positively affected the over-fitting concerning the fewer numbers of the weights in the network. Authors in [14] proposed a CNN that exploits more global contextual features and local features concurrently. In addition, this new network differs from a majority of the conventional applications of CNNs. It uses a final layer that has been a convolutional implementation of the fully connected layer allowing 40 fold acceleration. Furthermore, they described a 2-phase training process allowing to resolve problems associated with the imbalance of the tumor labels.
Researchers in [57] built a successful deep-learning technique via integration of the fully CNNs (FCNNs) and Conditional Random Fields (CRFs) in a united framework for obtaining segmentation outcomes with spatial consistency and appearance. They trained a deep learning-based segmentation pattern with the use of the two-dimensional image patch as well as the image slices. Another research in [18] proposes a new fabricated brain tumor segmentation technique according to a multi-cascaded convolutional neural network (MCCNN) and FCRFs. This segmentation procedure included two phases. Firstly, they designed a multi-cascaded network architecture via a combination of the intermediate outputs of numerous connected elements for considering the local dependency of the labels and utilizing the multi-scale features for coarse segmentation. Secondly, they applied the CRFs for considering spatial contextual information and eliminating some fake outcomes for segmentation. Researchers in [5] proposed a new DCNN that combines symmetry to segment brain tumors automatically. These neural networks that are also known as the Deep Convolutional Symmetric Neural Networks (DCSNN), extended the DCNN-based segmentation network via the addition of the symmetric masks in numerous layers.
Using other DCNN in [40], authors devised their network architecture known as the residual cyclic un-paired encoder-decoder network (RescueNet) using the mirroring and residual principles. RescueNet uses the un-paired opposed training for segmenting the whole-tumor and then core and enhanced areas in a brain MRI scan.
Another approach has been proposed based on the lightweight deep model in [58] called One-pass Multi-task Network (OM-Net) for solving the class imbalance that requires just one-pass calculation to segment the brain tumors. In [28], an innovative end-to-end segmentation of brain tumors was proposed that is a new fully CNN by modifying the U-Net architecture. In this model an up skip connection is applied between the encoding and decoding path. Also, an inception structure is adopted in each block of the network to help the model learn richer representations, and an efficient cascade training paradigm is used to segment brain tumor sub-regions sequentially. In opposite to patch-wise approaches, this model can automatically generate maps of segmentation for sequences of 2D slices. An automatic framework was proposed in [59] that benefits a three-stepped model: first, a 3D dense connectivity architecture is applied to create the backbone for feature extraction. Second, a novel feature pyramid module using 3D convolutional layers is used to fuse multi-scale contexts. Third, a 3D deep supervision procedure is equipped along this network to enhance the training phase.
For overcoming deficiencies and disadvantages of the CNN [7], a new machine learning structural design called the Capsule network (known as CapsNet) and is appropriate for rapid and deep learning of the data image, has been proposed in [48]. CapsNet has been considered to be powerful in the access to data transformation and rotation, while it needs fewer data-sets to train with a lower learning curve [25].
The most prominent characteristic of the Capsule Network has been known as the routing by agreement. This implies the fact that lower-level capsules could anticipate outputs of the higher level capsules. Hence, activating the capsules at higher levels corresponded to the agreement of just these anticipations. Moreover, the CapsNet learns suitable weights for the feature extraction and image categorization. In addition, it learns the way of inference of the spatial pose parameters from the images. In this regard, a capsule network would learn the determination if a plane is in the image, but also if the plane is situated to the right or left or if it is rotated or not. Such a condition has been called Equi-Variance that has been considered as one of the properties of the human 1-shot learning kind of vision [48]. Therefore, it creates a visual pattern for the more acceptable explanation of the learned features.
Based on several automatic procedures, images are segmented according to the voxel’s intensity and neighborhood data. Therefore, the mentioned methods have been flexible and provided reliable outputs. However, another groups of methods that are based on the object detection or local limits are viewed as less efficient than the earlier category of the procedures. Moreover, they enjoy lower flexibility on diverse information and various kinds of tumors.
As mentioned earlier, the present research proposes a rapid automatic framework with merits, including both groups of methods. Put differently, the proposed method utilizes features such as direction and intensity of each pixel, neighborhood information, and voxel along with the object detection approach. Therefore, for reducing space and time complexity of final CapsNet phase and providing a distribution with the greater balance for the training data, the research uses an active contour algorithm to detect the course region of the tumor. This method uses Flair image, because tumor region is lighter than other regions; hence its vector form is more suitable for ROI detection. The ROI method is introduced as one of the rapid algorithms that need no parameter adjustment.
3 Proposed method
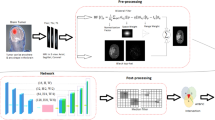
The present research introduces an automatic procedure to detect brain tumors in three-dimensional images. Figure 2 shows the block diagram of the segmentation procedure.
3.1 Preprocessing
The first step of the method prepares the MRI images for ROI extraction. The raw MRI images generally contain the artifacts like un-even brightness and other tissues; for example, skulls, eye, and so on, that diminish the precision of the outputs. Also, there are various methods to remove noise and normalize the distribution of gray levels in MR images.
Here, we utilize the features of Del vector space [23] to omit unnecessary steps of preprocessing. As we will explain in the next section, this new space has two important properties. Firstly, it is a work with finite differentiation of the input image and will give us a space, in which all values are relative and have direction, while removing the effect of different distributions of values in different images. Secondly, the Del vector space is noise resistant due to the nature of the space based on the research conducted by authors [23] who proposed this space. In addition, segmentation can be performed using this space on the image containing a high rate of noise, where our images do not have structural noise [23]. Hence, it allows us to perform the course segmentation phase (extract ROI) efficiently.
3.1.1 Transform to vector space
It is well known that Del, as a vector differential operator, has a widespread utilization in mathematics. Gradient and standard derivation have been considered two major issues in calculus, which are described respectively in applying to a 1D function and a field. Finally, with the use of the partial derivative operators in \({R}^{n}\) with the coordinate (\({x}_{1},\dots ,{x}_{n}\)) Del’s definition is as follow:
If we suppose D as one of the rectangle territories, \(F=F\left(x,y\right)i+Q\left(x,y\right)\)j could be the gradient field on D. Also, if \(\nabla\) represents the gradient, a \( U\left(x,y\right) \)would be on D, and \(F\) would be conservative [36]:
Notably, the finite differences have been considered the reasonable estimation of the derivatives in discrete cases. However, three kinds of finite differences have been introduced that include forward, central, and backward directions. Considering its benefits, experts in the field utilized the central one for transforming the images with the Del operator. Thus, as continuous mode, the presence of \(U\) that \({D}_{U}={P}_{U}i+{Q}_{U}j\) could be confirmed in the discrete cases, wherein \({P}_{U}\) and \({Q}_{U}\) would be described as follows:
here \(h\), \(k\to 0\). As seen in the digital images, the nearest neighbor of a pixel is in a distance as the same as the pixel width. On the other hand, the least possible values for \(h\), \(k\) in the digital images are equal to 1. Therefore, linear transformation conserved the homogeneous as well as additive conditions. Therefore, these conditions have been met in the central direction. Thus, if we assume \(h\), \(k=1\), the mentioned equations would be:
Therefore, the images could be converted into a vector space domain via running Eqs. (5) and (6) for all pixels individually.
3.2 Region of interest
After transforming the image to the vector space, an active contour based algorithm is employed for extracting the region of interest. It would possess no variables for adjustment, and thus, would be one of the quick techniques for selecting the image ROI. Finally, for achieving with higher accuracy, the algorithm is used on all slices of the three-dimensional images from channel T1.
To perform tumor segmentation, we use the mean value guided contour (MVGC) model that utilizes the mean value theorem for the exterior forces [23]. Then, the 3-phase repetitions are applied in methodology. Therefore, the first phase creates novel contour points based on the mean value theorem. Moreover, the second phase uses the exterior forces for guiding the points toward the object boundaries. In addition, the 3rd phase refines the contour points for addressing more precise information. In fact, MVGC is differentiated from each earlier snake formulation so that energy minimization is not formed, and it is determined immediately through the mean value theorem.
Figure 3 demonstrates an example of the ROI extraction phase on a slice of the Flair channel. Although the ROI result is not entirely match to expert’s annotation, we can see, the extracted ROI by MVGC contains all annotated regions by the expert, so we can say this method will not miss any sub-regions of the tumor.
3.3 Inhomogeneous voxels
In this step, we form the input of the proposed CapsNet as a prior knowledge. It has been already known that the tumor pixels have relations only with the neighbors. Therefore, we use this fact to define the different sizes of the voxels to give as input to the network, which is normalized between 0 and 1. Hence, it is not necessary to add an extra step to normalize data for CapsNet training and testing.
The output of the previous section is a 3D gray level image. Each pixel on the image has neighbors in six different directions: above, behind, left, right, up, and down. For each pixel in ROI, we extract 15 × 15 × 15 and 11 × 11 × 11 ordinary voxels. These voxels with the corresponding pixel in the center of them would be extracted from Flair and T1c images in the Del vector space. Then, the voxels, which did not appear in ROI, are removed. This causes the voxels to become inhomogeneous, as some voxels are complete and the rest are not. Moreover, inhomogeneous voxel architectures are chosen for two reasons. First, we aim only input voxel values of the region, which contribute to shape the ROI of each case and remove non-ROI voxels that may be noise or miss leading information. Second, we chose to size the voxels in order to have global and local information, which led us to a better deep model of classes.
3.4 Proposed capsule network
As shown in Fig. 4, the network’s inputs are 3D voxels extracted from Flair and T1 images. In order to use them, we define a new network architecture based on the CapsNet standard algorithm. Our approach uses two paths, one from local and the other from global voxels.
Concerning Fig. 4, the first path has two inputs: Flair and T1 global voxels. These 15 × 15 × 15 voxels pass through a convolutional layer consisting of 64 filters (kernels) in 5 × 5 × 5 dimensions with no zero paddings and stride 1, so the results will be 11 × 11 × 11 × 64 for each voxel. After the first convolutional layer of the global pathway, we will have 11 × 11 × 11 × 128 tensors as a result of concatenation Flair and T1 voxels. For this layer, we use ReLu activation function.
Tensors of size 11 × 11 × 11 × 128, which are produced by the first convolutional layer, will pass through the second convolutional layer using 512, 5 × 5 × 5 filters. Hence, after a reshape, we will have 7 × 7 × 7 × 1024 tensors.
In addition, the same procedure happens in the second path (named local pathway) where we use our local voxels of size 11 × 11 × 11 to pass through a convolutional layer consisting of 64 filters (kernels) in 3 × 3 × 3 dimension with no zero paddings and stride 1; therefore, the results would be 9 × 9 × 9 × 64 for each voxel. After the first convolutional layer of the local pathway, we will have 9 × 9 × 9 × 128 tensors as a result of the concatenation of Flair and T1 voxels as the same as the global pathway. At this point, like the global pathway, we use tensors from the first convolutional layer and pass them through the second convolutional layer using 512, 3 × 3 × 3 filters. Hence, after a reshape, we have a 7 × 7 × 7 × 1024 tensor as an output.
Before going to the next layer, tensors, which are produced by two pathways, are merged and become a single input for the next layer. At this point, we use the batch normalization for the first normal tensors from different sources and creating a standard view as well as reducing the risk the model overfitting.
The next level of the network is the Primary Caps layer. The layer possesses thirty-two primary capsules responsible for taking the secondary features determined by the second convolutional layer and generates a combination of the features. Notably, this layer possesses 32 primary capsules with great similarity with the convolutional layer in their nature. Hence, every capsule will apply 16, 3 × 3 × 3 convolutional kernels of the stride 1 to the 7 × 7 × 7 × 2048 input volume and consequently generates 5 × 5 × 5 × 16 output tensor. Finally, 32 capsules exist so that the shape of each output volume is 5 × 5 × 5 × 16 × 32.
The fourth layer of our CapsNet based architecture is named ClassCaps layer. This layer has five class capsules so that every class of the tumors has one capsule. Therefore, every capsule will take a 5 × 5 × 5 × 16 × 32 tensor as an input. Since each inner working of the capsule of the input vectors achieve its own 16 × 32 (16 is the dimension of each capsule of the previous section and 32 is the dimension of each ClassCaps layers capsules) of the weight matrix, they map the 16-dimensional input space to 32-dimensional capsule output space. At this level, we use dropout with the 0.25 drop rate to control overfitting.
Finally, the last layer of the proposed architecture transforms the ClassCaps layer of size 5 × 32 into a 5 × 1 layer of outputs. At this level, each row of nodes (1 × 32) associates with one corresponding output node. To produce output, we use a simple average of one row.
3.4.1 Details of the proposed model
In this section, we describe some details of the proposed network, such as loss function, training algorithm, and reconstruction of the output image.
3.4.2 Loss function
Here, the length of the instantiation vector is utilized for representing the probability of the existence of a capsule’s entity. Therefore, it is expected that the top-level capsule, the four-tumor segment class C, to had a long instantiation vector, if and only if that digit is existed on the image. Hence, for allowing for numerous classes, a distinct margin loss, \({L}_{C}\) is utilized for all class capsules C as in Eq. (7):
Where the class capsule \({v}_{c}\) has the margin \({m}^{-}=0.1\) and \({m}^{+}=0.9\). Moreover, \(\lambda\) down weighting of the loss for the absent tumor segment classes avoids the initial learning from the shrinkage of the length of the activity vectors of each class capsule. Thus, \(\lambda\) = 0:5 has been used, and total loss is easily the sum of the losses of each class capsule [39].
For understanding the key notion of the way of working the loss function, it should be remembered that the output of ClassCaps layer is 5 × 32 (or 5 × 64) dimensional vectors. However, in the course of training, for all training examples, a loss value would be computed for all five vectors based on Eq. (7), then the five values would be added for the calculation of the resulting loss. As a result of the discussion of the supervised learning, all training examples would possess the true label, and thus, it would be a five-dimensional 1-hot encoded vector with four zeros and one at the true positions. According to the loss function formula, the true label determines \({T}_{C}\) value, and it will be one if the true label corresponds with the class of this specific ClassCap and otherwise it will be zero.
3.4.3 Training process
In order to train the network, we use 20 epochs of the training process using standard Adam optimizer. We also utilize the early stopping method to avoid over- and under-fitting. Based on the model of input data, we apply the data augmentation phase, which contains two different methods and hybrid usage of them; first, the method flips an image of the random axis and the second one adds random noise of 10-20% to the image. The usage of the hybrid method allows to have a dataset around four times larger than the original dataset. In addition, we generally set the batch size equal to 75% of the training data (after holding out the validation set). It was memory-consuming, but the results show excellent results in much smaller epochs. The starting learning rate is 0.01 as it works almost excellent in lots of training processes. Figure 5 shows an example of the training process for the Brats2013 dataset based on the mentioned setting.
4 Results
To evaluate the proposed method, we utilize two datasets, Brats2013 and Brats2015. In the following sections the obtained results on these two datasets are presented and discussed.
4.1 Results on Brats 2013
The first set of experimentations is performed on the real patients’ data in 2013 and the brain tumor segmentation challenge (BRATS2013) as a part of the MICCAI meeting [35]. In fact, BRATS2013 dataset contains three sub-datasets. Training dataset contains 30 patients with the pixel-accurate ground truth (10 LG &20 HG tumors). In order to train the model, we use 75% of images for training (voxels of images) and 25% for validation. After augmentation, we have 120 MR images (including original images).
Moreover, the test dataset contains 10 (all HG tumors), and the leaderboard dataset consists of 25 patients (4 LG and 21 HG tumors). It should be mentioned that any ground truth is not presented for leaderboard and test datasets. Each brain in the dataset had similar orientations. Therefore, for all brains, four modalities of T1C, T1, Flair, and T2 are observed that are co-registered. Consequently, the training brains come with the ground truth, for which five labels of segmentation are created, such as necrosis, non-tumor, non-enhancing and enhancing tumors, and edema. Figure 6 displays instance data, ground truth, and segmentation results of the model.
It is possible to quantitatively evaluate the model functions on the test-set via uploading segmentation outputs to the online BRATS evaluation system [50]. This system provides quantitative outputs. For example, the structure of the tumors are classified into three distinct tumor regions, which is fundamentally caused by functional clinical utilizations. Authors in [35] defined the tumor regions as:
-
1.
A complete tumor region such as each of the four tumor structures,
-
2.
A core tumor region such as each tumor structure with the exception of edema, and.
-
3.
An enhancing tumor region such as the structure of the enhanced tumor.
Therefore, for each tumor region, the Dice (equal to F measure), Sensitivity as well as Positive Predictive Value (PPV) are computed as follows:
Here P stands for the model prediction. T refers to the ground truth label. In addition, \({T}_{pos}\) and \({T}_{Neg}\) stand for the sub-set of the voxels anticipated as positive and negative for tumor regions under study. A similar definitions for \({P}_{pos}\) ,\({ P}_{Neg}\) and P are used. Moreover, the Dice Coefficient represents overlapping between the automatic, and manual segmentation. Sensitivity refers to the rate of true positive (tumor regions, based on a, b, and c definition, which are mentioned above) predictions compared to the exact number of the tumor pixels. Specificity measures the rate of true negative compared to the exact number of the non-tumor pixels. Finally, PPV indicates the rate of the true declaration of the tumor regions in comparison with the declaration of the tumor regions (true or false). The above values equaled 0 to 1 and greater value implied the segmentation with higher accuracy. Besides, we can say, that dice shows how much we reach the results similar to the human expert segmentation, Sensitivity indicates that how precise we detect positive classes (tumor classes). In addition to two previous metrics, PPV shows the impact of miss classification of positive areas.
The online evaluation mechanism provides a rank for all methods introduced for assessment, which includes the techniques from 2013 BRATS challenge reported by [35] and the anonymized un-published techniques, for which any references are not provided.
The segmentation outputs obtained from a sample brain in an axial view are shown in Fig. 7. The results of the proposed model are visualized in Fig. 8 on the same subject as Fig. 7 from BRATS-2013 (from different meaningful slices to clarify the issue further). Figure 8 depicts the outputs in the axial views. The results show that the proposed model improves all four measures on each tumor region. Based on the proposed architecture, this study could gain the third rank on BRATS 2013 scoreboard.
Tables 1 and 2 present the numerical and detailed results of the proposed method. In order to achieve this result, we evaluated our method on HG and LG images, separately. Both tables show numerical evaluation results on average. The main reason behind this result would be the feature extraction and learning phase of the CapsNet model, where the proposed model uses two parallel convolutional layers that merge the first-order features and made a more suitable one. In addition, the size of the convolutional layers helps the model to have a better insight into the problem.
To train the proposed network, an Nvidia 2080ti GPU, 64GB of RAM and an Intel Xeon E7-8860 CPU are used. The training phase takes 68 h for 20 epochs (each epoch contains 75%) of training data. The inference time in the testing phase is 21ms for each set of input voxels. Based on the training and inference details, the proposed model needs approximately 86.2 s to segment a 3D MR image. Results and the experiments on BRATS datasets indicate that the proposed model needs 77.28 s to classify and reconstruct voxels (368 set of voxels in average) and almost 9 s for ROI detection phase (in a 3D MR image).
The assessment outputs of the top-ranked participants of BRATS2013 and outputs of the present research technique and the other six latest methods are summarized in Table 3. Based on the testing datasets ranking, the presented method in [45] reaches the first rank and [57] achieves the second rank. Finally, our procedure achieves the third rank. Therefore, any approach cannot generate the optimum segmentation outputs for each sub-tumoral region. Nevertheless, the proposed method achieves the most important and competitive performance in comparison with the latest strategies.
As reported in Table 3, the outputs of the proposed method are considerably superior to the proposed technique in [14], except for a partly lower PPV-value of the complete tumor regions.
Additionally, in [57] the brain tumor segmentation technique of combining the CRFs and FCNNs is presented. A distinct point in their research is that the present investigation employs two-level segmentation procedure, hence it could obtain the competitive performance and saves the time overhead. Therefore, they possess a training duration of approximately 12 days, while our process lasts nearly three days on a typical GPU server. As seen in Table 3, our method outputs are partially less than the results of the proposed method in [57] in a number of indices because we just used segmentation without any extra post-processing unit and they performed some complicated post processing operations.
Moreover, though their post processing technique had higher performance, it has mainly been hand-made and could not be readily transported to other associated segmentation tasks.
Based on what is mentioned above, the proposed method in [18], inspired by [14] and [57], also shows better results in some measures like the Dice of complete tumor. However, like [57], this work also uses a complex post-processing phase, which may be challenging be implemented on other tasks, and it has a time and space consuming process, including the multi cascade training and fusion of different directions of the segmentation results. Moreover, as the study conducted by Zhao et al. [57], their published research segmentation results has no post processing on BRATS 2013 testing dataset for the core, complete, and the enhancing tumor equal 0.81, 0.88, and 0.76 for Dice, 0.86, 0.81, and 0.69 for PPV as well as 0.90, 0.84, and 0.86 for Sensitivity. However, this study outputs, as they are shown in Table 3, are better than the presented method in [18] in most parameters.
Table 4 illustrates comparison results on the BRATS 2013 leaderboard dataset. Among the most advanced techniques, which are presented in Table 3, three of them plus (Zhou et al., 2020) [59] reported results for the leaderboard dataset. Results illustrate that our method has around 0.86 for Dice score in the case of complete tumor, and it is almost equal to the results reported in [57]. Moreover, for the sensitivity parameter, our method shows 0.78 as the best answer among other works. In other cases, the performance of the proposed method is among the top two in the mentioned results. Considering the time and space complexity of the other methods, our work demonstrates acceptable performance.
Finally,
Table 5 shows the comparison between the detailed results of BRATS 2013. As we can see, our method works better than [4] in all sub-classes.
4.2 Results on Brats 2015
The second set of experiments are performed on the real patients’ data achieved by the brain tumor segmentation challenge (BRATS2015), as a section of the MICCAI seminar [35]. BRATS2015 dataset contains two sub-datasets. This training dataset consists of 274 patients with the pixel-accurate ground truth (220 HG and 54 LG tumors). Same as Brats 2013, we use data augmentation to generate a larger dataset (4 times bigger) for the training process and use a similar portion of the augmented training data (75% for training and 25% for validation) for the training process.
The test dataset contains 110 cases of the high grade and low grade, though the grade has not been shown. For all brains, four co-registered modalities of T1C, T1, Flair, and T2. The training brains come with the ground truth, for which five segmentation labels have been developed that are necrosis, non-tumor, non-enhancing tumor, enhancing tumor, and edema. For evaluation, four anticipated labels would be merged into diverse sets of the whole tumor (each of the four classes of tumor), enhancing tumor (class 4), and core (classes 1,3,4). The datasets were subsequently pre-processed by organizers, and presented as the skull-stripped, recorded to a shared space and re-sampled to the isotropic one millimeter resolution. Finally, each volume has the size 240 × 240 × 155.
The experimental outputs are presented for our distinct CapsNet architecture on BRATS 2015. Table 6 demonstrates the numerical and detailed results of the proposed method. In order to achieve this result, we evaluated our method on all images from the test dataset of BRATS 2015. Table 6 illustrates the numerical evaluation results on average. As we can see, like experiments on BRATS 2013, the first architecture outperformed the second one in all three sub-compartments. The results are visualized in Fig. 9, which shows how our model works in 3D and 2D manners.
The proposed method has been compared with some other researches based on the BRATS2015 test dataset. Concerning Table 7, assessment outputs of the top-ranked participants and outputs of the proposed method as well as other eight latest strategies, are provided. According to the ranks for the testing dataset, the presented method in [28] ranked the first, [2] ranked the second, [18] achieved the third rank, and our proposed method achieved the fourth rank (Dice score). None of the approaches could generate optimum segmentation outputs for each sub-tumoral region. Nevertheless, it is shown that our method achieves the most important and competitive performance in comparison with the latest strategies. Finally, the proposed method achieves the most reasonable values for Sensitivity of the complete tumor region and DSC of the complete tumor.
5 Discussion
The present research assesses one of the newly designed CapsNet-based segmentation methods to delineate various brain tumor compartments that have been started from extracting ROI in the vector space using the contrast-enhanced T1-weighted MRI followed by extracting the inhomogeneous voxels from T1c and Flair channels. Finally, it uses two paths of the CapsNet based deep neural network to segment the images. As far as we know, a few brain tumor segmentation methods (like [4]) have been provided with just two MRI sequences. For example, multiple investigations illustrated complementary of the T1- and T2-weighted scans to segment the brain tumor. Deep convolutional neural network-based methods like [2, 14, 57] use all four channels of MRI, and in the special case, [18] in addition to four channels, uses super slices from three different orientations.
We use Del vector space to process MRI. In order to use vector space, we transform T1c using transform formulation presented by [23]. MVGC, presented by [23], is then used to perform the first segmentation on the slices of transformed T1c and extracted ROI. After this phase, we use this ROI and extracted inhomogeneous voxels to prepare the final data for the final part of the method. Finally, we use two CapsNet based architectures to classify the voxels and reconstruct input MRI.
According to the studies, the latest techniques on brain tumor segmentation fundamentally use deep learning strategies like CNNs [14, 18, 32, 45, 54, 57]. Several investigations obtained the Dice-scores estimating 90% with 4 MRI sequences (T1, T1c, Flair, T2). Mentioned techniques require a lot of annotated data and robust hardware to train. The CNNs took one image as the input and return a label or the segmentation mask as the output. Moreover, a complicated network of the hidden layers exists between them, which could be pooling, convolutional, activation, or the fully connected layers having numerous weights, and should be adjusted in the course of training. In fact, deep learning for the segmentation could be categorized into two distinct strategies. According to the first strategy, the features have been extracted from a local patch for every voxel that used the convolutional layers. After that, the features have been divided with a FCNN for obtaining a label for each voxel. The second strategy uses the FCNs like the U-Net [10], in which local information has been combined with the upsizing and downsizing step.
With respect to image fusion, our method can be considered in this category, unlike [27, 29] which does image fusion explicitly, our method using deep architecture method fuse two modalities of MRI implicitly and benefits new space which has suitable features of both input images.
Our technique applies the same steps as the first strategy, where we use CapsNet as an improvement for CNN, which could learn features better and result better in fewer depth networks. Nonetheless, it is assumed that it is possible to improve accuracy with the deep learning that has been investigated in the follow-up. Therefore, visual inspection of the current outputs frequently shows over-segmentation, is a healthy tissue allocated a tumor label, causing the lower Dice score. The reason behind over segmentation is the first MVGC phase, we narrow the task down to ROI for declining the space and time complicatedness but our method extracts the non-tumor regions as an ROI and left us with unbalanced data (ROIs contain a few non-tumor pixels). Consequently, the reduced performances are seen on the test-set in comparison with the training set, reflecting the model over-fitting, even though we explored the methods for avoiding dropout layer as well as batch normalization. As a result, it is possible to improve this new procedure via modeling the bigger spatial context of a single voxel in a highly developed manner. Another limitation of our work came from the lack of mathematical and computational sources to input vector space images directly to the network. Due to the limitation of Keras and Tensorflow frameworks and large dimensionality of the nature of our method, using direct vector space input would cause the input size to be four times larger than what it is.
6 Conclusion and future work
A novel automated brain tumor segmentation algorithm has been introduced by the present research and its validity has been confirmed. Brats 2013 and Brats2015, containing the Flair MRI and contrast-enhanced T1, has been utilized to create and evaluate the method. In fact, the CapsNet based method using the inhomogeneous voxels alongside preprocessing in the Del vector space for ROI extraction has been utilized to perform the brain tumor segmentation. Results showed a performance of 0.89 for Dice score in Brats 2013 and 0.87 for Dice score for Brats 2015 (whole tumor); thus, our method performance could compete with the latest deep learning techniques.
The main reason behind this model is to examine three theories; First using vector space-based active contour model can effectively segment ROI, which is achieved based on visualized and numerical results. Second, using 3D inhomogeneous patches would help to capture more information about a pixel. It will lead us to better segmentation considering various places which each class of tumor cells is placed. In this case, results show our method at least reach comparable results. Finally, we check that using two channels of MRI in a two-path multilevel CapsNet based deep neural network can properly learn the pattern of tumor cells, which is a success where our results outperform similar CNN-based architectures.
Respect to image fusion, unlike [12, 51] which do image fusion explicitly, our method uses deep architecture method to fuse two modalities of MRI implicitly and benefits new space which have suitable features of both input images.
The proposed method suffers from some limitations: although in standard tests ROI detection phase demonstrates noticeable results, but if this phase causes error it will effect final segmentation results. Also, as we mentioned in time complexity result, the network is light and it is faster than most of state of the art models, but converting an MRI to voxels and reconstruction causes some overheads in computation. Finally, do not explicitly taking whole ROI or MR image into the account would cause information loss, while directly effects system precision.
To continue the research, the fellow researchers are suggested to complete architecture by adding post-processing modules for reducing the amount of the false positive rate. Moreover, modification of the deep learning method, which could get an image from the vector space as in input, could change the result dramatically because of the embedded information in the vector space images.
References
Aslian H, Sadeghi M, Mahdavi R (n.d.) Magnetic resonance imaging-based target volume delineation in radiation therapy treatment planning for brain tumors using localized region-based active contour. Radiat Oncol Biol 87:195–201. https://doi.org/10.1016/j.ijrobp.2013.04.049
Bal A, Banerjee M, Sharma P, Chaki. R, Multi-class A (2020) Image classifier for assisting in tumor detection of brain using deep convolutional neural network. Springer, Singapore. https://doi.org/10.1007/978-81-322-2653-6
Bauer S, Nolte L, Reyes M (2011) Fully Automatic Segmentation of Brain Tumor Images Using Support Vector Machine Classification in Combination with Hierarchical Conditional Random Field Regularization. In: Int. Conf. Med. Image Comput. Comput. Interv., Springer, Berlin, Heidelberg, pp 354–361
Bonte S, Goethals I, Van Holen R (2018) Machine learning based brain tumour segmentation on limited data using local texture and abnormality. Comput Biol Med 98:39–47. https://doi.org/10.1016/j.compbiomed.2018.05.005
Chen H, Qin Z, Ding Y, Tian L, Qin Z (2019) Brain tumor segmentation with deep convolutional symmetric neural network. Neurocomputing. https://doi.org/10.1016/j.neucom.2019.01.111
Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J (2012) Deep neural networks segment neuronal membranes in electron microscopy images. In Adv. Neural Inf. Process. Syst., pp 2843–2851
Dargan S, Kumar M, Ayyagari MR, Kumar G (2020) A survey of deep learning and its applications: a new paradigm to machine learning. Arch Comput Methods Eng 27:1071–1092. https://doi.org/10.1007/s11831-019-09344-w
Deepak S, Ameer PM (2019) Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med 111:103345. https://doi.org/10.1016/j.compbiomed.2019.103345
Devi CN, Chandrasekharan A, Sundararaman VK, Alex ZC (2015) Neonatal brain MRI segmentation: A review. Comput Biol Med 64:163–178. https://doi.org/10.1016/j.compbiomed.2015.06.016
Dong H, Yang G, Liu F, Mo Y, Guo Y (2017) Automatic brain tumor detection and segmentation using U-net based fully convolutional networks. Commun Comput Inf Sci 723:506–517. https://doi.org/10.1007/978-3-319-60964-5_44
Geremia E, Menze B, Ayache N, Geremia E, Menze B, Ayache N, Adaptive S, Forest R (2013) Spatially adaptive random forest. In: IEEE 10th Int Symp Biomed Imaging, IEEE, pp 1344–1347
Gupta N, Bhatele P, Khanna P (2019) Glioma detection on brain MRIs using texture and morphological features with ensemble learning. Biomed Signal Process Control 47:115–125. https://doi.org/10.1016/j.bspc.2018.06.003
Havaei M, Dutil BF, Chris P, Hugo L, Jodoin P (2015). In: BrainLes (ed) A Convolutional Neural Network Approach to Brain Tumor Segmentation. Springer, Cham, pp 195–208. https://doi.org/10.1007/978-3-319-30858-6
Havaei M, Davy A, Warde-farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proc IEEE Conf Comput Vis Pattern Recognit, pp 770–778
Hoseini F, Shahbahrami A, Bayat P (2018) An Efficient Implementation of Deep Convolutional Neural Networks for MRI Segmentation. J Digit Imaging 31:738–747. https://doi.org/10.1007/s10278-018-0062-2
Hsieh TM, Liu YM, Liao CC, Xiao F, Chiang IJ, Wong JM (2011) Automatic segmentation of meningioma from non-contrasted brain MRI integrating fuzzy clustering and region growing, BMC Med Inf Decis Mak 11. https://doi.org/10.1186/1472-6947-11-54
Hu K, Gan Q, Zhang Y, Deng S, Xiao F, Huang W, Cao C, Gao X (2019) Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 7:92615–92629. https://doi.org/10.1109/ACCESS.2019.2927433
Irem Ersöz K, Ayça Çakmak P, Sekizkardes EG, Ibrikci T (2017) PCA based clustering for brain tumor segmentation of T1w MRI images, Comput. Methods Programs Biomed 140:19–28. https://doi.org/10.1016/j.cmpb.2016.11.011
Juan-Albarracín J, Fuster-Garcia E, Manjón JV, Robles M, Aparici F, Martí-Bonmatí L (2015) García-Gómez, Automated glioblastoma segmentation based on a multiparametric structured unsupervised classification. PLoS ONE 10:1–20. https://doi.org/10.1371/journal.pone.0125143
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78. https://doi.org/10.1016/j.media.2016.10.004
Kanas VG, Zacharaki EI, Davatzikos C, Sgarbas KN, Megalooikonomou V (2015) A low cost approach for brain tumor segmentation based on intensity modeling and 3D Random Walker, Biomed. Signal Process Control 22:19–30. https://doi.org/10.1016/j.bspc.2015.06.004
Kiaei AA, Khotanlou H (2017) Segmentation of medical images using mean value guided contour. Med Image Anal 40:111–132. https://doi.org/10.1016/j.media.2017.06.005
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In Proceeding of Conference on Neural Information Processing Systems (NIPS2012), pp 1106–1114
Kruthika KR, Maheshappa HD (2018) CBIR system using capsule networks and 3D CNN for Alzheimers disease Diagnosis. Inform Med Unlocked. https://doi.org/10.1016/j.imu.2018.12.001
Lee C, Wang S, Murtha A, Brown MRG, Greiner R (2008) Segmenting brain tumors using pseudo – Conditional random fields. In: Int. Conf. Med. Image Comput. Comput. Interv., Springer, Berlin, Heidelberg, pp 359–366
Li Y, Lv Z, Zhao J, Pan Z (2019) Improving performance of medical image fusion using histogram, dictionary learning and sparse representation. Multimed Tools Appl 78:34459–34482. https://doi.org/10.1007/s11042-019-08027-9
Li H, Li A, Wang M (2019) A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput Biol Med 108:150–160. https://doi.org/10.1016/j.compbiomed.2019.03.014
Li Y, Zhao J, Lv Z, Li J (2021) Medical image fusion method by deep learning. Int J Cogn Comput Eng 2:21–29. https://doi.org/10.1016/j.ijcce.2020.12.004
Liang Z-P, Lauterbur PC (2000) Principles of magnetic resonance imaging- a signal processing perspective. SPIE Optical Engineering Press, Bellingham
Lin T, Doll P, Girshick R, He K, Hariharan B, Belongie S, Ai F, Tech C (2017) Feature pyramid networks for object detection. In: Proc. IEEE Conf. Comput. Vis. Pattern Recognit, pp 2117–2125
Meier R, Bauer S, Slotboom J, Wiest R, Reyes M (2013) A hybrid model for multimodal brain tumor segmentation. Multimodal Brain Tumor Segmentation 31:31–37
Meier R, Bauer S, Slotboom J, Wiest R, Reyes M (2014)Appearance-and context-sensitive features for brain tumor segmentation. Proc MICCAI BRATS Chall. https://doi.org/10.13140/2.1.3766.7846
Menze BH, Van Leemput K, Lashkari D, Weber MA, Ayache N, Golland P (2010) A generative model for brain tumor segmentation in multi-modal images. Lect Notes Comput Sci (Including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 6362 LNCS 151–159. https://doi.org/10.1007/978-3-642-15745-5_19
Menze BH, Jakab A, Bauer S, Kalpathy-cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber M, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin H, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M (2015) The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Mercer PR (2014) More calculus of a single variable. Springer, Berlin
Mirajkar G, Barbadekar B (2010) Automatic segmentation of brain tumors from MR images using undecimated wavelet transform and gabor wavelets. 2010 IEEE Int Conf Electron Circuits Syst ICECS 2010 - Proc 702–705. https://doi.org/10.1109/ICECS.2010.5724609
Mohan G, Subashini MM (2018) MRI based medical image analysis: Survey on brain tumor grade classification. Biomed Signal Process Control 39:139–161. https://doi.org/10.1016/j.bspc.2017.07.007
Naser MA, Deen MJ (2020) Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput Biol Med 103758. https://doi.org/10.1016/j.compbiomed.2020.103758
Nema S, Dudhane A, Murala S, Naidu S (2020) Biomedical signal processing and control RescueNet: An unpaired GAN for brain tumor segmentation. Biomed Signal Process Control 55:101641. https://doi.org/10.1016/j.bspc.2019.101641
Nyúl LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19:143–150
Odland A, Server A, Saxhaug C, Breivik B, Groote R, Vardal J, Larsson C, Bjørnerud A (2014) Volumetric glioma quantification: comparison of manual and semi-automatic tumor segmentation for the quantification of tumor growth. Acta Radiol 56:1396–1403. https://doi.org/10.1177/0284185114554822
Özyurt F, Sert E, Avcı D (2020) An expert system for brain tumor detection: Fuzzy C-means with super resolution and convolutional neural network with extreme learning machine. Med Hypotheses 134. https://doi.org/10.1016/j.mehy.2019.109433
Pei L, Bakas S, Vossough A, Reza SMS, Davatzikos C, Iftekharuddin KM (2020) Longitudinal brain tumor segmentation prediction in MRI using feature and label fusion. Biomed Signal Process Control 55:101648. https://doi.org/10.1016/j.bspc.2019.101648
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35:1240–1251. https://doi.org/10.1109/TMI.2016.2538465
Pinto A, Correia H, Oliveira J, Rasteiro DMLD, Silva CA (2015) Brain tumour segmentation based on extremely randomized forest with high-level features. In: 37th Annu. Int. Conf. IEEE Eng Med Biol Soc. IEEE, pp 3037–3040
Ronneberger O, Fischer P, Brox T (2015) Convolutional networks for biomedical image segmentation. In: Int. Conf. Neural Inf. Process. Springer, Cham, pp 174–181
Sabour S, Hinton GE (2017) Dynamic routing between capsules. In proceeding of Conference on Neural Information Processing Systems (NIPS 2017), pp 3856–3866
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X (2017) Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assist Radiol Surg 12:183–203. https://doi.org/10.1007/s11548-016-1483-3
Szilágyi L, Lefkovits L, Barna IL, Iclanzan D, Benyó B (2015) Automatic brain tumor segmentation in multispectral MRI volumetric records. Int Conf Neural Inf Process 174–181. https://doi.org/10.1007/978-3-319-26561-2_21
Tong J, Zhao Y, Zhang P, Chen L, Jiang L (2019) MRI brain tumor segmentation based on texture features and kernel sparse coding. Biomed Signal Process Control 47:387–392. https://doi.org/10.1016/j.bspc.2018.06.001
Tseng K, Lin Y, Hsu W, Huang C (2017) Joint sequence learning and cross-modality convolution for 3D biomedical segmentation. In: Proc IEEE Conf Comput Vis Pattern Recognit
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: Improved N3 Bias Correction. IEEE Trans Med Imaging 29:1310–1320
Tustison NJ, Max KLS, Christopher W, Kandel BM, Gee JC, Grossman MC, Avants BB (2015) Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (Simplified) with ANTsR. Neuroinformatics 13:209–225. https://doi.org/10.1007/s12021-014-9245-2
Zhao L, Jia K (2015) Deep feature learning with discrimination mechanism for brain tumor segmentation and diagnosis. In: Int. Conf. Intell. Inf. Hiding Multimed. Signal Process., IEEE, pp 306–309. https://doi.org/10.1109/IIH-MSP.2015.41
Zhao X, Yihong W, Guidong S, Li Z, Yong F, Yazhuo Z (2016) Brain tumor segmentation using a fully convolutional neural network with conditional random fields. Int. Work. Brainlesion Glioma, Mult. Sclerosis, Stroke Trauma. Brain Inj. Springer, Cham, pp 75–87
Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111. https://doi.org/10.1016/j.media.2017.10.002
Zhou C, Ding C, Wang X, Lu Z, Tao D (2020)One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Process 29:4516–4529. https://doi.org/10.1109/TIP.2020.2973510
Zhou Z, He Z, Shi M, Du J, Chen D (2020) 3D dense connectivity network with atrous convolutional feature pyramid for brain tumor segmentation in magnetic resonance imaging of human heads. Comput Biol Med 103766. https://doi.org/10.1016/j.compbiomed.2020.103766
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aminian, M., Khotanlou, H. CapsNet-based brain tumor segmentation in multimodal MRI images using inhomogeneous voxels in Del vector domain. Multimed Tools Appl 81, 17793–17815 (2022). https://doi.org/10.1007/s11042-022-12403-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-022-12403-3










 edema
edema  enhanced tumor
enhanced tumor  necrosis
necrosis  non-enhanced tumor
non-enhanced tumor 

