Abstract
Automated tumor characterization has a prominent role in the computer-aided diagnosis (CAD) system for the human brain. Despite being a well-studied topic, CAD of brain tumors poses severe challenges in some specific aspects. One such challenging problem is the category-based classification of brain tumors among glioma, meningioma, and pituitary tumors using magnetic resonance imaging (MRI) images. The emergence of deep learning and machine learning algorithms have addressed image classification tasks with promising results. But an associated limitation with the medical image classification is the small sizes of medical image databases. This limitation, in turn, limits the availability of medical images for training deep neural networks. To mitigate this challenge, we adopt a combination of convolutional neural network (CNN) features with support vector machine (SVM) for classification of the medical images. The fully automated system is evaluated using Figshare open dataset containing MRI images for the three types of brain tumors. CNN is designed to extract features from brain MRI images. For enhanced performance, a multiclass SVM is used with CNN features. Testing and evaluation of the integrated system followed a fivefold cross-validation procedure. The proposed model attained an overall classification accuracy of 95.82%, better than the state-of-the-art method. Extensive experiments are performed on other MRI datasets for the brain to ascertain the improved performance of the proposed system. When the amount of available training data is small, the SVM classifier is observed to perform better than the softmax classifier for the CNN features. Compared to transfer learning-based classification, the adopted strategy of CNN-SVM has lesser computations and memory requirements.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Computer-aided diagnosis (CAD) of diseases has made considerable progress over the years. Improved imaging techniques, better image processing tools, advanced machine learning theories, the advent of deep learning concepts have made it possible. Automatic disease detection, disease category classification, medical image retrieval based reference, disease progress monitoring are popular research topics in medical image processing and analysis. The conventional approaches to disease detection and disease classification problems using medical images adopted various feature extraction and classification algorithms. Finding the most suited combination of features and a classifier is considered as a challenging task. The choice of this combination often relies on heuristics.
To some extent, the adoption of deep learning architecture, namely the convolutional neural network (CNN) addresses this issue. A CNN model acts as a combined unit, consisting of a feature extractor and a classifier. Currently, there is a significant interest in the implementation of CAD systems using CNN. The CAD applications using CNNs have been highly successful, achieving remarkable results. Recent work applies CNN on histological images for breast cancer classification into benign and malignant (Bardou et al. 2018). The work is extended to multiclass problems as well. One work formulates and explains tissue characterization as part of lung disease diagnosis (Anthimopoulos et al. 2016). CNN with modified activation functions forms the crux of the design in this work. The paper provides details on the tuning of hyper-parameters, and exhaustive analysis of results obtained. A comparison with the state-of-the-art method reveals the eminence of the designed architecture. The challenges faced during the design and implementation of automated CAD systems using CNN are diverse. First, the design choice of layers in CNN determines the feature extraction capabilities of the CNN. Second, the available amount of training data (medical images) determines the generalization capability of CNN on the data. The above factors, in turn, determine the accuracy of the classification of test data by CNN. Hence different strategies are considered for the classification problems.
A leading strategy that has gained popularity is the use of pre-trained deep networks via transfer learning. Researchers have designed and developed deep neural networks for the classification of natural images. Such deep networks can classify images in other domains where data is limited using transfer learning. A fine-tuned GoogLeNet was successfully employed to analyze computed tomography (CT) scans for the lung cancer detection problem (Fang 2018). A transfer learned AlexNet model was fine-tuned to detect pathological brain magnetic resonance imaging (MRI) images (Lu et al. 2019). Tavakoli et al. (2019) used CNN features to detect abnormalities in mammograms. Agarwal et al. (2018) fine-tuned the ResNet-50 CNN architecture for their study on mammograms. The authors conducted classification experiments for mass detection in mammograms and the classification of mammograms with masses into benign and malignant.
Improving accuracies in classification by combining CNN features with proven classifiers is another strategy to address the practical challenges. The ensemble arrangement of CNN for feature extraction and a separate classifier has been tested successfully on various applications. The deep features extracted from hyperspectral images were used for classification by a support vector machine (SVM) (Liu et al. 2017). The ensemble arrangement of CNN (for features) and SVM, resulted in a better performance than with the sole use of the CNN model having a softmax classifier (Bardou et al. 2018). Combining two machine learning techniques has proved useful in the field of bioinformatics (Cao et al. 2020). Classification of extensive volumetric data in bioinformatics could further be improved using meta-heuristic approaches (Sampathkumar and Vivekanandan 2019) and advanced optimization algorithms (Sampathkumar et al. 2020). The hybrid CNN-SVM models were successfully used to analyze the biosignals, electroencephalogram (EEG) for epileptic seizure prediction (Agarwal et al. 2018). Use of a long short term memory (LSTM) model further improved the EEG classification performance (Nagabushanam et al. 2019). Xue et al. (2016) applied the idea of combining CNN with SVM to their problem on microvascular morphological classification problem for oesophagal cancer detection. Despite providing an augmented training data, the CNN-SVM combination achieved higher accuracy than that produced by the stand-alone CNN classifier. However, there have been instances where this strategy was not useful. CNN (with softmax layer) was able to achieve a higher test accuracy than the hybrid CNN-SVM for the multi-class classification problem on MNIST and Fashion MNIST datasets (Agarap 2017). Notably, the amount of available training data was large in these instances.
This paper focuses on CAD systems for brain tumors. Computer-aided health analysis of the human brain is a well-researched topic and is still active. The availability of several open datasets for brain images has been supporting the research. The studies on brain tumor commonly use MRI images considering the soft tissue representing capability of MRI. Studies have proposed different methods for fully automated and semi-automatic detection of tumors in the brain from MRI images (Mahesh and Renjit 2018; Abbasi and Tajeripour 2017). The fully automated tumor detection algorithms extract features from MRI images, and then the features are used for classification into normal and tumorous. The semi-automatic tumor detection procedures require manual selection of the suspected tumor region to form the region of interest (ROI). The tumor detection relies on the analysis of the ROI. J. Amin et al. (2017) used an ensemble of features, including texture, shape, and intensity. The features were used to classify MRI images into cancerous and non-cancerous using SVM classifier. The researchers used more than one dataset to evaluate their experiments. The results were further improved by the preprocessing of images before feature extraction (Amin et al. 2018). The preprocessing included image enhancement using histogram matching and a segmentation technique for skull removal. Seetha and Raja (2018) proposed a CNN based classification for images with tumors and images without tumors. The deep networks could combine the feature extraction and classification steps of the tumor detection problem. Another class of fully automated tumor detection methods uses different clustering algorithms to partition the brain MRI into different regions. The segmented regions become the ROI for further analysis (Abbasi and Tajeripour 2017).
Tumor segmentation is a research problem that is closely associated with tumor detection problems. Many tumor detection algorithms follow a two-stage analysis. In the first stage, brain tumor images are classified into normal and tumorous. In the second stage, segmentation of tumor regions is performed on the tumorous images. Rajagopal (2019) considered the research problem of brain tumor detection and segmentation, specifically for the case of glioma diseases in their work. The researchers used a combination of grey level co-occurrence matrix (GLCM) features and random forest (RF) classifier to detect glioma in brain MRI images. The glioma images were then segmented using morphological operations. Nabizadeh and Kubat (2015) proposed an algorithm for brain tumor detection and segmentation in MRI images using Gabor features and an SVM classifier. The classification was done for each pixel in the images. The pixel-level classification forms the concept of semantic segmentation. Semantic segmentation plays a significant role in recent research works on brain tumor segmentation. The performance of semantic segmentation improves significantly if deep learning architectures could be applied. Dong et al. (2017) adopted the UNET architecture for brain tumor detection and segmentation. Their design choice of Soft Dice Metric as the cost function, to train the UNET, produced better results.
Once the tumor is detected in MRI images, the characterization of the tumor is significant for the therapeutic planning of tumor. The characterization of the brain tumor represents another set of classification sub-problems. Classifying brain MRI images into normal and tumorous is an important research problem. The task of characterizing brain MRI images having tumors into malignant and benign is a well-researched topic (Kumar et al. 2017). Malignant tumors are tumors that spread to other parts and are called the cancerous ones. They are prone to recurrence even after removal. In contrast, benign tumors do not spread to other parts of the body and respond well to medical care. A prominent work used texture features from CT scan images to discriminate benign and malignant tumors (Nanthagopal and Sukanesh 2013). The work evaluated the classification performance with an SVM classifier and with a probabilistic neural network classifier. Among the two, the SVM classifier produced higher accuracy. Huang et al. (2019) studied the problem of identifying the optimal machine learning technique for diagnosing brain metastases. Further, the classification of malignant tumors into low-grade and high-grade tumors (Dong et al. 2017; Mohan and Subashini 2018) is also an extensively investigated research problem. A prominent work classifies between high-grade and low-grade glioma (Kaur et al. 2017). The study used Hilbert transform-based analysis of texture features from tumor regions. Tumorous brain images were taken from BRATS and Harvard medical repositories for the research. Mzoughi et al. (2020) used a 3D CNN model for the grading of glioma. Their methodology relied on data augmentation procedures for higher performance).
Classification of a brain tumor based on its subtype is another significant related problem. Brain tumors of the same class can have variations concerning structure, size, and shape based on different patient-specific factors. In contrast, tumors from different categories might show similarities in appearance. This behaviour makes the problem more challenging. Comparatively, fewer works have been reported in categorizing brain tumors into different pathological types. Cheng et al. (2015) considered a 3-class classification of brain tumors into glioma, meningioma and pituitary tumors. While glioma is malignant, meningioma and pituitary tumors are usually benign. Glioma is a life-threatening brain tumor. It originates from glial cells, surrounding the nerve cells in the brain. Meningiomas arise from membranes surrounding the brain and develop very slowly. Yet, a few cases of meningioma can be aggressive. Pituitary tumors are benign tumors affecting the pituitary gland. They change the bodily functions due to unregulated hormone production by the affected pituitary gland. A thorough inspection of the MRI can aid in the diagnosis of the three tumor types. However, the study requires numerous scans of multi-view MRI images. The tumor classification problems could be extended to include more specific cases of tumors. Mohsen et al. (2018) formulated a four-class classification problem. The dataset included MRI images from three categories of malignant tumors and MRI images of normal brain. The authors used the discrete wavelet transform (DWT) features with the CNN classifier. In a closely related work, Zhou et al. (2018) used an ensemble of DenseNet and a recurrent neural network (RNN) for a 4-class brain tumor classification problem.
In this paper, we propose a fully automatic classification algorithm for brain tumors using MRI images. The defined objective is a 3-class classification problem for classifying brain images having three types of tumors:- glioma, meningioma, and pituitary tumor. These are the three most incident types of brain tumors (Cheng et al. 2016). A deep CNN is designed for extracting image features and a multiclass SVM is employed for the classification process. The evaluation uses the open dataset from Figshare. The following are factors that motivate our work. First, the need for improved accuracies in the classification problem involving glioma, meningioma and pituitary tumors. An accurate computer aided automatic classification system for the three specific tumor types would be of assistance to medical practitioners in their treatment plans. Second, high accuracies achieved in the recent classification tasks that used deep learning strategies and robust classifiers. Third, medical image data are not available in large numbers. Advanced design strategies are required to deal with this practical limitation. The following are the contributions from this work.
-
Designed a CNN model to extract features from brain MRI images with tumor.
-
The ensemble arrangement of CNN and SVM is found to produce an improved classification performance on small medical image datasets.
-
Produced an overall classification accuracy of 95.82% (on Figshare dataset), which is higher than the state-of-the-art methods.
-
A comparison of the proposed methodology with a transfer learning-based approach is provided in terms of computational complexity.
2 Dataset and related works
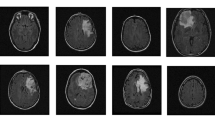
All the existing works on brain tumor classification into glioma, meningioma, and pituitary tumor have been tested on the dataset from Figshare (Dataset 1 2018). It is an open data set extensively used for the research problems in medical image classification and medical image retrieval (Cheng et al. 2015, 2016). The dataset is a collection of 3064 T1-weighted contrast-enhanced (CE) MRI slices belonging to 233 patients. The 2D slices are labelled and correspond to one among the three tumor classes of glioma, meningioma and pituitary tumor. The dataset is unbalanced, consisting of 1426 MRI images with glioma, 708 and 930 images corresponding to meningioma and pituitary tumor, respectively. The dataset contains multi-view slices of axial, sagittal and coronal sections. Each slice (image) is of size \(512 \times 512\) and is available as a .mat file. Table 1 provides details of the dataset and shows a few MRI images in axial-view from the dataset.
Some of the reported works on the 3-class tumor classification problem, using Figshare data, have followed fivefold cross-validation for evaluation. Some others have reported classification accuracy after performing a hold-out validation. If cross-validation is followed, every sample image in the data set gets tested during evaluation. In hold-out validation, only a fixed number of image samples gets tested during the assessment. The performance evaluation performed by following a cross-validation scheme is considered to be more reliable. On the other hand, the results of a hold-out validation scheme are dependent on the split rule that constituted the test set and the training set. The first and foremost significant work on brain tumor classification into glioma, meningioma and pituitary tumor, based on the dataset from Figshare, is by Cheng et al. (2015). The work made use of tumor masks available in the dataset to extract the ROI. Morphological dilations augmented the ROI. The features, specifically:- intensity histogram, GLCM and the bag of words (BOW) were extracted from the augmented ROI. The experiments included testing with different classifiers. A combination of BOW features and SVM classifier achieved the best performance. Evaluation of experiments followed fivefold cross-validation and achieved an overall accuracy of 91.28%. The other performance indicators used are sensitivity and specificity.
Another research paper proposed a combination of Gabor and DWT features and a neural network-based classifier to attain a classification accuracy of 91.90% (Ismael and Abdel-Qader 2018). The work by Pashaei et al. (2018) reported an accuracy of 93.68% with the use of a combination of CNN features and extreme learning machine (ELM) for classification. The CNN architecture consisted of four convolutional layers and a total of 240 filters, each of size \((3 \times 3)\). The authors used ELM with a radial basis kernel as the classifier. A modified CNN architecture in the form of Capsule networks was proposed in another work (Afshar et al. 2018). The Classification experiments were carried on brain MRI images as well as tumor segmented MRI images. Better classification results were achieved on the tumor segmented images. Capsule networks based design is followed in another work that considered spatial relations of tumor segmented regions for the classification task (Afshar et al. 2018). The design included measures to overcome the high sensitivity of capsule networks to image backgrounds. A CNN based architecture, utilized for differentiating brain tumors, achieved a validation accuracy of 84.19% (Abiwinanda et al. 2018). The Figshare dataset images were also used in the prominent project work on brain tumor classification (Paul 2016). One of the subproblems of the project work was to classify brain tumor images into three categories. Average fivefold cross-validation results were published in the report. Swati et al. (2019) proposed the use of deep transfer learning for the automatic brain tumor classification. The authors experimented with different pre-trained networks, including AlexNet, VGG-16, and VGG-19. To their observation, the design using VGG networks achieved better performance compared to that obtained using AlexNet. The fine-tuning of different layers in the network resulted in classification accuracy as high as 94.8%.
3 Methods
The proposed classification scheme uses CNN to extract features in brain MRI and an SVM for classification. Figure 1 illustrates the proposed scheme for classification.
3.1 The CNN architecture
The proposed classification scheme uses CNN to extract features in brain MRI. The input layer of the proposed CNN model is of size \(256 \times 256\). The CNN model consists of five convolution (Conv) layers and two fully connected (FC) layers, as shown in Fig. 2. The weights of convolution filters and the weights associated with the full connections form the learnable parameters in a CNN model. For a convolution layer, multiple filters are used in the design to capture multiple activations for the same input image. Padding of ‘1’ pixel thickness is incorporated to preserve the borders of the images. Different kernel sizes are chosen at different layers to capture feature representations at various resolutions. The kernel size for each convolution layer is provided in Table 2.
The size of volume of each layer is indicated in Fig. 2. The dimensions of a layer follows the rule given below. For an input volume of dimensions \((X_{1},Y_{1},Z_{1})\), convolved with K filters of size (F, F), size of the output is \((X_{2},Y_{2},K)\). \(X_{2}\) and \(Y_{2}\) are calculated as,
where P and S are padding and striding values. They are unity and two, respectively, in our design. Following each convolution layer, there is a batch normalization layer. The layer normalizes the previous layer outputs for the training samples in the batch of size 128. ReLU activation function is provided after each batch normalization layer. Max pooling is provided after each ReLU function. The purpose is to reduce the dimensions of the output between successive stages. The typical pooling operation applied in this design uses a max pool filter of size (2,2), stride (2,2) and no padding. The FC_1 layer has ten neurons, and FC_2 layer has three neurons.
3.1.1 Computational complexity of CNN
The factors considered in the design of the proposed CNN include the computational complexity and memory requirements. This consideration in design limited the CNN to have two FC layers and restricted it to have smaller convolution filters. The convolution stages and the dense connections of FC layer(s) account for most of the computations in a deep CNN model. The convolution operation is multiplications, followed by an addition. The computations, therefore correspond to multiplication and accumulation (MAC) operations. The number of MAC operations in a convolution layer (\(Ops_{ conv}\)) depends on the filter dimension (F\(_{1}\), F\(_{2}\), K) and the dimension of output feature map (X, Y, Z) by the relation,
The number of MAC operations in an FC layer (\(Ops_{FC}\)) is equal to the number of parameters (weights) of the layer. The total number of MAC operations for the entire network is the sum of the number of MAC operations for all the layers of the network.
3.1.2 Memory requirements of CNN
The memory requirements for the implementation of deep CNN models depend on the number of parameters. The learnable parameters are contributed by the convolution layers and the FC layers. The number of parameters for a convolution layer (Param\(_{conv}\)) is calculated as,
where Z\(_{in}\) represents the number of input channels for the layer. The number of parameters for the FC layer (Param\(_{FC}\)) is calculated as,
where X\(_{prev}\), Y\(_{prev}\) and Z\(_{prev}\) represent the dimension of the layer previous to FC layer. N\(_{FC}\) represents the number of output nodes for the FC layer. The computational overhead and the memory requirements determine the training time and test time required for a CNN.
3.2 Classifier
The designed CNN has a softmax layer at the output of the final FC layer. CNN, with a softmax layer, forms a stand-alone classifier. The performance of CNN as a classifier gets affected due to the phenomenon of overfitting. With the CNN model being complex, the effect of overfitting gets pronounced when the availability of training data is limited. In a way to improve the performance, we propose the use of SVM on the CNN features.
3.2.1 Softmax
CNN uses a softmax activation function after the final FC layer. The function is defined as,
where x represents the set of N variables. With the softmax function, the output becomes a measure of probability. The whole training process in a CNN classifier is driven by the loss function, Cross-Entropy and is defined by \(L_c\),
where q and q’ are true and predicted measures, respectively for output class. The loss returns a lower value when the predicted and true values are closer. Cross-entropy loss, which measures closeness between the true measure and predictions, is considered better for training and gradient calculations (in backpropagation). On the other hand, a mean square error (MSE) loss penalises heavily on incorrect predictions. Again, being suited for numerical outputs, MSE is not suited for a probabilistic interpretation.
3.2.2 SVM
SVM is a maximum margin classifier defined as an optimization problem (Cortes and Vapni 1995) as given below. Given the feature vector \(x_{i}\), weight vector w and class label \(y_{i}\)
where i represents the no. of samples, C is the cost parameter controlling the maximization of margin and minimization of classification error. and \(\xi \) represents the training errors.
SVM can be easily extended to multi-class problems. In practice, common approaches to multiclass classification are one-versus-all method and one-versus-one method (Rocha and Goldenstein 2013). For a c-class classification task, the former approach requires c binary SVM classifiers. The requirement is \(\frac{{c}({c} - 1)}{2}\) binary classifiers, when the latter approach is followed. The proposed work uses a 3-class classifier model with three SVM binary classifiers, following a one-versus-all approach. The loss function used for the SVM classification algorithm is Hinge Loss \((L_h)\). The function is mathematically defined as,
where q is the target label and \(q'\) is the output measure based on classifier decision. The function is convex and has a subgradient for the model parameters.
4 Experimental set-up
The experiment is successfully conducted on a computer system with 32 GB RAM, Intel Xeon CPU E3-1245-v6 @3.70GHz. The software used for the experimentation is Matlab 2018a.
4.1 Preprocessing
MRI images in the dataset are resized to (\(256 \times 256\)), and grey level values are normalized to values between [0 1]. The experimentation followed the fivefold cross-validation approach. The set of 233 patients in the dataset corresponds to a total of 3064 images. The dataset is randomly split into five subsets of roughly similar sizes. Across the subsets, the number of patients belonging to a specific category of a tumor is made approximately equal. Figure 3 shows the steps followed in preprocessing. Indicators ‘1’ to ‘5’, in the figure, represent five disjoint subsets formed to conduct the fivefold cross-validation. During each round of validation, one subset is assigned as a test set while the others form a training set. After five rounds of validation, every MRI image in the dataset gets tested and classified by the designed model. The validation results of each round are combined to get the overall performance of the classifier.
4.2 Feature extraction
The input training data images, including their class labels, are given to the CNN model. Training images are augmented using random rotations, scalings and translations. This aids in making the learning process more effective. The network learns general information rather than assimilating specific details of each image. The expected effect is a better classification for the unseen MRI image data. Training accuracy and loss functions are monitored during the training phase. A convergence of loss function to zero indicates adequate training of CNN, shown in Fig. 4. It indicates the CNN has learned to classify training data correctly. The response to an input image at a layer is termed as activation of the layer in CNN. We conducted experiments using the activations of the Conv_5 layer and FC_1 layers. Activations from FC_1 is a vector of length ten and activations of Conv_5 is vector of length 3136. Separate and independent experiments are conducted on FC_1 and Conv_5 feature sets. No feature selection (or dimension reduction) technique is used in this work. There is a set of user-chosen parameters, that aid in the proper training of the CNN model. They are known as hyper-parameters of CNN. Table 3 provides the values of hyper-parameters used in the design. Adam was selected as the optimizer because it has an adaptive learning rule. Adam was found to learn the data better than sgdm, the other popular optimizer. Batch normalization and L2 regularization were included to avert the undesirable overfitting phenomenon. The practical choice of the batch size is a trade-off between faster training and available memory space. In our design, the batch size for normalization was chosen as 128. The other hyper-parameter values were heuristically chosen after rounds of relevant experimentation on training data from Figshare.
4.3 Classification
The features extracted from training images, along with their class labels, are given to train the multiclass SVM model. The features extracted from test images are then given to the trained SVM. The predicted labels for the test features are obtained as output from the SVM model. Predicted labels are compared with true class labels to evaluate the performance of the classifier. The proposed work uses a three-class ECOC model with three binary SVM classifiers, following a one-versus-all approach. Each binary SVM applies a linear kernel function. The linear kernel was fixed after experimenting with linear and radial basis function (RBF) kernels. The linear kernel was observed to give slightly better results. The loss function used for SVM classification algorithm is Hinge Loss. Also, the model incorporates L2 regularization to enable modelling with a soft-margin. The SVM model uses bfgs as a solver for optimization. Improved classification accuracy is obtained for models included with error correcting output codes (ECOC). Table 3 lists the hyperparameters corresponding to the adopted SVM-ECOC model.
5 Evaluation
The average elapsed time during the training of CNN, following one complete trial of fivefold approach, is 2 h 06 min. However, the testing has been fast with the average time per image is less than a second. In this section, the performance of the classifier is evaluated and is compared with that of related works.
5.1 Comparison with related works in terms of accuracy
The most significant performance metric for a classifier is classification accuracy, which gives percentage of true predictions made by the classifier. We present the classification accuracies obtained under three different settings in the experiment.
-
The accuracy of designed CNN, when used itself as a standalone deep learning classifier, is 94.26%.
-
Activations from Conv_5 layer of CNN, as features to SVM classifier, produced an accuracy of 93.83%.
-
Activations from FC_1 layer to SVM classifier achieved an accuracy of 95.82%.
Figure 5 shows the progress of training and validation sets over iterations. Despite data augmentations, there is a limitation in achieving higher levels of validation performance for a CNN classifier. To some extent, this limitation is addressed by the use of SVM on CNN features. However, the performance is dependent on the feature set. CNN features in the form of activations from FC_1 layer achieve higher accuracy compared to that produced by activations from Conv_5 layer. This improvement is because the activations, of deeper layers, provide a better representation for an image. The FC_1 layer accumulates the responses from earlier layers and gives a more characteristic representation for the three classes of tumor images.
The proposed methodology is compared with all the works on this specific classification problem. Classification accuracy is used as the metric for comparison because of two reasons. First, it is the standard metric used in all the related works. Second, all the related works are evaluated using the same dataset. Table 4 summarises the works and gives a performance comparison. Among the works using hand-crafted designs, SVM on the BoW model of feature set was an effective approach (Cheng et al. 2015). With the adoption of deep learning concepts and the use of CNN features, the results improved (Pashaei et al. 2018; Swati et al. 2019). According to our experiments, the results further improved when CNN features are classified using an SVM model. Our methodology that uses a combination of features from the designed CNN and multiclass SVM for classification achieved the highest accuracy.
5.2 Other performance metrics
As the data set is unbalanced for the category samples, we evaluate the results with the help of a confusion matrix. Table 5 represents confusion matrix obtained for the experiment.
From the confusion matrix, the following key evaluation metrics are calculated for each class. For any class C, Precision (\(P_{C}\)) is the proportion of samples classified as C, that actually belong to C. Recall (\(R_{C}\)) gives the proportion of actual samples of C, that are being correctly identified as C. It is also known as the sensitivity of a classifier. Specificity (\(S_{C}\)) is the fraction of non-members of C, that is correctly detected. The expressions for calculating the metrics are as follows,
where TP represents the number of true positive cases while FP is the number of false positive cases in classification. TN is number of true negatives, and FN represents false negative samples of classification.
The performance measures for each category of disease is shown in Table 6. Concerning specificity for all the classes, the designed disease categorizer is superlative. Specificity is an important measure in disease classification because it gives the fraction of samples free of the specific disease class. Again, with reference to precision and recall measures, the classifier is considered as a high-performance system.
5.3 Validation using more datasets
Figshare is the only database available for the specific three classes of tumors discussed in this work. The proposed classifier arrangement is extended to some closely related problems. The objective is to permit the evaluation of the proposed classification scheme using other datasets.
5.3.1 Using Radiopaedia repository
Detection of tumor can be considered as a 2-class classification problem between tumorous and normal images. The final FC layer in the proposed CNN architecture is modified to have two output neurons instead of the original three neurons. Eighty samples of brain MRI images are taken from Radiopaedia repository (Dataset 2 2019) for the evaluation. Forty images are normal and the remaining forty are tumorous, as considered in a recent work (Seetha and Raja 2018). Evaluation using fivefold cross-validation resulted in a classification accuracy of 99.0% when CNN features were classified using SVM. The accuracy of the softmax classifier of CNN was 95.7%. Table 7 provides a comparison of the proposed method with the state-of-the-art method. The related work (Seetha and Raja 2018) used CNN with softmax for classification of images. The proposed work attained the improvement, using SVM on CNN features.
5.3.2 Using Harvard repository
The classifier arrangement is extended to a 4-class problem. The final FC layer of the proposed CNN architecture is modified to have four output neurons. 66 T2-weighted axial plane brain MRI images, of size \(256 \times 256\) from Harvard medical repository (Dataset 3 2019), are used for the experiment. 22 MRI images belong to the category normal and the remaining 44 images belong to one among the three malignant tumors, namely glioblastoma, sarcoma and metastatic bronchogenic carcinoma. Hence the four classes for the problem are normal, glioblastoma, sarcoma and metastatic bronchogenic carcinoma. Evaluation using fivefold cross-validation resulted in a classification accuracy of 98.78% when CNN features were classified using SVM. The accuracy of the softmax classifier of CNN was 94.54%. The performance was compared with the state-of-the-art method (Mohsen et al. 2018), and comparison results are provided in Table 8. Considering the small size of the dataset, the related work (Mohsen et al. 2018) applied the domain knowledge to extract features in the wavelet transform domain. According to the proposed work, the CNN features are used to characterize images and are classified using an SVM model.
The experimental results (given in Table 7 and Table 8) validate the improvements in medical image classification using the hybrid CNN-SVM arrangement.
6 Discussions
Features extracted by CNN have proven to be more capable in describing brain image data than the conventional hand-crafted features. However, an SVM classifier is observed to be more efficient than the softmax based classifier of CNN, in classifying the feature data.
6.1 Comparison with transfer learning
Research studies have successfully applied transfer learning to medical image classification problems, considering the scarcity of medical data. However, there is an associated drawback in it. The pre-trained models are intense such that the computational overhead and memory requirements are significantly high. A comparison of two works on the 3-class brain tumor classification problem is provided in Table 9. The previous work [33] applies different pre-trained networks such as VGG-16 on the Figshare dataset to solve the classification problem. The computations involved in such deep networks are large (Sze et al. 2017). The latter is the proposed method which uses the designed CNN to extract features. The proposed method produces an improved performance than the other method with lesser computations.
6.2 Improved performance with SVM
The softmax classifier of CNN classifies data based on the probability measures of output classes. The disadvantage with CNN as a classifier is overfitting. Figure 6 shows that the training loss goes to zero, but the validation loss does not. Beyond 200 iterations, the validation loss increases. This behaviour indicates that overfitting occurred with the CNN model. SVM, on the other hand, classifies data by transforming them into higher dimensional space. It is a maximum margin optimization problem based on Hinge loss. The constrained maximum margin model of SVM is less prone to overfitting. This feature of SVM is a significant consideration for medical image classification problems, where the availability of training data is limited. Besides, two factors make SVM less sensitive to outlier data. First, the Hinge loss diverges slower than the cross-entropy loss of softmax classifier of CNN. Second, the classification of test features relies only on the support vectors. Table 10 summarizes the experimental results. The observations emphasize that the CNN-SVM can reach better performance over CNN-softmax when the data size is limited.
6.3 Regarding misclassifications
Analysis using receiver operating characteristics (ROC) provides an insight into the misclassifications made by the classifier. ROC curve is the graph plotting true positive rate and false positive rate for a disease category. Figure 7 shows the ROC curve for the proposed SVM classifier on the CNN features. The area under the curve (AUC) indicates the ability of the model to distinguish between classes. Higher the AUC, better is the classifier is at predicting the disease category correctly. The AUC values for the diseases meningioma, glioma and pituitary tumor are 0.990, 0.993, and 0.996, respectively. The AUC value for meningioma is slightly lower than that of the other categories. As a result, comparatively there is poor performance with respect to precision and recall for the case of meningioma (Table 6). The accuracy of the 3-class brain tumor classification problem is affected by the following reasons. The MRI images of the three classes of tumors considered are observed to exhibit similar characteristics. Another significant reason is the imbalance of the Figshare dataset concerning the class samples. There are lesser samples for meningioma in the dataset. It is an essential factor as CNN training is dependent on the characteristics of a dataset.
7 Conclusion
The article primarily focused on developing an accurate algorithm for brain tumor classification. The CNN-SVM arrangement produced better classification results compared to the CNN with a softmax classifier. This fact has significance in medical image classification problems, where the number of samples available for any class could be minimal. This scheme is advantageous to transfer learning strategies, which requires exhaustive fine-tuning and high memory. The extended training time, needed for CNN before feature extraction, may be considered as a drawback. Future research will focus on speeding up of the training process for the CNN model, without compromising on performance. The developed system may be modified for image retrieval applications, using the same dataset. Extensive data augmentation procedures shall produce further improvements in performance. The classifier model may be tuned further to test on real images, to ascertain its applicability as a tool in medical diagnosis.
References
Abbasi S, Tajeripour F (2017) Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing 219:526–535
Abiwinanda N, Hanif M, Hesaputra ST, Handayani A, Mengko, TR (2019). Brain tumor classification using convolutional neural network. In: World Congress on Medical Physics and Biomedical Engineering 2018, Springer, Singapore, pp 183-189
Afshar P, Mohammadi A, Plataniotis KN (2018) Brain tumor type classification via capsule networks. In: 2018 25th IEEE International Conference on Image Processing (ICIP), IEEE, pp 3129-3133
Afshar P, Plataniotis KN, Mohammadi A (2019) Capsule networks for brain tumor classification based on mri images and coarse tumor boundaries. In: ICASSP 2019–2019 IEEE international conference on acoustics, speech and signal processing (ICASSP), IEEE, pp 1368–1372
Agarap AF (2017) An architecture combining convolutional neural network (CNN) and support vector machine (SVM) for image classification. arXiv preprint arXiv:1712.03541
Agarwal R, Diaz O, Lladó X, Martí R (2018) Mass detection in mammograms using pre-trained deep learning models. In: 14th International workshop on breast imaging (IWBI 2018), vol 10718, International Society for Optics and Photonics, p 107181F
Agarwal P, Wang HC, Srinivasan K (2018) Epileptic seizure prediction over EEG data using hybrid CNN-SVM model with edge computing services. In: MATEC Web of Conferences, vol 210, EDP Sciences, p 03016
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett. https://doi.org/10.1016/j.patrec.2017.10.036
Amin J, Sharif M, Raza M, Yasmin M (2018) Detection of brain tumor based on features fusion and machine learning. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-018-1092-9
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5):1207–1216
Bardou D, Zhang K, Ahmad SM (2018) Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access 6:24680–24693
Cao Y, Geddes TA, Yang JYH, Yang P (2020) Ensemble deep learning in bioinformatics. Nat Mach Intell 2:1–9
Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, Feng Q (2015) Enhanced performance of brain tumor classification via tumor region augmentation and partition. PloS One 10:e014449
Cheng J, Yang W, Huang M, Huang W, Jiang J, Zhou Y, Chen W (2016) Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. PloS One 11:e0157112
Cortes C, Vapni V (1995) Support-vector networks. Mach Learn 20(3):273–297
Dataset 1 (2018) Figshare brain tumor dataset’. https://do.org/10.6084/m9.figshare.1512427.v5. Accessed Dec 2018
Dataset 2 (2019) Radiopaedia dataset. https://radiopaedia.org. Accessed Nov 2019
Dataset 3 (2019) Harvard medical dataset. http://med.harvard.edu/AANLIB. Accessed Nov 2019
Dong H, Yang G, Liu F, Mo Y, Guo Y (2017). Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In: Annual conference on medical image understanding and analysis, Springer, Cham, pp 506–517
Fang T (2018) A novel computer-aided lung cancer detection method based on transfer learning from GoogLeNet and median intensity projections. In: 2018 IEEE international conference on computer and communication engineering technology (CCET), IEEE, pp 286–290
Huang S, Yang J, Fong S, Zhao Q (2019) Mining prognosis index of brain metastases using artificial intelligence. Cancers 11(8):1140
Ismael MR, Abdel-Qader I (2018) Brain tumor classification via statistical features and back-propagation neural network. In: 2018 IEEE international conference on electro/information technology (EIT), USA, pp 252–257
Kaur T, Saini BS, Gupta S (2017) Quantitative metric for MR brain tumour grade classification using sample space density measure of analytic intrinsic mode function representation. IET Image Process 11(8):620–632
Kumar S, Dabas C, Godara S (2017) Classification of brain MRI tumor images: a hybrid approach. Procedia Comput Sci 122:510–517
Liu B, Yu X, Zhang P, Yu A, Fu Q, Wei X (2017) Supervised deep feature extraction for hyperspectral image classification. IEEE Trans Geosci Remote Sens 56(4):1909–1921
Lu S, Lu Z, Zhang YD (2019) Pathological brain detection based on AlexNet and transfer learning. J Comput Sci 30:41–47
Mahesh KM, Renjit JA (2018) Evolutionary intelligence for brain tumor recognition from MRI images: a critical study and review. Evol Intell 11(1–2):19–30
Mohan G, Subashini MM (2018) MRI based medical image analysis: Survey on brain tumor grade classification. Biomed Signal Process Control 39:139–161
Mohsen H, El-Dahshan ESA, El-Horbaty ESM, Salem ABM (2018) Classification using deep learning neural networks for brain tumors. Future Comput Inform J 3(1):68–71
Mzoughi H, Njeh I, Wali A, Slima MB, BenHamida A, Mhiri C, Mahfoudhe KB (2020) Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J Digit Imaging. https://doi.org/10.1007/s10278-020-00347-9
Nabizadeh N, Kubat M (2015) Brain tumors detection and segmentation in MR images: Gabor wavelet vs. statistical features. Comput Electr Eng 45:286–301
Nagabushanam P, George ST, Radha S (2019) EEG signal classification using LSTM and improved neural network algorithms. Soft Comput 24:1–23
Nanthagopal AP, Sukanesh R (2013) Wavelet statistical texture features-based segmentation and classification of brain computed tomography images. IET Image Process 7(1):25–32
Pashaei A, Sajedi H, Jazayeri N (2018) Brain tumor classification via convolutional neural network and extreme learning machines. In: 2018 8th international conference on computer and knowledge engineering (ICCKE), Mashhad, pp 314–319
Paul J (2016) Deep Learning for brain tumor classification. PhD diss, Vanderbilt University
Rajagopal R (2019) Glioma brain tumor detection and segmentation using weighting random forest classifier with optimized ant colony features. Int J Imaging Syst Technol 29(3):353–359
Rocha A, Goldenstein SK (2013) Multiclass from binary: expanding one-versus-all, one-versus-one and ECOC-based approaches. IEEE Trans Neural Netw Learn Syst 25(2):289–302
Sampathkumar A, Vivekanandan P (2019) Gene selection using parallel lion optimization method in microarray data for cancer classification. J Med Imaging Health Inform 9(6):1294–1300
Sampathkumar A, Rastogi R, Arukonda S, Shankar A, Kautish S, Sivaram M (2020) An efficient hybrid methodology for detection of cancer-causing gene using CSC for micro array data. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-01731-7
Seetha J, Raja SS (2018) Brain tumor classification using convolutional neural networks. Biomed Pharmacol J 11(3):1457
Swati ZNK, Zhao Q, Kabir M, Ali F, Ali Z, Ahmed S, Lu J (2019) Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph 75:34–46
Sze V, Chen YH, Yang TJ, Emer JS (2017) Efficient processing of deep neural networks: A tutorial and survey. Proc IEEE 105(12):2295–2329
Tavakoli N, Karimi M, Norouzi A, Karimi N, Samavi S, Soroushmehr SR (2019) Detection of abnormalities in mammograms using deep features. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-019-01639-x
Xue DX, Zhang R, Feng H, Wang YL (2016) CNN-SVM for microvascular morphological type recognition with data augmentation. J Med Biol Eng 36(6):755–764
Zhou Y, Li Z, Zhu H, Chen C, Gao M, Xu K, Xu J (2018). Holistic brain tumor screening and classification based on densenet and recurrent neural network. In: International MICCAI Brainlesion workshop, Springer, Cham, pp 208–217
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deepak, S., Ameer, P.M. Automated Categorization of Brain Tumor from MRI Using CNN features and SVM. J Ambient Intell Human Comput 12, 8357–8369 (2021). https://doi.org/10.1007/s12652-020-02568-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-020-02568-w