Abstract
Compound P131 has been established to inhibit Cryptosporidium parvum’s inosine monophosphate dehydrogenase (CpIMPDH). Its inhibitory activity supersedes that of paromomycin, which is extensively used in treating cryptosporidiosis. Through the per-residue energy decomposition approach, crucial moieties of P131 were identified and subsequently adopted to create a pharmacophore model for virtual screening in the ZINC database. This search generated eight ADMET-compliant hits that were examined thoroughly to fit into the active site of CpIMPDH via molecular docking. Three compounds ZINC46542062, ZINC58646829, and ZINC89780094, with favorable docking scores of − 8.3 kcal/mol, − 8.2 kcal/mol, and − 7.5 kcal/mol, were selected. The potential inhibitory mechanism of these compounds was probed using molecular dynamics simulation and Molecular Mechanics Generalized Poisson Boltzmann Surface Area (MM/PBSA) analyses. Results revealed that one of the hits (ZINC46542062) exhibited a lower binding free energy of − 39.52 kcal/mol than P131, which had − 34.6 kcal/mol. Conformational perturbation induced by the binding of the identified hits to CpIMPDH was similar to P131, suggesting a similarity in inhibitory mechanisms. Also, in silico investigation of the properties of the hit compounds implied superior physicochemical properties with regards to their synthetic accessibility, lipophilicity, and number of hydrogen bond donors and acceptors in comparison with P131. ZINC46542062 was identified as a promising hit compound with the highest binding affinity to the target protein and favorable physicochemical and pharmacokinetic properties relative to P131. The identified compounds can serve as a basis for conducting further experimental investigations toward the development of anticryptosporidials, which can overcome the challenges of existing therapeutic options.

P131 and the identified compounds docked in the NAD+ binding site of Cryptosporidium parvum IMPDH.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidiosis is an opportunistic parasitic disease and a significant cause of diarrhea in humans [1, 2]. Immunocompromised populations such as infants, HIV, and T-cell immunodeficient patients are primarily at risk [3,4,5]. Clinical manifestation of the infection includes abdominal pain, nausea, watery diarrhea, and low-grade fever. If not managed properly, the symptoms can be life threatening [6,7,8,9]. Cryptosporidiosis takes the second position after rotavirus as the leading cause of diarrheal related death in children less than 5 years old [10]. In sub-Saharan Africa, where the disease is highly endemic, it accounts for around 2.4 million deaths in children young of > 24 months [11]. Most human cryptosporidiosis is caused by Cryptosporidium hominis and Cryptosporidium parvum [4, 12].
Presently, there is a therapeutic challenge in treating cryptosporidiosis [13]. The only FDA-approved drug for the disease is nitazoxanide, a compound that contains both a thiazole ring and a benzamidine ring [4]. Although nitazoxanide is well tolerated and does not present with a significant adverse drug reaction, it is not effective in immunodeficient individuals [14]. Other non–FDA-approved drugs administered in treating cryptosporidiosis but with limited success include paromomycin, azithromycin, and rifaximin [13, 15,16,17]. The side effects of these drugs include nephrotoxicity, hepatotoxicity, hearing loss, seizures, etc. [18, 19]. In light of the inadequacies of the current anticryptosporidials, it is therefore urgent to discover therapeutic compounds that have the propensity to overcome the deficiencies of these drugs.
Hedstrom et al.’s research group tackled this by synthesizing several thiazole-based ligands inhibiting Cryptosporidium growth [20,21,22,23,24,25,26,27,28,29,30]. These efforts produced a potential drug molecule, P131, which demonstrated excellent anticryptosporidial activity in vivo. In the in vivo experiment using a murine model, paromomycin was used as a control as nitazoxanide is not bioavailable. P131 at a single dose had the same therapeutic outcome when administered at 250 mg/kg body weight compared to the control group, which was treated with paromomycin at 2000 mg/kg body weight. With a thrice-daily administration of both drugs at the same concentration stated above, P131 elicited a superior parasiticidal activity when compared to paromomycin [22].
P131 potentiates its therapeutic action by inhibiting C. parvum’s inosine monophosphate dehydrogenase (CpIMPDH). It does this by binding to the NAD+ site (co-factor binding site) of the enzyme [31,32,33,34]. Inosine monophosphate dehydrogenase (IMPDH) is an essential enzyme for almost all organisms [33]. It catalyzes the conversion of inosine monophosphate (IMP) to xanthine monophosphate (XMP), which is the first and the rate-limiting step in the biosynthesis of guanine nucleotides [33]. Guanine nucleotides function as precursors for glycosylation, RNA, DNA, and tetrahydrobiopterin synthesis [35, 36]. Therefore, the inhibition of IMPDH hinders proliferation and eventuates in cell death.
Pharmacophore modeling is gaining ground as one of the paths to discovering novel drug candidates with favorable pharmacokinetic and pharmacodynamic properties [37]. These properties will altogether function to overcome the drawbacks limiting the efficacy of existing drug molecules. Among many others, these downsides include cross-resistance, toxicity, and adverse drug reaction [38]. The primary application of pharmacophore modeling is virtual screening, which is a shorter but efficient route to otherwise capital-intensive and time-consuming processes of drug discovery [39].
Although an abstract concept, pharmacophore modeling is an approach that elucidates and identifies the common chemical moieties of a set of ligands, which are crucial in eliciting a biological function [40,41,42,43,44,45,46,47]. In the present study, the per-residue energy decomposition (PRED) method was employed in the pharmacophore modeling of P131 coupled with a virtual screening of the ZINC database. The screening is to generate compounds with the potential to treat cryptosporidiosis. We examined the physicochemical and therapeutic suitability of the compounds as anticryptosporidials. Finally, to understand the molecular mechanism of these new drugs’ action, molecular dynamics simulation of the new compounds was carried out using the parent compound P131 as a comparative reference. The conclusions from this might give the experimental research groups potential drug leads that probably would overcome the limited efficacy of nitazoxanide.
Computational methods
Pharmacophore model generation and virtual screening
At the initial step, we obtained Cryptosporidium parvum IMPDH (CpIMPDH) co-crystallized with P131 at the co-factor site and inosine monophosphate at the active site from Protein Data Bank (PDB code 4RV8) [31]. Although a tetramer, only chains A and D with 652 residues were used to reduce computational resources. The target protein structure (CpIMPDH) was prepared using the graphical user interface (GUI) of UCSF Chimera [48], which involved the removal of ions, crystal waters, and non-standard residues. Missing residues were added using MODELLER [49]. A molecular dynamics simulation run of 20 ns was carried out to stabilize the pose of P131 in the co-factor pocket of CpIMPDH and serve as the cornerstone for the generation of a P131 pharmacophoric model. The total binding free energy of P131 to CpIMPDH was computed. After that, the PRED was analyzed by manually adding the binding residues of P131 to the MMPB/SA.py script integrated with Amber18Tools. PRED estimates the binding free energy each binding site residue contributes to the ligand’s overall binding free energy to the protein. Extrapolating from the PRED, the residues which contributed most to the binding of P131 and the moieties these residues interacted with in P131 were used to build a pharmacophore model (Fig. 1a–d). The pharmacophore query was uploaded in ZINCPharmer [50] to screen the ZINC database [51] for hits that possess these pharmacophoric moieties. The inclusion criteria were as follows: molecular weight of < 500 g/mol, hydrogen bond acceptors < 10, hydrogen bond donors < 5, and rotatable bonds < 5. The output was further filtered down to exclude non–drug-like hits by further screening with Lipinski’s rule of five and ADMET properties [52, 53].
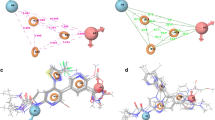
P131 pharmacophore model creation. a 2D layout of binding site residue interaction with P131 after 20-ns MD simulation. b The pharmacophoric moieties selected in P131, which interacted with the lowest energy contributing binding site residues. c Per-residue energy decomposition showing different energy components; each binding site residue contributed to the total binding free energy of P131. d Per-residue plot showing only the total binding free energy contributed by each binding site residue.
Molecular docking
With the prior definition of the P131 binding site in CpIMPDH, we performed molecular docking of the hits generated through virtual screening. Molecular docking was done to ascertain the complementarity of the hits to the co-factor binding site in CpIMPDH [54]. With Autodock Vina, the ligand docking estimations were carried out [55]. Employing the Autodock Graphical user interface by MGL tools, Gasteiger partial chargers were assigned, and atom types were defined [56]. The docked conformations were produced using the Lamarckian Genetic Algorithm [57]. In converting the ligand SDF file to the mol2 format, Maestro software was used [58], and to pdbqt format, we employed Raccoon integrated with AutoDock suite [56]. Autodock Vina was used in defining the dimensions and coordinates of the grid box. The grid box was defined as center (X = 2.649, Y = 21.997, Z = 77.399) and size (X = 11.165, Y = 9.462, Z = 7.323. During the docking process, a maximum of eight conformers was considered for each compound. After screening, molecular docking, and filtering, we selected the top three ligands with the best binding affinities toward CpIMPDH. The docked conformation of the compounds complexed with CpIMPDH was generated and visualized in the ViewDock plugin-integrated Chimera [48]. After that, MD simulation and post-MD analysis were carried out on the docked complexes.
Physicochemical properties and bioactivity screening of identified compounds
The pharmacokinetic (ADMET) properties of the identified compounds and the parent compound P131 were evaluated using validated online prediction tools, which have also been used extensively in other studies [59,60,61]. These include SWISSADME [52], which was used to predict/compute molecular weight, synthetic accessibility, lipophilicity, hydrogen bond acceptor and donor, bioavailability score, water solubility, gastrointestinal absorption, and blood–brain barrier permeability. Molinspiration Cheminformatics [62] was employed in predicting the topological surface area (TPSA) and the number of rotatable bonds. Likewise, oral toxicities and LD50 of the identified compounds and P131 were predicted using the ProTox web server [63]. This presents a faster approach for determining doses that are or have the potential to be toxic. Finally, DataWarrior [64] was used to predict ligand efficiency (LE), ligand lipophilic efficiency (LLE), and ligand efficiency–dependent lipophilicity (LELP). All these web servers were used to determine the pharmacokinetic (ADMET) properties of the ligands and to evaluate how well they adhered to Lipinski’s rule of five, which are a set of rules that predict the drug-likeness of a molecule [53].
Molecular dynamics (MD) simulation
Sequel to the docking of the identified hits in the co-factor site of CpIMPDH, we prepared the docked complexes for MD simulation. Each system was subdivided into apo (IMPDH bound to IMP in its active site) and complexes, composed of apo bound by P131 and apo bound by the hits in the co-factor site. Afterward, these were set up for a 200-ns MD simulation according to previously reported protocols [65,66,67]. MD simulation was performed using the Graphical Processor Unit (GPU) version of the Particle Mesh Ewald Molecular Dynamics (PMEMD) engine in the AMBER18 suite coupled with integrated modules [68]. FF14SB forcefield was used in defining protein parameters. P131 parameterization was done using the ANTECHAMBER module [69]. Afterward, the topology and parameter files for P131 and the identified compound complexes were generated with the LEAP module, which neutralizes the complexes by adding counter ions and solvates them in a 10-Å TIP3P water box. The systems were minimized partially at 2500 steps, with the restraint potential set at 500 kcal/mol Å2. A full minimization followed this for 5000 steps without energy restraints. Heating of the systems took place for 50 ps from 0 to 300 K in an NVT canonical ensemble using a Langevin thermostat [70] and at a harmonic restraint of 5 kcal/mol Å2. The systems were then equilibrated at 300 K for 1000 ps without energy restraints with Berendsen barostat, keeping the atmospheric pressure at 1 bar [71]. The MD production run was after that carried out for 200 ns [72]. The trajectories obtained were analyzed by the integrated CPPTRAJ and PTRAJ modules [73]. Origin data analytical tool was used in creating the needed plots [74]. 3D visualization of the structures and corresponding analyses were carried out on the GUI of UCSF Chimera.
Thermodynamic calculations
This calculation estimates the binding free energy (BFE) of P131 and the identified compounds to CpIMPDH. BFE gives information about the stability and the binding free energy of a ligand bound to a protein [75]. In our study, Molecular Mechanics Generalized Poisson Boltzmann Surface Area (MM/PBSA), which evaluates ligand interaction in biological macromolecules, was employed in estimating the BFE of the compounds [75, 76]. It is a very robust, widely used, and reliable analytical tool [77, 78]. MM/PBSA was calculated via AmberTools18-integrated MM/PBSA.py python script. This script employs continuum solvent models to automatically analyze binding free energies of snapshots from MD simulation [79].
Mathematically, binding free energy is depicted by the following equation;
Therefore,
∆Eint is given by the summation of ∆Eangle, ∆Ebond, and ∆Etorsion. MD simulation was run on the complex only, and a single trajectory was used. This approach minimizes error and noise; therefore, ∆Eint was canceled between receptor, ligand, and complex [76].
In the equation presented above, ∆Eele and ∆EvdW represent electrostatic and van der Waals energy contributions, respectively. ∆EMM is gas-phase energy. The solvation energy contribution is ∆Gsol, which is constituted by polar solvation energy contribution (∆Gpol (PB)) and non-polar solvation energy contribution (∆Gnp). ∆Eint represents the internal energy contribution, and T∆S, the conformational entropy change. To estimate the different energy each binding site residues contributes to the stabilization and affinity of the ligand, we analyzed the PRED, which, as earlier stated, was investigated by manually adding the binding residues of P131 and the hits generated to MMPB/SA.py script integrated with Amber18Tools.
Results and discussion
PRED upon P131 binding leads to the creation of a pharmacophore model for hit search
In constructing our PRED-based pharmacophore model, a 20-ns MD of P131 bound to CpIMPDH was performed, and the total binding free energy was computed. The overall binding free energy was subsequently decomposed on a per-residue basis to determine the energy contributed by each binding site residue to the stability of the protein–ligand complex [80]. The residues contributing considerably lower energy to the binding and stability of P131 were analyzed vis-a-vis the structural features of P131 they interacted with. These residues were deemed crucial to the interactions of P131 in CpIMPDH. These residues and the respective binding free energies they contributed were ASP252 (− 9.82 kcal/mol), SER164 (− 2.62 kcal/mol), ALA165 (− 1.65 kcal/mol), and MET302 (− 1.73 kcal/mol), and in the second chain (chain D), PRO26 (− 1.28 kcal/mol) and TYR358 (− 1.7 kcal/mol). The interaction between these residues and P131 is shown in Fig. 1. The moieties interacting with the residues contributing these energies made up the chemical scaffold that was used in screening the ZINC database for potential compounds that might portend a better therapeutic outcome than P131 (Fig. 1b).
Pharmacophore-based virtual screening
Virtual screening has evolved as a rapid and dynamic approach to produce potential drug-like compounds that may be optimized to interact efficiently with its therapeutic target [81]. It provides an important starting point in drug discovery called lead identification [82, 83]. The moieties in P131 interacting with the critical amino acid residues were selected on the ZINCPharmer platform. These included two aromatic rings, four hydrogen bond acceptors, and three hydrophobic rings. This pharmacophore scaffold was submitted to ZINCPharmer for pharmacophore pattern matching. The parameters of the hits to be generated were set to have the following: rotatable bond of ≤ 5 molecular weight of ≤ 500 Da. This was necessary to filter out non–drug-like hits that deviated from Lipinski’s rule of 5. With the pharmacophoric moieties selected as well as the filtering parameters, a total of eight ZINC compounds were outputted. Subsequently, these compounds were docked into the NAD+ binding site of CpIMPDH, and their binding affinity was scored. The top three of the identified compounds having the best docking scores were selected for further analysis. These compounds and their respective docking scores were ZINC46542062 (− 8.3 kcal/mol), ZINC58646829 (− 8.2 kcal/mol), and ZINC89780094 (− 7.5 kcal/mol). ZINC46542062 and ZINC58646829 docking scores were better than P131, which was − 7.9 kcal/mol. The 2D representation of the identified hits and their respective docking scores are presented in Table 1.
The differential binding free energy profiles of P131 and the identified compounds in CpIMPDH
Binding free energy calculations have evolved as a dynamic, inexpensive, and extensively used tool in estimating, at the atomic level, the interactions between a therapeutic compound and its biological target. It elucidates the role of each binding site residues in the ligand's stability and binding. The binding free energies of the identified compounds were calculated after MD simulation. We excluded the conformational entropy effect as it is computationally expensive to calculate by normal mode analysis [84] and its effect on binding free energy is debatable [77, 85,86,87]. Snapshots from 100 to 200 ns exhibited relative stabilities and were sampled for the energy calculation to minimize conformational entropy. All the compounds presented with favorable binding free energies (Table 2). However, ZINC46542062 had the lowest binding free energy of − 39.52 kcal/mol, which was better than the reference compound P131 (− 34.61 kcal/mol). This was followed by ZINC89780094 (− 32.68 kcal/mol) and ZINC58646829 (− 23.15 kcal/mol). Per-residue energy contribution to the overall total binding free energies of the identified compounds and prominent energy contributing residues for each complex are presented in Fig. SI-3 and Table S1.
Exploring the interaction dynamics of P131 s the identified compounds across the simulation period
To further understand what could have accounted for the favorable interactions of P131 and the identified compounds evidenced by the binding free energies (Table 2), we visualized the molecular interactions taking place at different time points of the MD simulation. The snapshots were selected at 50 ns, 100 ns, 150 ns, and 200 ns, representing the initial, intermediate, and final time points. In P131, at the initial stage of simulation (50 ns), three conventional hydrogen bonds were formed by GLU329, SER164, and MET302. This number thinned out as the simulation went on, leaving only GLU329 as the only consistent hydrogen bond–forming residue (Fig. 2). ASP252 and ASP163 also were consistent in forming an attractive charge interaction during the period of simulation (Fig. 2). ASP252 and SER164 were among the lowest energy contributing residues to the binding and stabilization of P131 in the NAD+ binding site in CpIMPDH (Table S1).
The binding sites’ multiple interactions with the three fluorine atoms of ZINC46542062 could have afforded it the lowest binding free energy value than P131 and other ZINC compounds. At each time point, the fluorine atoms were engaged in forming a conventional hydrogen bond, fluorine, pi-alkyl, and pi-sigma interactions at almost the same time. TYR358D, ASP163, LYS73, and CYS219 formed a steady conventional hydrogen bond throughout the simulation while MET302 maintained a constant pi-sulfur interaction.
The interaction dynamics of ZINC58646829 and ZINC89780094 are presented in Fig. 3. In ZINC89780094, consistent conventional hydrogen bonds were formed by SER164, SER169, and ASN171 in the entire course of the simulation. A favorable pi-sigma bond mediated by HIS 166 was also observed all through. On the converse, in ZINC58646829, weaker interactions were observed when compared to other screened compounds. Alkyl and pi-alkyl interactions dominated the simulation period. The foregoing could have accounted for ZINC58646829 having the highest binding free energy than other compounds examined (Table 2).
It is also important to note that Cryptosporidium parvum is a eukaryotic organism; however, through lateral gene transfer, it possesses a prokaryotic IMPDH [20, 88]. This permits anticryptosporidials selective inhibition of CpIMPDH, leaving the eukaryotic host’s IMPDH unaffected. There are two residues in the NAD+ site of CpIMPDH and prokaryotic IMPDH, responsible for anticryptosporidial drugs’ selectivity. These are ALA165 in chain A and TYR358 in chain D. These residues have been substituted in eukaryotic IMPDH [89]. We examined the consistency of these residues' interaction with P131 and the identified compounds at different time points selected. Despite their constant motion, the ligands interacted with at least one of these critical residues (Figs. 2 and 3). This interaction further underpins their importance in the potentiation of the therapeutic action of potential CpIMPDH inhibitors.
Evaluation of the drug-likeness and pharmacokinetics of P131 and the identified compounds
Early in drug discovery stages, drug leads’ pharmacokinetic properties must be determined to funnel down to compounds that can be optimized to a favorable range of optimal absorption, distribution, metabolism, excretion, and toxicity (ADMET). Sixty percent of drug failures have been attributed to ADMET challenges [90]. The conventional rules of 5 by Lipinski summarily posits that for a drug to be orally active, it must violate not more than two criteria from the following: not more than 10 hydrogen bond acceptors and 5 hydrogen bond donors, octanol–water partition effect of not greater than 5, lipophilicity not greater than 5, and a molecular weight of less than 500 g/mol [53, 91]. Going by the rule above, the molecular weight of P131 and the identified compounds were within the range of acceptable limits (≤ 500). ZINC46542062 had the highest (435.37 g/mol), while ZINC89780094 had the lowest (351.38 g/mol).
One of the vital indices in determining the pharmacokinetic properties of drugs is lipophilicity (logP). For a drug to potentiate its action, it must interact with its specific target protein. In doing this, it must cross cell membranes, which are almost always lipophilic; therefore, the drug must be soluble in non-polar solvents, fats, and lipids [92]. Lipophilicity impacts drug potency, solubility, permeability, selectivity, and toxicology [93,94,95,96,97]. Lipinski places the logP benchmark as 5 [91]. Compounds with logP higher than 5 have higher toxicity, poorer absorption, lower solubility, and excretion [98, 99]. Also, a very low logP value of compounds presents with ADMET challenges [92]. Generally, drug leads having a logP ranging from 2 to 3 are favorable [100]. ZINC89780094 had the lowest value of logP of 2.45; however, the parent drug P131 and the other two drug leads had > 3 value of logP (Table 3). This implies that ZINC89780094 has a higher propensity for cell membrane permeability and bioavailability. Beyond Lipinski's rules, other parameters have been established to play a role in estimating the drug-likeness of compounds. These parameters include ligand efficiency (LE), ligand lipophilic efficiency (LLE), and LELP [101]. Both LLE and LELP take logP into account while LE does not [92]. LE is used in estimating the affinity of a ligand to its therapeutic target [102]. It also calculates and compares the potency of compounds vis-a-vis their molecular size [103, 104].
Although it can account for the potency irrespective of their molecular sizes, however, not factoring their lipophilicity has its drawbacks as employing LE as an efficiency index alone can lead to the selection of compounds with higher potency but low ADMET characteristics [105,106,107]. From our results, P131 and the identified compounds are within the LE range, which is > 0.3 kcal/mol/heavy atom (Table 3). This depicts that they have desired potency at the right weight, and their ADMET properties may further be optimized without losing their potencies.
LLE measures both the potency of the compounds and its lipophilicity while excluding its molecular size [94, 108]. It exploits how the potency of compounds can be improved while at the same time maintaining low lipophilicity.[94] With the optimal value set at > 5 [94], P131 had the highest LLE value of 10.31, followed by ZINC89780094, ZINC46542062, and ZINC58646829, which had 6.36, 5.4, and 5.09, respectively (Table 3). The import of the result is that the compounds can serve as a suitable starting material for further optimization and development.
LELP, on the other hand, considers the molecular size, potency, and lipophilicity of compounds as a composite unit [101]. It overcomes the lipophilicity exclusion of LE and the molecular size exclusion of LLE. It has been found that drugs having a suboptimal value of LELP did not go far in the drug development process as their drug-likeness is negatively affected [101]. P131 and the identified compounds all had LELP value within the acceptable range of LELP (Table 3), with the lowest being P131 (− 2.7 kcal/mol) and the highest as ZINC46542062 (8.7 kcal/mol).
Synthetic accessibility score (SA score) is another parameter that evaluates how easy or difficult it is to synthesize a compound [109]. SA score must be determined early as a compound that might have been assessed to have excellent ADMET properties might be challenging to synthesize. SA score ranges from 1 (easy to synthesize) to 10 (difficult to synthesize) [109]. From the four compounds assessed, P131 was the hardest to synthesize with an SA score of 3.85, followed by ZINC46542062, ZINC58646829, and ZINC89780094 with SA score of 3.34, 3.06, and 2.89, respectively (Table 3). Moving forward is evaluating the topological polar surface area (TPSA) metric of the compounds that measure the contribution of oxygen, nitrogen, and hydrogens to the compound’s molecular surface area [110]. It gives information about the ability of compounds to permeate the cells. Therefore, it has been postulated that the lower the TPSA value, the easier will be the assimilation of the compounds into the cell [111,112,113]. Although all the compounds fell within the range of allowable TPSA (140 A2) [114], P131 will have some difficulty in permeating the cell when compared to the hits as it has the highest TPSA (117.5 A2) while ZINC58646829 has the lowest (69.16 A2) (Table 3). The value of TPSA of P131 is not far-fetched from the higher number of hydrogen bond donors and acceptors it possesses compared to other identified compounds as the number of hydrogen bonding bears a correlation with TPSA [113].
Finally, the toxicological assessment of the compounds was done. This parameter is presented as LD50. It has been established that the lower the LD50, the more toxic the drug and otherwise [63]. Extrapolating this to P131 and the identified compounds, P131 has the lowest LD50 (2000 mg/kg), making it the most potentially toxic of all the hits, closely followed by ZINC46542062, which has 2662 mg/kg. Both ZINC58646829 and ZINC89780094 appear safer, having an LD50 value of 10750 mg/kg and 11500 mg/kg, respectively (Table 3). All the physicochemical properties that have been evaluated and their established upper limits, both P131 and the identified compounds, have favorable ADMET properties but can still benefit from some improvements.
Conformational perturbations induced by the binding of P131 and the identified compounds
When ligands bind to proteins, they induce conformational changes that that may consequently affect the functions of the proteins [115, 116]. The structural changes that occur when P131 and the identified compounds bind to CpIMPDH were evaluated by calculating the root mean square deviation (RMSD), the root mean square fluctuation (RMSF), and the radius of gyration (RoG) of the generated MD trajectories.
RMSD has evolved as a parameter for comparing protein structure, characterizing related proteins conformation, and stability. It also assesses the quality of MD simulation [117, 118]. It uses the alpha carbon of each amino acid residues that make up the protein to estimate the conformational changes during the simulation with reference to its initial structural conformation [117]. From the RMSD graph (Fig. 4a), all systems attained equilibrium at around 30 ns and exhibited variations in their stability throughout the simulation time. The binding of P131 and the identified hits stabilized the structure of CpIMPDH when compared to the apo. The RMSD of the unliganded CpIMPDH was 2.92 Å; the binding of P131 to CpIMPDH minimally stabilized the system with RMSD value of 2.84 Å. However, ZINC46542062, ZINC58646829, and ZINC89780094 stabilized it more than P131 with an RMSD value of 2.47 Å, 2.24 Å, and 2.48 Å, respectively.
The root mean square fluctuations estimate the flexibility of the protein structure [119]. This is done by measuring the fluctuation of the C-alpha atom of each of the amino acid residues that make up the protein. It is interesting to note that in the apo and complexes, the highest residue fluctuation occurred at chain B (Fig. 4b). The unbound system had an average RMSF value of 12.57 Å, while P131-CpIMPDH had 8.57 Å. The average RMSF values of CpIMPDH complexed with the identified compounds followed that of the apo closely. CpIMPDH–ZINC46542062 had 12.40 Å while CpIMPDH–ZINC89780094 had 12.79 Å. However, CpIMPDH–ZINC58646829 had the highest average residue fluctuation of 13.89 Å. The discrepancy in the RMSF values of P131 relative to the identified compounds might stem from the size of P131, which is bigger than the other ligands, and the fact that it bends at the linker region, which affords it more allowance to interact with more residues in the adjacent chain of CpIMPDH (chain B), thereby reducing the fluctuation of that particular chain.
Closely related to the RMSD is the RoG, which also provides insight into the protein structure’s stability by determining how compact or loose the protein structure is. This is estimated by measuring the distance of the individual atom of the protein to their centroid. A high RoG value indicates that the protein structure is packed loosely while a lower value portrays the otherwise. The binding of the P131 and the identified compounds made CpIMPDH a bit loose when compared with the apo (27.13 Å). The binding of P131 to CpIMPDH increased the protein’s RoG to 28.01Å. ZINC58646829 impacted the RoG of CpIMPDH the most, with an average value of 28.13 Å. ZINC46542062 had 28 Å, while ZINC89780094 had the least among the ligands with an average value of 27.96 Å (Fig. 4c). The slight difference in the RoG values of the apo and the complexes shows an insignificant structural deviation of the complexes from the native structure. This conclusion also lends credence to the information given by the average RMSD values of the apo and the complexes. Therefore, we can safely postulate from the RMSD and RoG indices that the identified compounds have almost the same effect on CpIMPDH compared to P131, which implies that they might also potentiate their action by the same structural inhibitory mechanism.
Conclusions
The limited efficacy of nitazoxanide in treating cryptosporidiosis led to the synthesis of P131, which inhibits CpIMPDH. Inferring from the total binding free energy of P131 and the identified compounds, ZINC46542062 had a better binding free energy to CpIMPDH than P131, suggesting that ZINC46542062 might have better inhibitory potential than P131. Also, from the evaluation of their pharmacokinetic parameters, the identified compounds were more therapeutically suitable than P131, especially in their toxicity (LD50) index, synthetic accessibility, lipophilicity, and the number of rotatable bonds, hydrogen bonds, and acceptors, which altogether impact the permeability of compounds into the cell. On the other hand, P131 and the identified compounds influenced the conformational dynamics of CpIMPDH, almost similarly with negligible variations. Insight from the present study could serve as a starting point in designing new inhibitors for CpIMPDH with improved therapeutic properties.
Data Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Current WL, Garcia LS (1991) Cryptosporidiosis. Clin Microbiol Rev 4:325–358
Clark DP (1999) New insights into human cryptosporidiosis. Clin Microbiol Rev 12:554–563
Shirley D-AT, Moonah SN, Kotloff KL (2012) Burden of disease from cryptosporidiosis. Curr Opin Infect Dis 25:555–563. https://doi.org/10.1097/QCO.0b013e328357e569
Checkley W, White Jr AC, Jaganath D (2015) A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15:85–94
Chappell CL, Okhuysen PC, Sterling CR et al (1999) Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg 60:157–164. https://doi.org/10.4269/ajtmh.1999.60.157
Mac Kenzie WR, Hoxie NJ, Proctor ME et al (1994) A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med 331:161–167. https://doi.org/10.1056/NEJM199407213310304
Molbak K, Hojlyng N, Gottschau A et al (1993) Cryptosporidiosis in infancy and childhood mortality in Guinea Bissau, West Africa. Br Med J 307:417–420. https://doi.org/10.1136/bmj.307.6901.417
Kotloff KL, Nataro JP, Blackwelder WC et al (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet 382:209–222. https://doi.org/10.1016/S0140-6736(13)60844-2
Newman RD, Sears CL, Moore SR et al (1999) Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J Infect Dis 180:167–175. https://doi.org/10.1086/314820
Khan A, Shams S, Khan S et al (2019) Evaluation of prevalence and risk factors associated with Cryptosporidium infection in rural population of district Buner, Pakistan. PLoS One 14. https://doi.org/10.1371/journal.pone.0209188
Squire SA, Ryan U (2017) Cryptosporidium and Giardia in Africa: current and future challenges. Parasit Vectors 10:1–32. https://doi.org/10.1186/s13071-017-2111-y
Vanathy K, Parija SC, Mandal J et al (2017) Cryptosporidiosis: a mini review. Trop Parasitol 7:72. https://doi.org/10.4103/TP.TP_25_17
Sparks H, Nair G, Castellanos-Gonzalez A, White AC (2015) Treatment of Cryptosporidium: what we know, gaps, and the way forward. Curr Trop Med Reports 2:181–187. https://doi.org/10.1007/s40475-015-0056-9
Amadi B, Mwiya M, Sianongo S et al (2009) High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect Dis:9. https://doi.org/10.1186/1471-2334-9-195
Amenta M, Dalle Nogare ER, Colomba C et al (1999) Intestinal protozoa in HIV-infected patients: effect of rifaximin in Cryptosporidium parvum and Blastocystis hominis infections. J Chemother 11:391–395. https://doi.org/10.1179/joc.1999.11.5.391
Hicks P, Zwiener RJ, Squires J, Savell V (1996) Azithromycin therapy for Cryptosporidium parvum infection in four children infected with human immunodeficiency virus. J Pediatr 129:297–300. https://doi.org/10.1016/S0022-3476(96)70258-5
Gathe JC, Mayberry C, Clemmons J, Nemecek J (2008) Resolution of severe cryptosporidial diarrhea with rifaximin in patients with AIDS. J Acquir Immune Defic Syndr 48:365–366
Paromomycin oral : uses, side effects, interactions, pictures, warnings & dosing—WebMD. https://www.webmd.com/drugs/2/drug-5160/paromomycin-oral/details#side-effects. Accessed 28 Jun 2020
Smith NH, Cron S, Valdez LM, Chappell CL, White AC Jr (1998) Combination Drug Therapy for Cryptosporidiosis in AIDS. J Infect Dis 178:900–903. https://doi.org/10.1086/515352
Umejiego NN, Li C, Riera T et al (2004) Cryptosporidium parvum IMP dehydrogenase: identification of functional, structural, and dynamic properties that can be exploited for drug design. J Biol Chem 279:40320–40327. https://doi.org/10.1074/jbc.M407121200
Gorla SK, Kavitha M, Zhang M et al (2012) Selective and potent urea inhibitors of cryptosporidium parvum inosine 5′-monophosphate dehydrogenase. J Med Chem 55:7759–7771. https://doi.org/10.1021/jm3007917
Gorla SK, McNair NN, Yang G et al (2014) Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob Agents Chemother 58:1603–1614. https://doi.org/10.1128/AAC.02075-13
Gorla SK, Kavitha M, Zhang M et al (2013) Optimization of benzoxazole-based inhibitors of Cryptosporidium parvum inosine 5′-monophosphate dehydrogenase. J Med Chem 56:4028–4043. https://doi.org/10.1021/jm400241j
Johnson CR, Gorla SK, Kavitha M et al (2013) Phthalazinone inhibitors of inosine-5′-monophosphate dehydrogenase from Cryptosporidium parvum. Bioorg Med Chem Lett 23:1004–1007. https://doi.org/10.1016/j.bmcl.2012.12.037
Kirubakaran S, Gorla SK, Sharling L et al (2012) Structure–activity relationship study of selective benzimidazole-based inhibitors of Cryptosporidium parvum IMPDH. Bioorg Med Chem Lett 22:1985–1988. https://doi.org/10.1016/j.bmcl.2012.01.029
Macpherson IS, Kirubakaran S, Gorla SK et al (2010) The structural basis of Cryptosporidium-specific IMP dehydrogenase inhibitor selectivity. J Am Chem Soc 132:1230–1231. https://doi.org/10.1021/ja909947a
Maurya SK, Gollapalli DR, Kirubakaran S et al (2009) Triazole inhibitors of Cryptosporidium parvum inosine 5′-monophosphate dehydrogenase. J Med Chem 52:4623–4630. https://doi.org/10.1021/jm900410u
Sharling L, Liu X, Gollapalli DR et al (2010) A screening pipeline for antiparasitic agents targeting Cryptosporidium inosine monophosphate dehydrogenase. PLoS Negl Trop Dis 4:e794. https://doi.org/10.1371/journal.pntd.0000794
Sun Z, Khan J, Makowska-Grzyska M et al (2014) Synthesis, in vitro evaluation and cocrystal structure of 4-oxo-[1]benzopyrano[4,3-c]pyrazole Cryptosporidium parvum inosine 5′-monophosphate dehydrogenase (CpIMPDH) inhibitors. J Med Chem 57:10544–10550. https://doi.org/10.1021/jm501527z
Umejiego NN, Gollapalli D, Sharling L et al (2008) Targeting a prokaryotic protein in a eukaryotic pathogen: identification of lead compounds against cryptosporidiosis. Chem Biol 15:70–77. https://doi.org/10.1016/j.chembiol.2007.12.010
Kim Y, Makowska-Grzyska M, Gorla SK et al (2015) Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity. Acta Crystallogr Sect F Struct Biol Commun 71:531–538. https://doi.org/10.1107/S2053230X15000187
Makowska-Grzyska M, Kim Y, Maltseva N et al (2015) A novel cofactor-binding mode in bacterial IMP dehydrogenases explains inhibitor selectivity. J Biol Chem 290:5893–5911. https://doi.org/10.1074/jbc.M114.619767
Hedstrom L (2009) IMP dehydrogenase: structure, mechanism, and inhibition. Chem Rev 109:2903–2928. https://doi.org/10.1021/cr900021w
Felczak K, Chen L, Wilson D et al (2011) Cofactor-type inhibitors of inosine monophosphate dehydrogenase via modular approach: targeting the pyrophosphate binding sub-domain. Bioorg Med Chem 19:1594–1605. https://doi.org/10.1016/j.bmc.2011.01.042
Allison AC, Kowalski WJ, Muller CD, Eugui EM (1993) Mechanisms of action of mycophenolic acid. Ann N Y Acad Sci 696:63–87. https://doi.org/10.1111/j.1749-6632.1993.tb17143.x
Allison AC, Eugui EM (2000) Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 47:85–118. https://doi.org/10.1016/S0162-3109(00)00188-0
Hassan Baig M, Ahmad K, Roy S et al (2016) Computer aided drug design: success and limitations. Curr Pharm Des 22:572–581. https://doi.org/10.2174/1381612822666151125000550
Kaalia R, Kumar A, Srinivasan A, Ghosh I (2015) An ab initio method for designing multi-target specific pharmacophores using complementary interaction field of aspartic proteases. Mol Inform 34:380–393. https://doi.org/10.1002/minf.201400157
Qing X, Lee XY, De Raeymaeker J et al (2014) Pharmacophore modeling: advances, limitations, and current utility in drug discovery. J Receptor Ligand Channel Res 7:81–92. https://doi.org/10.2147/JRLCR.S46843
Kaserer T, Beck KR, Akram M et al (2015) Pharmacophore models and pharmacophore-based virtual screening: concepts and applications exemplified on hydroxysteroid dehydrogenases. Molecules 20:22799–22832. https://doi.org/10.3390/molecules201219880
Dror O, Schneidman-Duhovny D, Inbar Y et al (2009) Novel approach for efficient pharmacophore-based virtual screening: method and applications. J Chem Inf Model 49:2333–2343. https://doi.org/10.1021/ci900263d
Richmond NJ, Abrams CA, Wolohan PRN et al (2006) GALAHAD: 1. Pharmacophore identification by hypermolecular alignment of ligands in 3D. J Comput Aided Mol Des 20:567–587. https://doi.org/10.1007/s10822-006-9082-y
Jones G, Willett P, Glen RC (1995) A genetic algorithm for flexible molecular overlay and pharmacophore elucidation. J Comput Aided Mol Des 9:532–549. https://doi.org/10.1007/BF00124324
Leach AR, Gillet VJ, Lewis RA, Taylor R (2010) Three-dimensional pharmacophore methods in drug discovery. J Med Chem 53:539–558
Simulations B (2019) Learning the footprints and fingerprints: pharmacophore modeling in the discovery of potential drug candidates. https://doi.org/10.3844/jobsp.2019
Kumalo HM, Soliman ME (2016) Per-residue energy footprints-based pharmacophore modeling as an enhanced in silico approach in drug discovery: a case study on the identification of novel β-secretase1 (BACE1) inhibitors as anti-Alzheimer agents. Cell Mol Bioeng 9:175–189. https://doi.org/10.1007/s12195-015-0421-8
Cele FN, Ramesh M, Soliman MES (2016) Per-residue energy decomposition pharmacophore model to enhance virtual screening in drug discovery: a study for identification of reverse transcriptase inhibitors as potential anti-HIV agents. Drug Des Dev Ther 10:1365–1377. https://doi.org/10.2147/DDDT.S95533
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Eswar N, Webb B, Marti-Renom MA et al (2006) Comparative protein structure modeling using Modeller. Curr Protoc Bioinforma Chapter 5:Unit-5.6. https://doi.org/10.1002/0471250953.bi0506s15
ZINCPharmer: pharmacophore search of the ZINC database. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3394271/. Accessed 29 Apr 2020
Irwin JJ, Shoichet BK (2005) ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177. https://doi.org/10.1021/CI049714
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:1–13. https://doi.org/10.1038/srep42717
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44:235–249. https://doi.org/10.1016/S1056-8719(00)00107-6
Ripphausen P, Nisius B, Peltason L, Bajorath J (2010) Quo vadis, virtual screening? A comprehensive survey of prospective applications. J Med Chem 53:8461–8467. https://doi.org/10.1021/jm101020z
Trott O, Olson A (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334.AutoDock
Forli S, Huey R, Pique ME et al (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905–919. https://doi.org/10.1038/nprot.2016.051
Morris GM, Ruth H, Lindstrom W et al (2009) Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Schrödinger Release 2020-4: Maestro, Schrödinger, LLC, New York, NY, 2020
Karthick V, Nagasundaram N, Doss CGP et al (2016) Virtual screening of the inhibitors targeting at the viral protein 40 of Ebola virus. Infect Dis Poverty 5. https://doi.org/10.1186/s40249-016-0105-1
Lawal M, Olotu FA, Soliman MES (2018) Across the blood–brain barrier: neurotherapeutic screening and characterization of naringenin as a novel CRMP-2 inhibitor in the treatment of Alzheimer’s disease using bioinformatics and computational tools. Comput Biol Med 98:168–177. https://doi.org/10.1016/J.COMPBIOMED.2018.05.012
Liao C, Sitzmann M, Pugliese A, Nicklaus MC (2011) Software and resources for computational medicinal chemistry. Future Med Chem 3:1057–1085
Molinspiration Cheminformatics. https://www.molinspiration.com/. Accessed 1 May 2020
Drwal MN, Banerjee P, Dunkel M et al (2014) ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res 42:3–8. https://doi.org/10.1093/nar/gku401
Sander T, Freyss J, Von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. https://doi.org/10.1021/ci500588j
Agoni C, Munsamy G, Ramhrack P, Soliman M (2020) Human rhinovirus inhibition through capsid “canyon” perturbation: structural insights into the role of a novel benzothiophene derivative. Cell Biochem Biophys 78:3–13
Agoni C, Salifu EY, Munsamy G et al (2019) CF3-pyridinyl substitution on anti-malarial therapeutics: probing differential ligand binding and dynamical inhibitory effects of a novel triazolopyrimidine-based inhibitor on Plasmodium falciparum dihydroorotate dehydrogenase. Chem Biodivers. https://doi.org/10.1002/cbdv.201900365
Olotu FA, Soliman MES (2019) Dynamic perspectives into the mechanisms of mutation-induced p53-DNA binding loss and inactivation using active perturbation theory: structural and molecular insights toward the design of potent reactivators in cancer therapy. J Cell Biochem 120:951–966. https://doi.org/10.1002/jcb.27458
Nair PC, Miners JO (2014) Molecular dynamics simulations: from structure function relationships to drug discovery. Silico Pharmacol 2. https://doi.org/10.1186/s40203-014-0004-8
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Grest GS, Kremer K (1986) Molecular dynamics simulation for polymers in the presence of a heat bath. Phys Rev A 33:3628–3631. https://doi.org/10.1103/PhysRevA.33.3628
Berendsen HJC, Postma JPM, Van Gunsteren WF et al (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. https://doi.org/10.1063/1.448118
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Roe DR, Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095. https://doi.org/10.1021/ct400341p
Seifert E (2014) OriginPro 9.1: scientific data analysis and graphing software—software review. J Chem Inf Model 54:1552. https://doi.org/10.1021/ci500161d
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discovery 10:449–461. https://doi.org/10.1517/17460441.2015.1032936
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51:69–82. https://doi.org/10.1021/ci100275a
Chaudhary N, Aparoy P (2017) Deciphering the mechanism behind the varied binding activities of COXIBs through molecular dynamic simulations, MM-PBSA binding energy calculations and per-residue energy decomposition studies. J Biomol Struct Dyn 35:868–882. https://doi.org/10.1080/07391102.2016.1165736
Gupta A, Chaudhary N, Aparoy P (2018) MM-PBSA and per-residue decomposition energy studies on 7-phenyl-imidazoquinolin-4(5H)-one derivatives: identification of crucial site points at microsomal prostaglandin E synthase-1 (mPGES-1) active site. Int J Biol Macromol 119:352–359. https://doi.org/10.1016/j.ijbiomac.2018.07.050
Case DA (2018) Amber 18. Univ California, San Francisco
Woods CJ, Malaisree M, Michel J et al (2014) Rapid decomposition and visualisation of protein–ligand binding free energies by residue and by water. Faraday Discuss 169:477–499. https://doi.org/10.1039/c3fd00125c
Shoichet BK (2004) Virtual screening of chemical libraries. Nature 432:862–865
Kenakin T (2003) Predicting therapeutic value in the lead optimization phase of drug discovery. Nat Rev Drug Discov 2:429–438
Huber W (2005) A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound–target interactions. J Mol Recognit 18:273–281. https://doi.org/10.1002/jmr.744
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R et al (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Yang T, Wu JC, Yan C et al (2011) Virtual screening using molecular simulations. Proteins Struct Funct Bioinform 79:1940–1951. https://doi.org/10.1002/prot.23018
Wallnoefer HG, Liedl KR, Fox T (2011) A challenging system: free energy prediction for factor Xa. J Comput Chem 32:1743–1752. https://doi.org/10.1002/jcc.21758
Weis A, Katebzadeh K, Söderhjelm P et al (2006) Ligand affinities predicted with the MM/PBSA method: dependence on the simulation method and the force field. J Med Chem 49:6596–6606. https://doi.org/10.1021/jm0608210
Striepen B, Pruijssers AJP, Huang J et al (2004) Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci U S A 101:3154–3159. https://doi.org/10.1073/pnas.0304686101
Hedstrom L, Liechti G, Goldberg JB, Gollapalli DR (2011) The antibiotic potential of prokaryotic IMP dehydrogenase inhibitors. Curr Med Chem 18:1909–1918. https://doi.org/10.2174/092986711795590129
Tetko IV, Bruneau P, Mewes H-W et al (2006) Can we estimate the accuracy of ADME–Tox predictions? Drug Discov 11:700–707. https://doi.org/10.1016/j.drudis.2006.06.013
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Arnott JA, Planey SL (2012) The influence of lipophilicity in drug discovery and design. Expert Opin Drug Discovery 7:863–875. https://doi.org/10.1517/17460441.2012.714363
Liu X, Testa B, Fahr A (2011) Lipophilicity and its relationship with passive drug permeation. Pharm Res 28:962–977
Leeson PD, Springthorpe B (2007) The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov 6:881–890. https://doi.org/10.1038/nrd2445
Waring MJ (2009) Defining optimum lipophilicity and molecular weight ranges for drug candidates—molecular weight dependent lower log D limits based on permeability. Bioorg Med Chem Lett 19:2844–2851. https://doi.org/10.1016/j.bmcl.2009.03.109
Gleeson MP, Hersey A, Montanari D, Overington J (2011) Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat Rev Drug Discov 10:197–208. https://doi.org/10.1038/nrd3367
Hughes JD, Blagg J, Price DA et al (2008) Physiochemical drug properties associated with in vivo toxicological outcomes. Bioorg Med Chem Lett 18:4872–4875. https://doi.org/10.1016/j.bmcl.2008.07.071
Greene N, Aleo MD, Louise-May S et al (2010) Using an in vitro cytotoxicity assay to aid in compound selection for in vivo safety studies. Bioorg Med Chem Lett 20:5308–5312. https://doi.org/10.1016/j.bmcl.2010.06.129
Price DA, Blagg J, Jones L et al (2009) Physicochemical drug properties associated with in vivo toxicological outcomes: a review. Expert Opin Drug Metab Toxicol 5:921–931
Ahmed SSSJ, Ramakrishnan V (2012) Systems biological approach of molecular descriptors connectivity: optimal descriptors for oral bioavailability prediction. PLoS One 7:e40654. https://doi.org/10.1371/journal.pone.0040654
Hopkins AL, Keserü GM, Leeson PD et al (2014) The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov 13:105–121. https://doi.org/10.1038/nrd4163
Hopkins AL, Groom CR, Alex A (2004) Ligand efficiency: a useful metric for lead selection. Drug Discov Today 9:430–431
Carr RAE, Congreve M, Murray CW, Rees DC (2005) Fragment-based lead discovery: leads by design. Drug Discov Today 10:987–992
Mortenson PN, Murray CW (2011) Assessing the lipophilicity of fragments and early hits. J Comput Aided Mol Des 25:663–667. https://doi.org/10.1007/s10822-011-9435-z
Bembenek SD, Tounge BA, Reynolds CH (2009) Ligand efficiency and fragment-based drug discovery. Drug Discov Today 14:278–283
Nissink JWM (2009) Simple size-independent measure of ligand efficiency. J Chem Inf Model 49:1617–1622. https://doi.org/10.1021/ci900094m
Reynolds CH, Tounge BA, Bembenek SD (2008) Ligand binding efficiency: trends, physical basis, and implications. J Med Chem 51:2432–2438. https://doi.org/10.1021/jm701255b
Ryckmans T, Edwards MP, Horne VA et al (2009) Rapid assessment of a novel series of selective CB2 agonists using parallel synthesis protocols: a lipophilic efficiency (LipE) analysis. Bioorg Med Chem Lett 19:4406–4409. https://doi.org/10.1016/j.bmcl.2009.05.062
Ertl P, Schuffenhauer A (2009) Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J Cheminform 1:8. https://doi.org/10.1186/1758-2946-1-8
Bahmani A, Saaidpour S, Rostami A (2017) A simple, robust and efficient computational method for n-octanol/water partition coefficients of substituted aromatic drugs. Sci Rep 7:1–14. https://doi.org/10.1038/s41598-017-05964-z
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717. https://doi.org/10.1021/jm000942e
Fernandes J, Gattass CR (2009) Topological polar surface area defines substrate transport by multidrug resistance associated protein 1 (MRP1/ABCC1). J Med Chem 52:1214–1218. https://doi.org/10.1021/jm801389m
Prasanna S, Doerksen R (2008) Topological polar surface area: a useful descriptor in 2D-QSAR. Curr Med Chem 16:21–41. https://doi.org/10.2174/092986709787002817
Congreve M, Carr R, Murray C, Jhoti H (2003) A “rule of three” for fragment-based lead discovery? Drug Discov. Today 8:876–877
Gulzar M, Ali S, Khan F et al (2019) Binding mechanism of caffeic acid and simvastatin to the integrin linked kinase for therapeutic implications: a comparative docking and MD simulation studies. J Biomol Struct Dyn 37:4327–4337
Machaba KE, Mhlongo NN, Soliman MES (2018) Induced Mutation Proves a Potential Target for TB Therapy: A Molecular Dynamics Study on LprG. Cell Biochem Biophys 76:345–356. https://doi.org/10.1007/s12013-018-0852-7
Pitera JW (2014) Expected distributions of root-mean-square positional deviations in proteins. J Phys Chem B 118:6526–6530. https://doi.org/10.1021/jp412776d
Brüschweiler R (2002) Efficient RMSD measures for the comparison of two molecular ensembles. Proteins Struct Funct Bioinform 50:26–34. https://doi.org/10.1002/prot.10250
Król M, Roterman I, Piekarska B et al (2005) Analysis of correlated domain motions in IgG light chain reveals possible mechanisms of immunological signal transduction. Proteins Struct Funct Genet 59:545–554. https://doi.org/10.1002/prot.20434
Acknowledgment
The authors appreciate the Centre for High Performance Computing (CHPC), Cape Town, South Africa, for making computational resources available.
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
Conceptualization, methodology, formal analysis and investigation, writing—original draft preparation: K.F.O. Methodology: E.A.I. Writing—review and editing: C.A. Writing—review and editing: F.A.O. Supervision: M.E.S.S.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 813 kb)
Rights and permissions
About this article
Cite this article
Omolabi, K.F., Iwuchukwu, E.A., Agoni, C. et al. A probable means to an end: exploring P131 pharmacophoric scaffold to identify potential inhibitors of Cryptosporidium parvum inosine monophosphate dehydrogenase. J Mol Model 27, 35 (2021). https://doi.org/10.1007/s00894-020-04663-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04663-3