Abstract
This study aimed to identify compounds that are capable of inhibiting polyphosphate kinase (PPK), the protein that shares a similar pathway with the target of isoniazid (INH) in the oxidative phosphorylation pathway of Mycobacterium tuberculosis (MTB). The three-dimensional structure of the PPK was predicted based on the homology modeling principle via Modeller9.16. Structural analysis revealed that the PPK has three active sites—metal ion-binding site (Arg431 and Arg461), ATP-binding site (Asn91, Tyr524, Arg624, and His652), and phosphohistidine intermediate active site (His491). The amino acids mentioned earlier play an essential role in the activity of the PPK, and their inhibition would block the function of the PPK. Ten thousand one hundred and six (10,106) ligands were obtained through virtual screening against Zinc database. These compounds were filtered by Lipinski rule of five and molecular docking analysis. A total of ten compounds with good AutoDock binding energy that varied between − 9.92 and − 8.01 kcal/mol which was lower than the binding energy of − 1.13 kcal/mol of Mg2+ (metallic cofactor) were selected. These compounds were further filtered for their pharmacokinetic and toxicity properties to further remove the compounds with unwanted properties. Three ligands—ZINC41125011, ZINC20318248, and ZINC20321877 with desired pharmacokinetic and toxicity properties were selected for molecular dynamic (MD) simulation and molecular generalized Born surface area (MM-GBSA) analysis. The result of the studies shows that all the three complexes are relatively stable in the binding site of the PPK, after 50 ns MD simulation. Therefore, these identified compounds are regarded as prospective inhibitors of MTB after positive experimental validation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tuberculosis (TB) is a disease caused by an infectious agent known as Mycobacterium tuberculosis (MTB). It is one of the most dominant and fatal infectious diseases, with the prevalence rate of 10.4 million cases and 1.4 million mortality rate in 2015 (WHO 2016). The disease poses a serious threat as a result of the emergence of multi-drug-resistant TB (MDR-TB). This resistance is caused by the strains of MTB that are resistant to rifampicin and isoniazid (first-line antibiotics), and it is not a recent phenomenon. Since 2006, more severely resistant strains of MTB known as extensively drug-resistant (XDR) TB was discovered (CDC 2006). This strain, being MDR, is also resistant to fluoroquinolone and at most a second-line drug (kanamycin, capreomycin or amikacin). Recently, more deadly strains have been discovered which are resistant to all antituberculosis medicines and they have been termed as total drug-resistant TB (TDR-TB) (Velayati et al. 2009). Resistance to antituberculosis drugs is attributed to many hereditary factors such as spontaneous chromosomal mutation during mycobacterial replications or amplification of genetic mutation as a result of human negligence which includes single drug therapy, prescription of incorrect medication by physicians and lack of strict adherence of the patients to drugs’ prescription (Vareldzis et al. 1994). Another essential feature of MTB is the lack of plasmid, which mediates the resistance process in several bacterial species. Hence, it is necessary to develop antituberculosis drugs that can minimize the drug-resistant mayhem.
Isoniazid has long been used for the treatment of tuberculosis. It is a prodrug that requires initiation or activation by catalase–peroxidase (KatG) enzymes encoded by KatG gene to produce its effects (Zhang et al. 1992). The antibiotic interferes with the biosynthesis of mycolic acid via protein [NADH-dependent enoyl-acyl carrier (ACP)-reductase] encoded by inhA in the oxidative phosphorylation pathway (Rawat et al. 2003). However, the drug becomes resistant to MTB when it undergoes mutation in some selected genes such as KatG, inhA, NDH, ahpC, and KasA (Palomino and Martin 2014). INH resistance is the most frequently occurring in MTB as compared to other tuberculosis drugs at a rate of 105−6 per base per generation frequency in vitro. Although, polyphosphate kinase (PPK) shared the same pathway with the target of isoniazid, it was proposed as an alternative target to inhibit the pathway (Lee et al. 2001; Johnson et al. 2006; Cloete et al. 2016). Therefore, inhibition of this enzyme could hinder the pathway and disrupt the growth of the MTB by limiting the availability of ATP during the dormancy (Murphy and Brown 2007).
Methods
Comparative modeling
The primary sequence of PPK (NP_217500.1) from Mycobacterium tuberculosis was obtained from NCBI (National Centre for Biotechnological Information). The resemblance of the PPK sequence was explored using BLASTP (Basic Local Alignment Search Tool) (Altschul et al. 1997) against the PDB (Protein Data Bank) (Bernstein et al. 1977). Four protein sequences (1XDO, 1XDP, 2O8R, and 3GGD) were obtained using query coverage, sequence identity, sequence similarity, and E-value. Though, 2O8R was further favored due to the high resolution of 1.5 Å. The pair-sequence alignment of the PPK and the template (2O8R) was performed via ClustalW (Thompson et al. 1994). The result of the sequence alignment indicated a high sequence similarity of 53.4%. Thus, the selected template is appropriate for comparative modeling (Xu et al. 2015). The 3D structure of PPK was modeled based on the protein structure modeling program Modeller9v16 (Pieper et al. 2010). The technique was used to build a 3D structure using data obtained from the alignment which include side chains, hydrogen bonding, main chain, and dihedral angles through spatial restraints. The process generates ten different 3D model structures and the model with Least DOPE (discrete optimized protein energy) value was chosen and subjected to MD simulation using AMBERTOOLS10 to minimize energy (Case et al. 2015). Lastly, the 3D model of the PPK, the minimized model, and the template were assessed via Ramachandran plot (Zhang et al. 2005), Verify_3D (Lüthy et al. 1992), and ERRAT (Colovos and Yeates 1993).
Virtual screening (VS)
VS was employed to identify good ligands which have an affinity to bind to macromolecule using ligand libraries. Virtual screening was performed to ascertain ligands with the preferred therapeutic outcome such as antimicrobial activity. The PPK complexes with ATP submitted the RASPD tool (Mukherjee and Jayaram 2013) to identified a compound with better docking pose. A total of ten thousand one hundred and six (10,106) ligands that can bind to PPK with minimum energies were found from PubChem (Kim et al. 2015) and Zinc (Irwin and Shoichet 2005) databases. These compounds were validated via PyRx program (Rigi et al. 2017), and further filtered for physicochemical properties such as molecular weight, lipophilicity, hydrogen bond donor and hydrogen bond acceptor via DataWarrior tool (Sander et al. 2015). The 3D structure of the ligands with suitable features was retrieved from the databases and used for the docking analysis to determine their free binding affinities with PPK.
Molecular docking analysis
Molecular docking analysis of PPK and the selected ligands that possessed the required drug-like properties was carried out to assess the free binding conformation of the protein–ligand complex via AutoDock4.2 program (Morris et al. 1998). This binding conformation would help to determine the binding energy of the protein and the ligand. The AutoDock4.2 is an essential tool for docking as it is swift and allows the springiness of the side chain of the macromolecule. During the process, polar hydrogen with known Kollman charges is used for the protonation of the PPK. The model of the PPK is obtained via parameterization of the AutoDock4.2. The PDBQT is derived from Protein Data Bank using the graphical interface of the AutoDock4.2. The PDBQT is a coordinate file which contained all the data related to the types of the atom, partial particle charges, and the torsional degree of freedom. Autogrid is used to calculate electrostatic energy, desolvation potentials and grid affinity of each atom such as carbon, hydrogen, nitrogen, and oxygen of the ligand. The value of x, y, and z of the grid center was set. The dimension of the grid was kept at 60 × 60 × 60 Å, with a spacing of 0.375 Å. The binding energy of the PPK–ligand complex was calculated using a Lamarckian genetic algorithm (La Motta et al. 2007; Gasteiger and Marsili 1980). Ten different free binding energies were generated for each ligand, and the least binding energy was selected. Lastly, the PPK–ligands complex was analyzed using Pymol (1.7.4.5 Edu) (DeLano 2002) and Ligplot+ tool (Laskowski and Swindells 2011; Wallace et al. 1996).
ADME and toxicity analysis
ADME and toxicity properties (for example, absorption, distribution, metabolism, excretion, and metabolism) were predicted via DataWarrior program (Sander et al. 2015), AdmetSAR program (Cheng et al. 2012), and ADME/TOX tool (Lipinski et al. 1997; Veber et al. 2002). This phase is vital because of several ligands unable to reach a clinical trial as a result of unwanted pharmacokinetic properties. In this study, the ligands with better free binding affinities were chosen and checked for ADME and toxicity properties. The properties comprise of “Blood–Brain Barrier (BBB) penetration, Human Intestinal Absorption (HIA), Cytochrome P450 (CYP450 2D6) Inhibitor, Plasma Protein Binding, mutagenicity, tumorigenicity, irritation, and reproduction.” These properties are essential due to their effects on the exposure of the inhibitor to the human body, which affects the pharmacological activity and performance of the molecule.
Molecular dynamic (MD) simulation analysis
The ligands which possessed acceptable pharmacokinetic features were selected and subjected to MD simulation using AMBERTOOLS10 package (Case et al. 2015). The entire complex prepared via the addition of explicit hydrogen to them using protonate 3D and the missing parameter of the ligands were added using the antechamber. TLeap contained molecular graphics component of the system topology, and it was used to construct the topology and coordinate file of the PPK and the ligand. The GAFF and ff12SB force field were allotted to protein and ligand, respectively. The whole system was placed into TIP3P water neutralized using sodium ions curbed in a buffer solution of 10 Å inside an octahedral box. The structural artifact that arose during edifice of the model was detached by exposing the system to a minimization cycle of 5000 steps of conjugate gradient and 5000 steps of steepest descent, with a constraint run at 544 kcal/mol/Å on the complex. Furthermore, the system was minimized for 2500 steps of steepest descent and with 2500 steps of conjugate gradients after the elimination of the restrained. Then, the system was heated with the initial and final temperature of 0.0 k and 300 k, respectively, for 100,000 steps. The collision frequency was fixed at 1 ps in the absence of pressure control. The production of MD simulation was performed at a steady temperature of 300 K and the pressure of 1 atm via Berendsen barostat for constant pressure simulation using the time step of 2 fs. The MD simulation of 50 ns long was carried out and SHAKE was used to restraint all the hydrogen bonds. Lastly, the stability of the complexes was examined by determining the root mean square deviation (RMSD). The deviation of the residues around their initial position was analyzed using root mean square fluctuation (RMSF). A radius of gyration was also examined to analyze the degree of how folded or unfolded the complex was. The entire analyses were carried out using Ptraj component of the AMBERTOOLS10 package.
Binding energy analysis using MM-GBSA
The MM-GBSA is an excellent technique used to ascertain the binding energy of a small molecule (mostly ligand) and the macromolecule (protein). It based on the trajectory of molecular dynamics simulations of the protein–ligand complex. The method attained intellectual achievement over the years concerning precision and computational strength among the empirical scoring (Genheden and Ryde 2015). The binding energy of PPK–ligands complex was determined using MM-GBSA techniques implemented in Amber 14 in this study. The binding energy was examined based on the average of 500 snapshots extracted at each 10 ps from the simulation trajectory of the last 5 ns. All the steps used are presented below:
where Gcomplex is the free binding energy of the complex, Greceptor is the free binding energy of the receptor, Gligand is the free binding energy of the ligand, T∆S is the sum of configurational entropy, EMM is the sum of molecular mechanical gas-phase free binding energy, Gsol is the sum of solvation binding energy, Eele is the gas-phase electrostatic energy, EvdW is the van der Waals energy, polar energy (Gpolar) and nonpolar energy (Gnonpolar) and ∆GMM-GBSA is the sum of free binding energy of the protein and ligand complex.
Result and discussion
Homology modeling of PPK
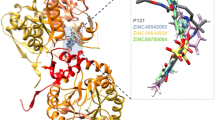
Comparative modeling was used to build the three-dimensional structure of polyphosphate kinase (PPK) using the reference sequence (NP_217500.1) obtained from NCBI. The 3D model was developed based on the principle implemented in Modeller9.16 (Webb and Sali 2014; Martí-Renom et al. 2000; Fiser and Do 2000). The query sequence (reference sequence) was searched against the PDB. Four template proteins (1XDO, 1XDP, 2O8R, and 3GGD) were obtained. However, 2O8R was further selected based on high resolution, and high statistical significance made it an essential template for the homology modeling. The pairwise alignment between the query sequence (PPK) and the template (2O8R) showed that their sequence share 35.0% (234/669 residues) sequence identity, 53.4% (357/669 residues) sequence similarity, and 6.9% (46/669 residues) gaps (see supplementary material). Besides, the query sequence and the template are highly conserved in the phosphohistidine intermediate active site (His491), ATP-binding site (Asn91, Tyr524, Arg624, and His652) and metallic ion (Mg2+)-binding site (Arg431 and Arg461) (Chen et al. 2017; Nightingale et al. 2017). During the modeling process, all the side chains, structurally conserved regions, structural variable regions, and main chains were transferred to the PPK based on spatial restraints. The process generated ten models and the one with least DOPE energy value and folded assessment score GA341 of one was selected. The selected model was then used for energy minimization via MD simulation using AMBERTOOLS10 package. The minimized model was superimposed on the template from Porphyromonas gingivalis with an RMSD of 0.448 Å (Fig. 1b). The validation of the structural model, the minimized model, and the template was performed using Verify_3D, stereochemistry quality (Ramachandran plot), and ERRAT Quality Factor. The analysis of the Ramachandran plot revealed that the stereochemistry quality of the 3D modeled had 83.7% of the entire amino acids in the most favorable region, 14.7% in the additional allowed region, and 1.1% in the generously allowed region, while it was 0.6% in the disallowed region. In contrast, the minimized 3D model and the template had 89.0% and 89.8% of their entire amino acids in the most favorable regions. However, 9.4% and 9.8% of their total residues were in the additional allowed regions, 0.8% and 0.4% were in the generously allowed region, and 0.9% and 0.3% were in the disallowed regions, respectively. Based on the analyses of the Ramachandran plot, the minimized 3D model structure had a good and satisfactory quality (Table 1). The quality factor of the modeled structure, template, and the minimized modeled structure was determined using ERRAT factor for protein structure verification algorithm implemented in the ERRAT. ERRAT results indicate the proportion of various atoms that underwent non-bonded interactions, with 95% statistical significance level. The ERRAT value of the 3D model before minimization was 55.216%, while after minimization the quality increased and had a score of 72.424%. Similarly, Verify_3D was used to determine the compatibility of the 3D atomic model with a primary sequence (1D), based on the structural class in its location and environment. The verify_3D score of the 3D model and the minimized 3D model was 81.40% and 82.08%, respectively, with an improvement in the 3D structure after the minimization. The total amount of the van der Waals repulsion energy and steric clashes before minimization of the 3D model was 1158 and 1277.32 kcal/mol, while after minimization it was 309 and 188.844 kcal/mol, respectively. The reduction in the values of van der Waals repulsion energy and steric clashes in the minimized 3D modeled was evidence of gaining stability after the minimization (Table 1).
The modeled structure of PPK and the structural superimposition to the template (2O8R). a The 3D model of PPK indicating phosphohistidine intermediate active site (His491), ATP-binding site (Asn91, Tyr524, Arg624 and His652) and metallic ion (Mg2+)-binding site (Arg431 and Arg461). b Structural superimposition of the Polyphosphate kinase (PPK) (blue) from Porphyromonas gingivalis and the model structure (red) (RMSD = 0.448 Å)
Virtual screening and docking analysis
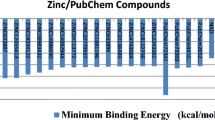
The analysis of polyphosphate kinase (PPK) from MTB revealed that the protein has three binding sites, which include “ATP-binding site, metal ion-binding site, and phosphohistidine intermediate active site.” The amino acids involved in the ATP-binding site are Asn91, Tyr524, Arg624, and His652. The amino acids that formed the metal ion-binding site are Arg431 and Arg461. Finally, the residue involved in the phosphohistidine intermediate active site is His491. Inhibition of any of the above residues that formed these binding sites impedes the function of the protein. VS of ligand database could offer prospective inhibitors which are possibly used as a drug in the future. Ten thousand one hundred and six (10,106) ligands were obtained based on virtual screening against Zinc database. These ligands were checked for Lipinski rule of five properties and docking analysis (Tables 2 and 3). The result of docking analysis showed that the selected compounds exhibited numerous bonding interactions (such as hydrophobic interaction, electrostatic interaction and hydrogen bonding interaction) with the PPK. Ten compounds (ZINC13400518, ZINC41125011, ZINC00095540, ZINC40478863, ZINC20318248, ZINC13400496, ZINC13400518, ZINC20321877, ZINC00095542 and ZINC40478862) with good binding affinities were selected. The binding affinities of the selected ligands varied between − 9.92 and − 8.01 kcal/mol which was lower than the binding affinity of − 1.13 kcal/mol for Mg2+ (Fig. 2). The amino acids which are involved in the hydrogen bonds with the selected ligands include Arg56, Trp60, Lys83, Asn91, Asn515, Lys519, Thr520, Arg522, Asp526, Ala521, Arg624, and Ile626. ZINC13400518 had the binding affinity of − 8.78 kcal/mol and formed three hydrogen bonds via interactions with a polar amide of Asn91 (distance = 2.70 Å), an amino group of Arg522 (distance = 2.90 Å), and Arg624 (distance = 3.17 Å) (Fig. 3a). ZINC41125011 had the best binding affinity of − 9.92 kcal/mol. This compound formed seven hydrogen bonds by accepting electrons from the amino group of Lys83 (distance = 3.31 Å), the polar amide of Asn91 (distance = 2.97 Å) and amino groups of Arg522 (distance = 3.25 Å, 2.87 Å and 3.22 Å) and Arg624 (distance = 2.92 Å and 2.92 Å) (Table 3). Interaction of the ligand with Asn91 is essential, since the amino acid is involved in the ATP-binding site of the PPK. These interactions would impede the catalytic function of the protein in MTB (Fig. 3b). Five compounds (ZINC13400518, ZINC41125011, ZINC40478863, ZINC13400518, and ZINC00095542) formed hydrogen bonds by accepting electrons from the amino group of Arg624, while two other ligands (ZINC20318248 and ZINC20321877) exhibited electrostatic interactions with the same residue (Fig. 3a–j). Arg624 is a vital amino acid which formed the ATP-binding site of the PPK. Formation of hydrogen bonds or electrostatic interaction between the amino acids mentioned above and the ligands help to prevent the catalytic function of the protein. Likewise, four ligands (ZINC13400518, ZINC41125011, ZINC40478863, and ZINC00095542) formed four hydrogen bonds by accepting electrons from polar amide of Asn91, while another four ligands (ZINC20318248, ZINC13400496, ZINC13400518, and ZINC40478862) presented electrostatic interactions with the Asn91. As mentioned earlier, Asn91 is essential in PPK, and it was involved in the ATP-binding site. Finally, three (ZINC00095540, ZINC20318248, and ZINC40478862) and 2 (ZINC00095540 and ZINC40478862) compounds exhibited electrostatic interactions with Tyr524 and His652, respectively. Interaction of the ligands with any of these amino acids would affect the catalytic function of the PPK. Hence, the entire ligands selected in this study are prospective inhibitors of PPK, based on the docking result.
ADME and toxicity analysis
The results of molecular docking revealed that, ten compounds (ZINC13400518, ZINC41125011, ZINC00095540, ZINC40478863, ZINC20318248, ZINC13400496, ZINC13400518, ZINC20321877, ZINC00095542 and ZINC40478862) had good binding affinities. These compounds were checked for ADME and toxicity properties to further eliminate the compounds with unwanted features which might be fatal to the human system. Most of the drug’s failure was associated with toxicity or unwanted pharmacokinetic features. The ADMET properties analyzed include “Blood–Brain Barrier (BBB) penetration, Human Intestinal Absorption (HIA), Cytochrome P450 (CYP450 2D6) Inhibitor, Aqueous Solubility, Plasma Protein Binding, mutagenicity, Ames test, tumorigenicity, carcinogenicity, reproducibility, and irritability.” The results showed that all the compounds passed via BBB. Similarly, all the ligands had positive HIA except ZINC00095540 and ZINC00095542 which rendered them harmful and therefore violated the ADME rules. None of the compounds were found to be a potential inhibitor of CYP450 2D6. Likewise, the ligands were partially attached to the plasma membrane and can diffuse via the membrane to exert pharmacological activity (Table 4). Also, the analysis of the toxicity was performed for all the selected compounds. The results of the toxicity studies revealed that the compounds were harmless except ZINC13400518, ZINC40478863, ZINC13400496, ZINC13400518, and ZINC40478862 which varied in their toxicity level (Table 4). Though two of these compounds (ZINC00095540 and ZINC00095542) were harmless, they disobey other ADME properties (HIA-). Therefore, three compounds (ZINC41125011, ZINC20318248, and ZINC20321877) that fulfilled all ADMET properties were selected for the MD simulation and MM-GBSA studies (Table 4).
MD simulation studies
The outcome of the molecular docking studies showed that ten compounds with good binding affinities were selected and checked for ADMET properties. This screening led to the selection of three compounds (ZINC41125011, ZINC20318248, and ZINC20321877) that had all the pharmacokinetics features. These compounds were subjected to MD simulation studies. The MD simulation of the three compounds was performed to assess their stability in the substrate-binding site of the PPK. Three protein–ligand complexes were simulated differently for 50 ns, and the complexes were named as protein-complex-1 (PPK–ZINC41125011), protein-complex-2 (PPK–ZINC20318248), and protein-complex-3 (PPK–ZINC20321877). RMSD was used to analyze the stability of the protein–ligand complexes. RMSF was used to determine the movement of the amino acid residues within the substrate-binding site during the 50 ns MD simulation. The firmness of the complex was checked using a radius of gyration to determine whether the complex is folded or not. The result of the MD simulation revealed that protein-complex-1 was equilibrated and stabilized at 1 ns of MD simulation, with an average value of 3.90 ± 0.02 Å, a maximum value of 4.64 Å, and a minimum value of 1.42 Å. It (protein-complex-1) achieved equilibrium and rigidity due to the interaction of the ZINC41125011 with the flexible loop regions of the PPK. This interaction augmented its rigidity and reduced its oscillation. Protein-complex-2 equilibrated and oscillated at 5 ns, with a mean value of 4.18 ± 0.02 Å, a maximum value of 5.08 Å and minimum value of 1.22 Å, although it was less stable when compared with the protein-complex-1. The equilibration and oscillation of the complex (protein-complex-2) were studied around its fixed site, signifying high rigidity and lesser instability of the complex. This rigidity probably occurred due to the interaction of the bound ligand and the flexible loop regions of the PPK. Similarly, protein-complex-3 equilibrated at 20 ns and had a mean value of 5.47 ± 0.04 Å, a maximum value of 6.82 Å, and a minimum value of 1.24 Å, although it was less than the average values of protein-complex-1 and protein-complex-2. Hence, the three complexes achieved a different level of rigidity during the 50 ns MD simulation (Fig. 4). The RMSF of the protein-complex-1, protein-complex-2, and protein-complex-3 was determined, and the result represents the degree of motion of amino acids within the substrate-binding site of the PPK. In the protein-complex-1 and PPK—protein-complex-3 complex, all the amino acids had RMSF values less than 10 Å (< 10 Å) except the residues between Gln300–Arg308, which had values > 10 Å. Likewise, all the amino acids in the protein-complex-2 had RMSF values < 10 Å excluding Gln300–Ala301 (Fig. 5). Lesser RMSF values in the majority of the sites of the entire complexes were not separated from the capability of the compounds to present both hydrogen and electrostatic interactions with the flexible loop region of the PPK. This is also proved by the RMSD result where the entire complex equilibrated and oscillated with low values (Fig. 4). Also, all the ligands had low residue fluctuation in the metal ion-binding site (Arg431 and Arg461), ATP-binding site (Asn91, Tyr524, Arg624, and His652) and phosphohistidine intermediate active site (His491) of the PPK. As shown in Fig. 6, all the residues in the binding site had RMSF values between 2 and 4 Å except residue Arg461 in protein-complex-2 which had the RMSF value of 6.5 Å. Also, the main secondary structural elements of the protein and the ligands (ZINC41125011, ZINC20318248, and ZINC20321877) in all the protein–ligand complexes remained close to their initial structures before the MD simulation. As indicated in Fig. 7, the final structures (after MD simulation) were superimposed to their corresponding initial structures (before MD simulation) and had RMSF values between 2 and 3 Å. These RMSF values showed that all the ligands remained close to their initial binding site after the 50 ns MD simulation. The radius of gyration of all the complexes was analyzed to check the level of compactness in the protein–ligand complex, based on how folded or unfolded the complex is. In the protein-complex-1, it equilibrated and moved in a relatively constant value between 29 and 29.5 Å throughout the 50 ns. Similarly, protein-complex-2 and protein-complex-3 complex equilibrated and moved with the average mean of 29.50 ± 0.01 Å and 29.09 ± 0.01 Å, respectively (Fig. 8). Therefore, the three complexes (protein-complex-1, protein-complex-2, and protein-complex-3) formed a relatively stable folded polypeptide structure after the 50 ns MD simulation.
Superposition of the initial complex structures before MD simulation and final complex structures after MD simulation of 50 ns. a The final structure of protein-complex-1 is indicated in blue, while the initial structure is indicated in red (RMSF = 2.885 Å). b The final structure of protein-complex-2 is indicated in green, while the initial structure is indicated in red (RMSF = 2.930 Å). c The final structure of protein-complex-3 is indicated in yellow, while the initial structure is indicated in red (RMSF = 3.160 Å)
MM-GBSA (free binding energy) analysis
This technique is a vital method for calculating the binding energy of a small molecule (mostly ligand) and the macromolecule (protein). In this study, the free binding energy of PPK and the ligands complex were determined via the MM-GBSA technique. The binding energy was determined using the values of the gas-phase electrostatic energy (Eele), van der Waals (EvdW), polar (Gpolar) and nonpolar (Gnonpolar) constituent of the PPK and the ligands complex. The results of the study showed that ZINC41125011 possessed the free binding energy of − 27.32 ± 0.93 kcal/mol, while ZINC20321877 had a greater free binding energy of − 16.60 ± 0.50 kcal/mol. This result strengthened the output of the docking analysis where ZINC41125011 had the minimum binding energy of − 9.92 kcal/mol (Table 5).
Conclusion
A total of ten thousand one hundred and six (10,106) compounds were retrieved through virtual screening against Zinc databases using PyRx and RASPD tools. These ligands were checked for the Lipinski rule of five and docking analysis. Ten ligands with better binding affinities were selected. The binding energies of the selected ligands varied between − 9.92 and − 8.01 kcal/mol which was lower than the binding energy of − 1.13 kcal/mol of Mg2+. These ligands were checked for ADMET properties to further eliminate the compounds with unwanted properties. Three compounds (ZINC41125011, ZINC20318248, and ZINC20321877) which possessed all the desired pharmacokinetic and toxicity properties were considered for MD simulation and MM-GBSA analyses. The outcome of the analysis showed that all the complexes form relatively stable folded polypeptide structure after the 50 ns MD simulation and so they were regarded as potential inhibitors of PPK from MTB after positive experimental validation.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402 (External Resources Pubmed/Medline (NLM) CrossRef (DOI) Chemical Abstracts Service (CAS) Cambridge Scientific Abstracts (CSA) ISI Web of Science)
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) The Protein Data Bank. FEBS J 80(2):319–324
Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE et al (2015) AMBER 2015. University of California, San Francisco
Centers for Disease Control and Prevention (2006) Emergence of MTB with extensive resistance to second-line drugs—worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep 55(11):301
Chen C, Huang H, Wu CH (2017) Protein bioinformatics databases and resources. In: Wu CH, Arighi CN, Ross KE (eds) Protein bioinformatics. Humana Press, New York, pp 3–39
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52:3099–3105
Cloete R, Oppon E, Murungi E, Schubert WD, Christoffels A (2016) Resistance related metabolic pathways for drug target identification in Mycobacterium tuberculosis. BMC Bioinform 17(1):75
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
DeLano WL (2002) The PyMOL user’s manual. DeLano Scientific, San Carlos, p 452
Fiser A, Do RKG (2000) Modeling of loops in protein structures. Protein Sci 9(9):1753–1773
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449–461
Irwin JJ, Shoichet BK (2005) ZINC: a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Johnson R, Streicher EM, Louw GE, Warren RM, Van Helden PD, Victor TC (2006) Drug resistance in Mycobacterium tuberculosis. Curr Issues Mol Biol 8(2):97–112
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2015) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
La Motta C, Sartini S, Mugnaini L, Simorini F, Taliani S, Salerno S, Marini AM, Da Settimo F, Lavecchia A, Novellino E, Cantore M, Failli P, Ciuffi M (2007) Pyrido [1, 2-a] pyrimidin-4-one derivatives as a novel class of selective aldose reductase inhibitors exhibiting antioxidant activity. J Med Chem 50(20):4917–4927
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786
Lee AS, Teo AS, Wong SY (2001) Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 45(7):2157–2159
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83
Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Šali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29(1):291–325
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Mukherjee G, Jayaram B (2013) A rapid identification of hit molecules for target proteins via physico-chemical descriptors. Phys Chem Chem Phys 15(23):9107–9116
Murphy DJ, Brown JR (2007) Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis 7(1):84
Nightingale A, Antunes R, Alpi E, Bursteinas B, Gonzales L, Liu W, Luo J, Qi G, Turner E, Martin M (2017) The proteins API: accessing key integrated protein and genome information. Nucleic Acids Res 45(W1):W539–W544
Palomino JC, Martin A (2014) Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 3(3):317–340
Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjölander K, Ferrin TE, Burley SK, Sali A (2010) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res 39(suppl_1):D465–D474
Rawat R, Whitty A, Tonge PJ (2003) The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc Natl Acad Sci 100(24):13881–13886
Rigi G, Nakhaei MVA, Eidipour H, Najimi A, Tajik F, Taher N, Yarahmadi K (2017) Virtual screening following rational drug design based approach for introducing new anti amyloid beta aggregation agent. Bioinformation 13(2):42
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–473
Thompson DJ, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Vareldzis BP, Grosset J, de Kantor I, Crofton J, Laszlo A, Felten M, Raviglione MC, Kochi A (1994) Drug-resistant tuberculosis: laboratory issues. World Health Organization recommendations. Tuber Lung Dis 75:1–7
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, ZiaZarifi AH, Hoffner SE (2009) Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136(2):420–425
Wallace AC, Laskowski RA, Thornton JM (1996) Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci 5(6):1001–1013
Webb B, Sali A (2014) Protein structure modeling with MODELLER. Method Mol Biol 1137:1–15
World Health Organization (2016) Global tuberculosis report. WHO, Geneva
Xu J, Yuan H, Ran T, Zhang Y, Liu H, Lu S, Xiong X, Xu A, Jiang Y, Lu T, Chen Y (2015) A selectivity study of sodium-dependent glucose cotransporter 2/sodium-dependent glucose cotransporter 1 inhibitors by molecular modeling. J Mol Recognit 28(8):467–479
Zhang Y, Heym B, Allen B, Young D, Cole S (1992) The catalase–peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358(6387):591
Zhang X, Zhang S, Hao F, Lai X, Yu H, Huang Y, Wang H (2005) Expression, purification and properties of shikimate dehydrogenase from Mycobacterium tuberculosis. J Biochem Mol Biol 38(5):624
Acknowledgements
The authors of this paper are very much grateful to Prof. Pawan Dhar (Jawaharlal Nehru University), Prof. B. Jayaram (Indian Institute of Technology Delhi), Dr. Kalaiarasan P. (Jawaharlal Nehru University), and Mr. Shashank Shekhar (Indian Institute of Technology Delhi) for providing facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflict of Interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alhaji Isa, M., Singh Majumdar, R. Computer-aided drug design based on comparative modeling, molecular docking and molecular dynamic simulation of Polyphosphate kinase (PPK) from Mycobacterium tuberculosis. J Proteins Proteom 10, 55–68 (2019). https://doi.org/10.1007/s42485-019-00006-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-019-00006-w