Abstract
The effectiveness of treatment for lung nodules is significantly impacted by early diagnosis of the nodules on CT scans. Radiologists are aided by the utilization of CAD technologies. The purpose of this paper is to present a methodology for identifying lung cancer: “(i) pre-processing, (ii) segmentation, (iii) feature extraction, and (iv) detection.” The GF methods commence with the input image being pre-processed. To segment the pre-processed images, a “improved cross-entropy-based active contour segmentation model” is implemented. Then, using a CNN that has been optimized, features such as LBP, entropy, and contrast are computed. The self-adaptive tunicate swarm algorithm (SATSA) is used to optimize CNN weights. For this study, the LIDC-IDRI dataset was used, and Python was used for experimentation. Because pulmonary nodules can differ significantly in size, shape, texture, and appearance, it’s crucial to include a wide variety of patterns in the training dataset. The authors hope to improve the generalization and robustness of the CNN model in identifying lung nodules by incorporating a large number of patterns. Comparing the CNN + SATSA model to existing models allowed researchers to assess its effectiveness in identifying lung nodules. The suggested model’s correctness is demonstrated by its high recall, false discovery rate, FNR, MCC, FPR, precision, FOR, accuracy, specificity, NPV, FMS, and sensitivity values.
Similar content being viewed by others

Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Nowadays, people are troubled with more diseases, particularly cancer. As per the predicted report, about 8.9 million people are died from cancer. In this world, every 6 death is due to the lung cancer that makes it as the second leading reason of death and the most deadly disease is lung cancer (Xie et al. 2019). The formation of malignant nodules in the lung or lung lobe is the primary cause of lung cancer. Early lung nodule diagnosis is more important (Zuo et al. 2019) to reduce the fatality rate from lung cancer. Since the nodules are little dots in CT images, the clinicians require to observe the image individually, which is a time-consuming task. In the CT consecutive images, the nodules may exist in four different slices. The nodules are classified further as part solid, GGO, and solid based on the different brightness degrees in the CT images (Li et al. 2019; Aresta et al. 2020). However, the GGO nodule is less clear than the bronchial structures and pulmonary vessels, and it is illustrated through hazy improved lung attenuation with no obscuration of the bronchial walls and underlying vascular markings. The part solid nodule includes both GGO and solid components that shows the central area of solid attenuation and the peripheral GGO. Many researchers have shown that the CAD approaches would assist the physicians for improving the pulmonary nodules detection rate. The most significant step in the CAD system is the nodule screening (Halder et al. 2020). The classifier and the feature value selection are more important to maintain the maximum accuracy and sensitivity of the system.
In this approach, the nodule detection of the applicant would provide the correct contour and minimize the misjudgements (Zhang and Kong 2020; Wang et al. 2020). The greyscale thresholds are used for the objects with maximum brightness including the solid nodules and blood vessels. Furthermore, the image superposition system is used for improving the GGO quickly with less brightness; therefore, the candidate points is acquired with a binarization threshold. The contour correction and decreased noise are removed for facilitating the succeeding classifications effectively by the morphological disconnects operation (Cao et al. 2019; Li et al. 2018). The iterative surface elimination methods and the 3D shape-based feature descriptors set are implemented for reducing the over-snatch and to alter the nodule profile associated with pleura or blood vessels, which enhances the overall sensitivity of SVM classifiers (Naqi et al. 2019; Veronica 2020).
The lung nodule detection includes two approaches, i.e. the VOI detection methods promise the maximum sensitivity of the succeeding phases and the classifier approaches to minimize the FPs (Saba et al. 2019). Moreover, the advance in the research is categorized into three periods. At the first period, neither the classifiers nor detectors implanted are based on the NN. The detection approaches are the threshold-based models like lung segmentation systems. However, the threshold-based approaches are more complex than the lung region segmentation approaches for selecting the VOIs since the lung nodules are more diverse edges and shapes (Feng et al. 2019; Wang et al. 2019). The easy classifiers are trained for determining the lung nodules obtained in the VOIs chosen. The CNNs (Arul et al. 2019; Sarkar 2020; Chandanapalli et al. 2019; Deotale et al. 2020) are employed for reducing the FPs count in most of the research works. Thus, the detector approaches and threshold-based were employed in this period; however, they are more complex (Naqi et al. 2018). Both the classifiers and detectors are the NN-based approaches (Fu et al. 2019; Bhagyalakshmi et al. 2018; Vinolin and Vinusha 2018; Srinivasa Rao et al. 2019). The major contributions are:
-
Introducing an improved cross-entropy-based active contour segmentation model for segmenting the pre-processed lung nodule images.
-
Proposing a new SATSA model for CNN weight optimization.
The rest of the paper is arranged as follows. Section 2 of this essay discusses the review on lung nodule detection. The overall design of the accepted methodology is described in Sect. 3. The suggested segmentation method and image pre-processing are illustrated in Sect. 3.1. In Sect. 3.2, the feature extraction process for lung nodule identification is illustrated. The categorization of lung nodules is shown in Sect. 3.3 using a CNN that has been optimized for training using a self-adaptive swarm approach. Section 4 details the outcome. At last, Sect. 5 finishes the work.
2 Literature review
In 2020, Zuo et al. (2020) have introduced a novel 3D CNN model for reducing FPs in the detection of lung nodule. Moreover, the new 3D CNN has included the implanted multiple branches in its framework. Every branch processed the feature map with various depths from the layer. Further, each branch was cascaded at its ends; therefore, the features from various depth layers were joined for predicting the candidate’s categories. The adopted model in the lung nodule candidate classification includes a competitive score on LUNA16 dataset with certain measures like 97.83% accuracy, 87.71% sensitivity, 94.26% precision, and 99.25% specificity, respectively. The adopted method was efficient in the lung nodules detection. Finally, the adopted scheme in lung nodule detection has attained the competitive score to reduce the FP and has used as the reference to classify the nodule candidates.
In 2020, Cao et al. (2020) have proposed a TSCNN model for detecting the lung nodule. At the initial stage, the proposed model was to determine an initial lung nodules detection based on the enhanced U-Net segmentation network. For obtaining the larger recall rate without determining the extreme FP nodules, a new sampling strategy and two-phase prediction method were proposed for training purpose during this stage. Then, the TSCNN architecture was constructed into classification networks. In addition, the generalization ability has enhanced the FP reduction approach through the ensemble learning process. The adopted architecture on the LUNA dataset was examined, and it has obtained the better detection outcomes.
In 2019, Li et al. (2019) have introduced a DL on the basis of detection approach in the lung nodule. The patch-based multi-resolution CN was utilized for feature extraction, and four different fusion approaches were also employed for the classification purpose. The adopted lung nodule detection scheme consists of two major parts such as training and testing. Moreover, the images were pre-processed via the rib suppression and lung field segmentation for obtaining the RoI and improved the lung nodules visibility. Next, the images were improved by the histogram operation. The adopted model has shown enhanced concert with robustness than other conventional models. The R-CPM and FAUC of the adopted model were 98.7% and 98.2%, correspondingly.
In 2020, Harsono et al. (2020) have suggested a new lung nodule classification and detection approach by the single-stage detector known as “I3DR-Net”. Moreover, the proposed approach in the multi-scale 3D Thorax CT-scan dataset was obtained through integrating the weight of I3D backbone pre-trained natural images. The I3DR-Net would generate the remarkable outcomes on the task of lung nodule texture detecting with mAP 22.86% and 49.61% and AUC of 70.36% and 81.84% for private and public dataset. Finally, the adopted scheme has altered the RetinaNet NN backbone, NMS, loss function, and weighted clustering approach of adopted detection box.
In 2019, Kuo et al. (2019) have proposed new approach to detect the GGO nodules, solid, and part solid in chest CT. Moreover, the adopted model has included nodule enhancement, image pre-processing, candidate detection, lung segmentation, and FPs reduction. The edge searching approach has replaced the “computing-intensive iterative hole-filling method” for the purpose of lung segmentation. In the nodule enhancement, the image accumulation was used for extracting the nodules with widely distributed grey levels and rapidly improved the individual nodules grey level. Further, SVM was applied twice for reducing the FPs. At last, the experimental outcomes have shown that the adopted rapid detection model has low FPs and maximum sensitivity to assist the clinicians’ diagnosis.
In 2019, Xu et al. (2019) published the “DeepLN Dataset”, a multi-resolution CT screening image dataset. The “semi-automatic annotation system and three-level labelling criterion” was also acquired to guarantee the efficacy and precision of lung nodule annotation. To locate pulmonary nodules, the multi-level characteristics were extracted using the NN-based detector. The severe negative mining and the modified focal loss function were utilized to address normal category imbalance issues. Numerous NN algorithms with varying resolutions were synthesized using the newly discovered “non-maximum suppression” technique. The simulation’s final results demonstrated that its predictions were accurate and that it was more effective than alternative approaches.
In 2018, Gu et al. (2018) introduced a new CAD method for identifying the lung nodules via the multi-scale prediction with 3D DCNN integrated approach to aid radiologists in accurately identifying lung nodules. The 3D CNN was employed with more spatial 3D contextual information than previous 2D CNN models, and after training with 3D samples of the lung nodule representation, more discriminative features were generated. This strategy included cube clustering and multi-scale cube prediction, which was developed to detect microscopic nodules with exceptional accuracy. 2D CNN demonstrated high sensitivity, a positive CPM score, and a high degree of assurance.
Jiang et al. (2018) developed an outstanding model for detecting lung nodules in 2018 based on multi patches derived from images and augmented by the Frangi filter. By combining the two classes of images, the channel-based CNN method was used to learn radiologists’ information. This was done to establish the four levels of the nodule. Due to the assets of two groups and the connected dataset, the adopted model reduced the amount of data compared to the conventional CNN framework and produced generally acceptable results. In comparison to other methodologies, the proposed model has demonstrated superior sensitivity. The simulation results demonstrated that this strategy improved lung nodule detection more effectively.
The review of lung nodule detection was performed. However, the training technique was not utilized as a reference with the same sophisticated frameworks for the quick convergence networks. Initially, a novel 3D CNN model was implemented in Zuo et al. (2020), which exhibits high accuracy, greater sensitivity, increased precision, and maximum specificity. TSCNN model was exploited in Cao et al. (2020) that offer false positive reduction, improved generalization ability, and better detection performance, but more complicated background information was obtained in the medical images than natural images. Moreover, CNN model was deployed in Li et al. (2019) that offer robustness and detection performance. Nevertheless, the proposed model was not employed in the large CXR database gathered from various hospitals. Likewise, I3DR-Net model was exploited in Harsono et al. (2020), which offers better AUC, high confidence score, improved sensitivity, and minimum FPR. However, the adopted model was not implemented in the real-time CT scan by combining the software interface, cloud computing, and suitable GPU. SVM model was exploited in Kuo et al. (2019) that have high sensitivity, good detection outcomes, and low false positives; however, the objects not in nodule shape were eliminated like blood vessels and lung tissues. In addition, 3D DCNN model was introduced in Xu et al. (2019), which offers accurate predictions, improved efficiency, and maximum sensitivity. However, the robustness of the detector was not improved. 3D deep CNN model was proposed in Gu et al. (2018) that offers high sensitivity, satisfied CPM score, and high confidence. However, there is a need to focus on automated classification of lung nodules and FP reduction. Finally, CNN model was implemented in Jiang et al. (2018), which offers sensitivity and good detection performance; however, all the black images in the CNN structure were not considered as the input images. The challenges of existing methods are explained in the Table 1.
3 Proposed system
This paper aims to introduce a new lung nodule detection approach: “(i) pre-processing, (ii) segmentation, (iii) feature extraction, and (iv) classification”. At first, GF is deployed for pre-processing, and then, novel active contour segmentation model is established for segmentation. Moreover, features like LBP, entropy, and contrast features are derived and then classified via optimized CNN model. Further, the CNN weights are optimized via adopted SATSA model. This enhances the detection accuracy of the proposed system. Figure 1 demonstrates the structural design of adopted method.
3.1 Image pre-processing and proposed segmentation process
The input image \({\text{Im}}\) from database is pre-processed using the filtering techniques, i.e. Gaussian filtering. In this process, the noise and artefacts are removed from the input data. Then, the pre-processed image is further used for segmentation process. Novel active contour segmentation algorithm is used for segmenting lung image regions.
3.1.1 Gaussian filtering techniques
Gaussian filtering would blur the images and take away the noises. The Gaussian function in one dimension is determined in Eq. (1).
In Eq. (1),\(\sigma\) denotes the distribution of SD. Moreover, the distribution is assigned with zero mean. The Gaussian function of the SD plays a significant role based on its behaviour.
The Gaussian function is applied in the following ways in several study fields: define the smoothing operator, mathematics, and probability distribution for data or noise. The fundamental characteristics of the Gaussian function are confirmed in relation to its integral as stated in Eq. (2).
Furthermore, the feasible value, which ranges from negative to positive values, probabilistically characterizes the entirety of a given space. In addition, the Gaussian function never equals zero. It behaves symmetrically. It is a symmetric function. The 2D Gaussian function is used while working with the images. Therefore, the two 1D Gaussian functions product is determined in Eq. (3).
The Gaussian filters worked as a point spread function via the 2D distribution. It is attained with the image through convolving the 2D Gaussian distribution function. Moreover, the discrete approximation is produced in the Gaussian function. Further, it requires an infinite large convolution kernel and as nonzero in all places of the Gaussian distribution. In addition, the distribution is very near to 0 that concerns 3 SD from the mean. Moreover, 99% of the distribution decreases within 3 SDs. The kernel size is normally limits to hold only values within 3 SDs of the mean.
Superior values of \(\sigma\) create a higher blurring, i.e. wider peak. The Gaussian nature of the filter is maintained by raising the kernel size. Similarly, the coefficients of Gaussian kernel depend on the \(\sigma\) value. Gaussian filters should not conserve the image brightness. This filtering is used to get rid of the noise. The pre-processed image is denoted as \({\text{Im}}^{{{\text{pre}}}}\).
Gaussian filtering is a popular image processing technique that uses a kernel based on the Gaussian distribution to smooth an image. It is effective at reducing high-frequency noise and blurring sharp edges, both of which can help to improve image quality and remove unwanted artefacts.
Gaussian filtering was chosen in the pre-processing stage because of its ability to suppress noise while preserving important image features. High-frequency noise components are attenuated by convolving the image with a Gaussian kernel, resulting in a smoother image. This can improve segmentation by reducing the impact of noise on subsequent analysis. Additionally, Gaussian filtering can function as a point spread function, which means it simulates the effect of blurring in the imaging system. This can be advantageous in medical imaging, where the acquired images may suffer from blurring due to various factors such as motion artefacts or limitations of the imaging equipment. By applying Gaussian filtering, the blurring effect can be approximated, leading to a more accurate representation of the underlying structures and making the subsequent segmentation algorithm more robust.
3.1.2 Improved cross-entropy-based active contour segmentation model
At first, \({\text{Im}}^{{{\text{pre}}}}\) the world is divided into ROI and non-ROI zones by the ACM. The ACM also divides the image into small local zones based on the foreground and background. Further, \({\text{Im}}^{{{\text{pre}}}}\) in the domain \(\vartheta\) is segmented in the maximum iterations setting as \(\max^{{\left( {{\text{iter}}} \right)}}\).
The proposed algorithm iterates 600 times to find the most cost-effective solution. Each iteration includes a specific set of operations or calculations on the image or its segmented regions. These iterations allow the algorithm to refine its segmentation results, gradually improving segmentation accuracy and quality. The algorithm aims to converge towards a solution that optimally separates the foreground and background regions by performing multiple iterations.
Likewise, the closed contour \(CL\) = 0, and it belongs to the distance function \(\kappa\). Moreover, \(CL\) is defined mathematically as in Eq. (4). The \(CL\) interior is \(CL_{{\text{int}}}\), and it can be seen through the rounded the determination of Heaviside function in Eq. (5). The exterior closed contour \(CL\) is numerically specified in Eq. (6).
Additionally, the AUC with the Dirac delta is mathematically smoothed as in Eq. (7).
Here, the domain \(\vartheta\), the distinctive spatial variables \(b\) and \(g\) are the single points. In Eq. (8), \(\rho\) indicates the radius parameters. The local regions masking takes place by the function \(H\left( {b,g} \right)\). Further, an energy function is determined with \(H\left( {b,g} \right)\) based on the generic force function \({\text{GFF}}\) in Eq. (9). Normally, \({\text{GFF}}\) refers to “generic internal energy measure”. By using the periodicity term, the curve is smoothed. The length of the curve’s arc is weighted by fixing the \(\varphi\) = 0.2. Equation (10) illustrates the overall energy computation, and the final evolution equation is related to the fundamental energy variation with respect to \(\kappa\), and it is expressed in Eq. (11).
The proposed active contour segmentation is determined as follows:
In Eq. (12), \(L\left( k \right)\) portrays the length of the curve \(k\), \(s\left( k \right)\) indicates the area inside the curve, \(f\left( \chi \right)\) depicts the greyscale value of the image, and \(\mu\), \(v\), \(\lambda_{1}\), and \(\lambda_{2}\) denote the weights of corresponding energy terms. Moreover, the cross-entropy is used to calculate the similarity among the segmented and the original regions.
The probability distribution of object region \(k_{b}\) and background region \(k_{a}\) is \(Q = \left\{ {q_{1} ,q_{2} , \ldots q_{h} } \right\}\) and \(P = \left\{ {p_{1} ,p_{2} , \ldots p_{h} } \right\}\). Kullback–Leibler distance utilizes the cross-entropy of \(P\) and \(Q\). Moreover, the obtained segmented image is denoted as \({\text{Im}}^{{{\text{seg}}}}\).
3.2 Feature extraction for lung nodule detection
Three features are extracted in this phase that include:
-
LBP features
-
Contrast features
-
Entropy features
3.2.1 LBP features
In addition, the segmented image undergoes LBP feature extraction. The LBP (Honeycutt and Plotnick 2008) is more discriminatory and easier to implement computationally. LBP, or local binary pattern, is a texture descriptor that is widely used in computer vision and image processing. It works by comparing the intensity values of pixels in a greyscale image’s neighbourhood around each pixel. LBP captures the image’s local structure and texture information by encoding the relationships between the central pixel and its neighbours into a binary pattern. The LBP algorithm involves determining the size and shape of the neighbourhood, comparing the intensity values of pixels within the neighbourhood to the central pixel, and generating a binary pattern based on these comparisons. This binary pattern is then converted into a decimal value that represents the central pixel’s local texture information. A concise representation of the texture distribution is obtained by calculating histograms of these decimal values across the entire image.
LBP is well known for its resistance to changes in lighting and ability to capture fine-grained texture details. It has found use in a variety of computer vision tasks such as texture classification, face recognition, and object detection. LBP is a powerful tool for characterizing and analysing local texture information in images due to its simplicity, efficiency, and potential for multi-scale analysis.
The LBP operator is used to assign decimal values to each image pixel. A negative number is also represented as 0, a positive number as 1, and a zero value as 0. The LBP codes are all the binary codes for obtaining the binary number, starting at the upper left. A vast number of local descriptions provided by the texture descriptor are combined to form the global description. In addition, the properties of these texture objects are extracted in accordance with their discernibility. In Eq. (14), \(SH_{{{\text{cl}}}}\) and \(SH_{{{\text{pl}}}}\) refer to the image centre pixel and the centre pixel intensities from the neighbour \({\text{pl}}\), respectively. The LBP descriptors are denoted as \({\text{LBP}}\left( \cdot \right)\), and \({\text{NE}}_{{{\text{Pl}}}}\) represents the count of neighbour. Further, the LBP descriptor function \(F_{{{\text{LBP}}({\text{pl}},{\text{cl}})}}\) is determined in Eq. (15). The obtained LBP features are denoted as \({\text{Im}}^{{{\text{LBP}}}}\).
3.2.2 Contrast features
Variations in luminance or colour can distinguish between two objects. The summation of square variance, or Honeycutt and Plotnick (2008) the contrast characteristics, is the calculation of the contrast in intensity between a pixel’s neighbouring pixels and the entire image pixel. It also calculates the total diversity of the greyscale images. Here, Eq. (16) represents the contrast value calculation. The obtained contrast features are denoted as \({\text{Im}}^{{{\text{con}}}}\).
3.2.3 Entropy features
Entropy and energy are both measures of the orderliness of an image. The entropy characteristic of an image is a statistical characteristic (Honeycutt and Plotnick 2008). These characteristics are computed utilizing the segmented regions. It computes the grey-level concentration intensity of the GLCM and returns its sum of squared elements. Equation (17) determines the energy value calculation. The obtained entropy features are denoted as \({\text{Im}}^{{{\text{En}}}}\). Finally, the obtained features are designated as \({\text{Im}}^{{{\text{Fea}}}}\), and it is expressed in Eq. (18).
Because of their ability to capture different aspects of the nodule’s characteristics, the extraction of LBP, contrast, and entropy features is critical in the context of lung nodule extraction. LBP features are important in analysing the local texture patterns found in lung nodules. LBP features can effectively capture nodule texture patterns, such as speculated edges or internal structures, and provide valuable information for nodule detection and classification. The ability to distinguish between lung nodules and surrounding tissues is aided by contrast features. Because nodules have higher contrast than surrounding lung tissue, the algorithm can focus on regions with significant intensity variations by incorporating contrast features. This can lead to better detection and differentiation of nodules from the background. Entropy characteristics reveal information about the texture complexity or heterogeneity of lung nodules. Because of irregular internal structures or heterogeneous tissue composition, malignant nodules frequently have higher entropy. The algorithm can capture the textural variations associated with malignant nodules by extracting entropy features, allowing for the potential differentiation of benign and malignant cases.
The combination of LBP, contrast, and entropy features enables a thorough examination of lung nodule characteristics such as texture patterns, intensity variations, and textural complexity. By incorporating these features into the analysis, the algorithm can use their discriminative power to improve nodule detection and classification accuracy, ultimately assisting in the early diagnosis and treatment of lung cancer.
3.3 Lung nodule classification: optimized CNN with optimal training via proposed self-adaptive tunicate swarm algorithm
3.3.1 Optimized CNN model
A CNN that has been tuned to classify lung nodules receives the obtained features (LeCun et al. 2010). Known classifier CNN is made up of “three layers, such as pooling, fully connected, and convolution layers.” The \(r{\text{th}}\) layers harmonized to \(z{\text{th}}\) feature map in position \(\left( {e,\,x} \right)\) is implied by \(S_{e,x,z}^{r}\), and it is implied by Eq. (19). The \(z{\text{th}}\) filter value is specified in \(r{\text{th}}\) layer and optimal bias term \(\to\) \(B_{l}^{r}\) and weight vector \(\to\) \(W_{l}^{r}\), \(I_{e,x}^{r}\) \(\to\) associated input patches. As a result, the adopted SATSA technique is employed to optimally tune the weight. Consider the “activation value \(\left( {{\text{AF}}_{e,x,z}^{r} } \right)\) and the nonlinear activation function as \({\text{AF}}\;\left( \cdot \right)\)” as defined in Eq. (20). The shift variance is calculated as given in Eq. (21). The pooling function \({\text{pool}}\;\left( {} \right)\) and local neighbourhood \(\left( {{\text{AF}}_{e,x,z}^{r} } \right)\) at neighbour position \(\left( {e\,,\,x} \right)\) \(\to\) \(J_{e,x}\).
In CNN, Eq. (22) determines the loss function. Further, the CNN output, \(t{\text{th}}\) input data and target values are determined as \({\text{OUT}}^{\left( t \right)}\), \(U^{\left( t \right)}\) and \(V^{\left( t \right)}\), correspondingly.
The pooling layer of CNN carried out the down sampling operations using the convolutional layers’ output. Additionally, there are two distinct types of pooling: average pooling and maximal pooling. While the average pooling represents the mean value, the maximum pooling has reached the greatest value.
The optimized CNN model used in the study had a total of 8 layers, 5 of which were hidden layers. The number of hidden layers is critical in determining the model’s robustness because it has a direct impact on the model’s ability to learn complex representations and extract meaningful features from the input data. The model can capture hierarchical representations of the input by using multiple hidden layers, with each layer learning increasingly abstract and high-level features. This network depth allows the model to handle intricate patterns and variations in data, resulting in improved performance and generalization. Furthermore, as more hidden layers are added, the model gains the ability to learn nonlinear relationships and model complex decision boundaries. This adaptability improves the model’s robustness by allowing it to handle a wide range of input variations and adapt to various lung nodule characteristics.
3.3.2 Solution encoding
As previously mentioned, the weights of CNN are adjusted using the SATSA model. Here, the total CNN weights are shown. Maximizing accuracy is the goal of the SATSA model \(\left( {{1 \mathord{\left/ {\vphantom {1 {{\text{loss}}}}} \right. \kern-0pt} {{\text{loss}}}}} \right)\), and it is shown in Eq. (14) (Fig. 2).
3.3.3 Proposed SATSA algorithm
Although the current TSA (Kaur et al. 2020) technique is capable of detecting food sources in the ocean, it is oblivious of any food sources in the search area. This study predicts that the SATSA model will outperform this scenario. In most cases, self-improvement in the present optimization models causes the algorithm to become even more effective at resolving optimization problems (Rajakumar 2013a, 2013b; Swamy et al. 2013; George and Rajakumar 2013; Rajakumar and George 2012). TSA defines the two tunicate behaviours for locating the food source (Susan and Aju 2022). Jet propulsion and swarm intelligence are the two TSA behaviours. The self-adaptive tunicate swarm algorithm (SATSA), an optimization algorithm inspired by nature, aims to address challenging optimization issues. It is based on the collective behaviour of marine invertebrates called tunicates, which are renowned for their extraordinary capacity to adapt to and survive in a variety of environments. SATSA uses self-adaptation mechanisms to get around the drawbacks of conventional optimization algorithms. Through the use of these mechanisms, the algorithm is able to dynamically modify the parameters and search tactics used during the optimization process, resulting in better performance and greater effectiveness. The handling of high-dimensional and nonlinear optimization issues is a key feature of SATSA. Multiple search agents simultaneously explore the solution space using a population-based approach. SATSA can explore a larger area of the search space and find a variety of solutions thanks to this parallel search.
Jet propulsion behaviour The tunicate must also adhere to the following three conditions: maintaining close proximity to the best searching agent, migrating towards the best searching agent’s position, and avoiding conflicts between searching agents. As a consequence of the optimal response, the swarm’s behaviour shifted the position of the other search agent. The three factors that influence the behaviour of jet propulsion are listed below. (i) Avoiding conflicts: To evade the conflicts among the searching agents, the \(\vec{G}\) vector is used for calculating the location of novel searching agents, and it is determined in Eq. (24).
In Eqs. (25) and (26), \(\vec{R}\) \(\to\) water flow advection, \(\vec{Y}\) \(\to\) social forces among the search agents, \(\vec{Z}\) \(\to\) gravity force, and the variables \(d_{1}\), \(d_{2}\), and \(d_{3}\) \(\to\) random number ranges. Conventionally, the \(\vec{G}\) vector calculates the position of novel search agents. However, as in SATSA technique, the positions are updated as shown in Eq. (27).
In Eq. (27), \(X_{best}\) and \(R_{worst}\) portray the best solution and worst solution, correspondingly.
(ii) Moving in the direction of the best neighbour: This is modelled as shown in Eq. (28), where \(u\) \(\to\) current iteration,\(\vec{D}\) \(\to\) distance amid tunicate and food source, \(\vec{M}\) \(\to\) position of food source, \(\vec{X}_{y} \left( u \right)\) \(\to\) tunicate position and \(rand\) \(\to\) random digit.
(iii) Converge towards the best search agent: This phase is modelled as shown in Eq. (29).
In Eq. (20) \(\vec{X}_{y} \left( u \right)\) indicates the updated position of tunicate at \(\vec{M}\) food source position. However, as per the adopted SATSA method, instead of \(rand\) function we used the chaotic map of circular map as per Eq. (30), in which \(w = 0.02\).
Swarm behaviour To simulate the tunicate’s swarm behaviour, it saves the 1st and 2nd optimal best solutions and the other search agent’s positions are updated as in Eq. (31).
The ultimate position is discovered within a random cylindrical or cone-shaped spot that represents the tunicate position. The SATSA model’s pseudocode is explained in Algorithm 1.
Algorithm 1: Proposed SATSA Model |
Step 1: Initialize populace \(\vec{X}_{y}\) |
Step 2: Select initial constraints and entire iterations |
Step 3: Compute the fitness value using Eq. (23) |
Step 4: The proposed best search agent is exposed in Eq. (30) after the fitness value calculation |
Step 5: Each search agent’s proposed updated position is provided in Eq. (31) |
Step 6: The agents are adjusted in a specified search space away from the boundary |
Step 7: Calculate the fitness and Update \(\vec{X}_{y}\) Only if there is a superior alternative to the preceding optimal solution |
Step 8: Exit; it satisfies the ending criterion |
Alternately, repeat steps 6& 7 |
Step 9: The most promising outcome has thus far been returned |
4 Results and discussion
4.1 Simulation procedure
Python was utilized to execute the adopted CNN + SATSA method for lung nodule identification, and the results were validated. The LIDC-IDRI dataset was utilized for this study (Armato et al. 2011). The LIDC-IDRI (Lung Image Database Consortium—Image Database Resource Initiative) dataset was used in this study. The collection of CT images and annotations of lung nodules in this dataset are well-known. The dataset used in this study specifically included 7371 lesions that at least one radiologist classified as “nodules.” We chose 500 samples for the experiment from this sizable dataset. We trained and tested their suggested method for identifying and categorizing lung nodules using these samples. Following an 80–20 split, the dataset was split into training and testing sets. The model was trained using 400 samples, or 80% of the samples, during the training phase. In order to optimize the model’s parameters and discover the patterns suggestive of lung nodules, this involved feeding the model the CT scan images and associated annotations. The training procedure was designed to give the model the ability to correctly identify nodules in unobserved data. The remaining 20% of the samples (100 samples) were set aside for testing the trained model’s performance. These samples served as a separate evaluation set and weren't used during the training phase. The dataset had a 60% abnormal sample distribution, representing cancerous cells, and 40% normal sample distribution. This distribution reflects a real-world scenario in which detecting and classifying abnormal nodules indicative of potential lung cancer is critical. We wanted to ensure the model’s robustness and accuracy in identifying cancerous nodules by including a large number of abnormal samples. These test samples were used to evaluate the model’s capability to recognize and classify lung nodules accurately. While it is possible that in this specific study, no irrelevant or unusable samples were encountered, it is important to acknowledge that in other scenarios, the presence of such samples can occur. The occurrence of irrelevant or unusable samples can vary depending on factors such as data collection methods, inclusion criteria, and potential issues during the data acquisition process.
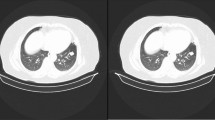
The efficacy of the CNN + SATSA model for lung nodule detection was determined by comparing it to current models such as DBN (Wang et al. 2016), SVM (Avci 2009), CNN + EHO (Elhosseini et al. 2019), CNN + WOA (Mirjalili and Lewis 2016), CNN + TSA (Kaur et al. 2020), and CNN + WTEEB (Kumar and Amalanathan 2022). The performance was also determined by adjusting the TP for metrics such as "accuracy, sensitivity, specificity, precision, recall, FMS, thread score, FDR, FNR, FPR, FOR, NPV, and MCC," respectively. The final images are shown in Fig. 3.
4.2 Performance analysis
Figures 4, 5 and 6 compare the CNN + SATSA model’s performance against that of DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB. Furthermore, the adopted CNN + SATSA model achieves outstanding accuracy (92); in contrast, the typical approaches for TP 60 in Fig. 4c produce lower accuracy values for DBN (70), SVM (79), CNN + EHO (83), CNN + WOA (84), CNN + TSA (87), and CNN + WTEEB (88). For TP 50, the suggested CNN + SATSA model has specificity that is 30%, 16.67%, 11.11%, 7.78%, 4.44%, and 2.22% higher than DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB, as shown in Fig. 4b. Furthermore, as shown in Fig. 4d, the CNN + SATSA algorithm beats DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB in terms of TP 90 sensitivity (98). For TP 40, the adopted model CNN + SATSA achieves higher precision (94) in detected results than DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB, as shown in Fig. 4a.
The given different measures are appropriate in proving the betterment of proposed work in detecting the nodule. This achievement is due to the empowerment of proposed segmentation process that segments the ROI and non-ROI regions effectively.
Figure 5 illustrates FPR, FNR, FDR, and FOR of CNN + SATSA and existing schemes. For the purpose of achieving the improved performance, the negative measures should be minimal. The adopted CNN + SATSA method holds minimum FPR value that is 92.30%, 90.90%, 86.6%, 80%, 66.67%, and 50% than “DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB”, respectively for TP 90 in Fig. 5a. Thus, the presented CNN + SATSA model is proved with less error (indicating high accuracy on nodule detection).
In Fig. 6, numerous metrics for the accepted and current models are examined, including NPV, recall, MCC, FMS, and thread score. The adopted CNN + SATSA model achieves maximum recall (91) with a training percentage of 80; in contrast, typical schemes like as DBN (73), SVM (80), CNN + EHO (83), CNN + WOA (84), CNN + TSA (86), and CNN + WTEEB (88) achieve low recall. In Fig. 6d, the CNN + SATSA model has a thread score that is 34.73%, 23.15%, 21.05%, 11.51%, 9.47%, and 5.26% higher than SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB. As a result, the adopted CNN + SATSA algorithm outperformed other contemporary models.
4.3 Analyses of the overall performance
Table 2 provides a summary of the accuracy experiments conducted on the proposed CNN + SATSA model for TPs 40, 50, 60, 70, 80, and 90. CNN + SATSA has demonstrated that it can identify all TPs with greater precision than DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB. In terms of TP 90 accuracy (9.55), Table 1 demonstrates that the CNN + SATSA model outperforms DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB. Additionally, for TP 70, the suggested CNN + SATSA model performs 3.97 per cent better than other traditional models (Pawar and Premchand 2023) including DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB. The outcome shows that the CNN + SATSA combination outperforms the traditional models.
4.4 Statistical analysis
Table 3 statistically compares the CNN + SATSA technique to other known models. As shown in the table, the acceptable CNN + SATSA model gave better outcomes for numerous case scenarios, including the best, worst, mean, standard deviation, and median values. Under the best-case scenario, the adopted model outperformed "DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB" by 22.21%, 12.74%, 11.70%, 4.86%, 2.91%, and 2.69%, respectively. In terms of accuracy, the adopted model outperforms other conventional models such as DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB (92.27347). The proposed model also has the highest median value (91.95007), while the existing schemes have the lowest accuracy values, which are DBN (71.79335), SVM (80.19017), CNN + EHO (84.08179), CNN + WOA (85.09699), CNN + TSA (89.24413), and CNN + WTEEB (89.77986).
The confusion matrix has been generated in this work and it describes the model’s predictions in detail, including true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The proportion of correctly classified samples out of the total number of samples is represented by accuracy, which is calculated using these values. The use of positive and negative values in the confusion matrix is critical in the context of lung nodule detection. Accuracy captures the overall performance of the detection system in correctly identifying lung nodules (TP) as well as correctly classifying non-nodules (TN) by taking both positive and negative values into account. The numericals in the table show how the adopted CNN + SATSA model outperforms other models in terms of accuracy, recall, and specific thresholds (TP 90, TP 70, TP 80). TP 90 represents the true positive rate when a 90% threshold is used, indicating how well the model detects lung nodules with high certainty. Similarly, TP 70 and TP 80 represent true positive rates at 70% and 80% thresholds, respectively. These values provide information about the model’s performance in identifying lung nodules at various confidence levels.
5 Conclusion
This work developed an effective lung cancer detection model. The GF methods commence with the input image being pre-processed. To segment the pre-processed images, a "improved cross-entropy-based active contour segmentation model" was implemented. Then, features such as LBP, entropy, and contrast were classified using an optimized CNN. The CNN weights were also SATSA model optimized. Thus, the output was detected. Furthermore, the adopted CNN + SATSA model obtains higher accurateness values (9.55 \(\times 10^{ + 01}\)) for TP 90 over DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB, respectively. Further, the proposed CNN + SATSA model holds 3.97% maximum accuracy value than DBN, SVM, CNN + EHO, CNN + WOA, CNN + TSA, and CNN + WTEEB, correspondingly for TP 70. The adopted CNN + SATSA model attained maximum recall (~ 91) for TP 80, whereas the traditional approaches only manage low recall, like DBN (~ 73), SVM (~ 80), CNN + EHO (~ 83), CNN + WOA (~ 84), CNN + TSA (~ 86), and CNN + WTEEB (~ 88). In summary, the CNN + SATSA model presented in the work outperforms other models in terms of accuracy and recall for lung cancer detection. However, a few limitations must be addressed, such as dataset limitations and the need for continuous improvement. The proposed method has the potential to make significant contributions to the field of lung cancer detection by taking these factors into account and exploring future directions.
Data availability
The data used in this research will be available on request.
Change history
20 August 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s00500-024-10085-7
Abbreviations
- AUC:
-
Area under curve
- NMS:
-
Non-maximum suppression
- 3D:
-
Three-dimensional
- GGO:
-
Ground-glass opacity
- FOR:
-
False omission rate
- DCNN:
-
Deep convolutional neural network
- FAUC:
-
Area under the FROC
- CPM:
-
Competition performance metric
- FP:
-
False positive
- CT:
-
Computed tomographic
- GF:
-
Gaussian filtering
- CN:
-
Convolutional network
- mAP:
-
Mean average precision
- I3D:
-
Inflated 3D ConvNet
- CNNs:
-
Convolutional neural networks
- NN:
-
Neural network
- TSCNN:
-
Two-stage convolutional neural networks
- TP:
-
Training percentage
- SVM:
-
Support vector machine
- NPV:
-
Net predictive value
- 2D:
-
Two-dimensional
- RoI:
-
Region of interest
- VOI:
-
Volume of interest
- FPR:
-
False positive rate
- WOA:
-
Whale optimization algorithm
- TSA:
-
Tunicate swarm algorithm
- FNR:
-
False negative rate
- WTEEB:
-
Whale with tri-level enhanced encircling behaviour
- MCC:
-
Matthews correlation coefficient
- R-CPM:
-
Refined competition performance metric
- DL:
-
Deep learning
- FDR:
-
False discovery rate
- FMS:
-
F-measure
- SATSA:
-
Self-adaptive tunicate swarm algorithm
- EHO:
-
Elephant herding optimization
References
Ali I, Muzammil M, Haq IU, Khaliq AA, Abdullah S (2020) Efficient lung nodule classification using transferable texture convolutional neural network. IEEE Access 8:175859–175870. https://doi.org/10.1109/ACCESS.2020.3026080
Aresta G et al (2020) Automatic lung nodule detection combined with Gaze information improves radiologists’ screening performance. IEEE J Biomed Health Inform 24(10):2894–2901. https://doi.org/10.1109/JBHI.2020.2976150
Armato SG 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY (2011) The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931. https://doi.org/10.1118/1.3528204
Arul V, Sivakumar VG, Marimuthu R, Chakraborty B (2019) An approach for speech enhancement using deep convolutional neural network. Multimed Res 2(1):37–44
Avci E (2009) A new intelligent diagnosis system for the heart valve diseases by using genetic-SVM classifier. Expert Syst Appl 36(7):10618–10626
Bhagyalakshmi V, Ramchandra GD, Geeta D (2018) Arrhythmia classification using cat swarm optimization based support vector neural network. J Netw Commun Syst 1(1):28–35
Cao H et al (2019) Multi-branch ensemble learning architecture based on 3D CNN for false positive reduction in lung nodule detection. IEEE Access 7:67380–67391. https://doi.org/10.1109/ACCESS.2019.2906116
Cao H et al (2020) A two-stage convolutional neural networks for lung nodule detection. IEEE J Biomed Health Inform 24(7):2006–2015. https://doi.org/10.1109/JBHI.2019.2963720
Chandanapalli SB, Reddy ES, Lakshmi DR (2019) Convolutional neural network for water quality prediction in WSN. J Netw Commun Syst 2(3):40–47
Deotale N, Kolekar U, Kondelwar A (2020) Self-adaptive particle swarm optimization for optimal transmit antenna selection. J Netw Commun Syst 3(1):1–10
Elhosseini MA, El Sehiemy RA, Rashwan YI, Gao XZ (2019) On the performance improvement of elephant herding optimization algorithm. Knowl Based Syst 166:58–709
Feng Y, Hao P, Zhang P et al (2019) Supervoxel based weakly-supervised multi-level 3D CNNs for lung nodule detection and segmentation. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-018-01170-5
Fu L, Ma J, Chen Y et al (2019) Automatic detection of lung nodules using 3D deep convolutional neural networks. J Shanghai Jiaotong Univ (Sci) 24:517–523. https://doi.org/10.1007/s12204-019-2084-4
George A, Rajakumar BR (2013) APOGA: an adaptive population pool size based genetic algorithm. In: AASRI Procedia—2013 AASRI conference on intelligent systems and control (ISC 2013), vol 4, pp 288–296. https://doi.org/10.1016/j.aasri.2013.10.043
Gu Y, Lu X, Yang L, Zhang B, Yu D, Zhao Y, Gao L, Wu L, Zhou T (2018) Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput Biol Med 103:220–231
Halder A, Chatterjee S, Dey D, Kole S, Munshi S (2020) An adaptive morphology based segmentation technique for lung nodule detection in thoracic CT image. Comput Methods Programs Biomed 197:105720
Harsono IW, Liawatimena S, Cenggoro TW (2020) Lung nodule detection and classification from Thorax CT-scan using RetinaNet with transfer learning. J King Saud Univ Comput Inf Sci (In Press)
Honeycutt CE, Plotnick R (2008) Image analysis techniques and gray-level co-occurrence matrices (GLCM) for calculating bioturbation indices and characterizing biogenic sedimentary structures. Comput Geosci 34(11):1461–1472
Jiang H, Ma H, Qian W, Gao M, Li Y (2018) An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J Biomed Health Inform 22(4):1227–1237. https://doi.org/10.1109/JBHI.2017.2725903
Kaur S, Awasthi LK, Sangal AL, Dhiman G (2020) Tunicate Swarm algorithm: a new bio-inspired based metaheuristic paradigm for global optimization. Eng Appl Artif Intell 90:103541
Kumar MK, Amalanathan A (2022) Automated lung nodule detection in CT images by optimized CNN: impact of improved whale optimization algorithm. Comput Assist Methods Eng Sci 29(1–2):7–31
Kuo CF, Huang CC, Siao JJ, Hsieh CW, Huy VQ, Ko KH, Hsu HH (2019) Automatic lung nodule detection system using image processing techniques in computed tomography. Biomed Signal Process Control 56:101659
LeCun Y, Kavukvuoglu K, Farabet C (2010) Convolutional networks and applications in vision. In: International symposium on circuits and systems, pp 253–256
Li C, Zhu G, Wu X, Wang Y (2018) False-positive reduction on lung nodules detection in chest radiographs by ensemble of convolutional neural networks. IEEE Access 6:16060–16067. https://doi.org/10.1109/ACCESS.2018.2817023
Li Y, Zhang L, Chen H, Yang N (2019) Lung nodule detection with deep learning in 3D thoracic MR images. IEEE Access 7:37822–37832. https://doi.org/10.1109/ACCESS.2019.2905574
Li X, Shen L, Xie X, Huang S, Xie Z, Hong X, Yu J (2019) Multi-resolution convolutional networks for chest X-ray radiograph based lung nodule detection. Artif Intell Med 103:101744
Mirjalili S, Lewis A (2016) The whale optimization algorithm. Adv Eng Softw 95:51–67
Naqi SM, Sharif M, Yasmin M (2018) Multistage segmentation model and SVM-ensemble for precise lung nodule detection. Int J CARS 13:1083–1095. https://doi.org/10.1007/s11548-018-1715-9
Naqi SM, Sharif M, Lali IU (2019) A 3D nodule candidate detection method supported by hybrid features to reduce false positives in lung nodule detection. Multimed Tools Appl 78:26287–26311. https://doi.org/10.1007/s11042-019-07819-3
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24(7):971–987
Pawar VJ, Premchand P (2023) Modified convolutional neural network for lung cancer detection: improved cat swarm-based optimal training. pp 37–59
Rajakumar BR (2013a) Impact of static and adaptive mutation techniques on genetic algorithm. Int J Hybrid Intell Syst 10(1):11–22. https://doi.org/10.3233/HIS-120161
Rajakumar BR (2013b) Static and adaptive mutation techniques for genetic algorithm: a systematic comparative analysis. Int J Comput Sci Eng 8(2):180–193. https://doi.org/10.1504/IJCSE.2013.053087
Rajakumar BR, George A (2012) A new adaptive mutation technique for genetic algorithm. In: Proceedings of IEEE international conference on computational intelligence and computing research (ICCIC), Coimbatore, India, pp 1–7. https://doi.org/10.1109/ICCIC.2012.6510293
Saba T, Sameh A, Khan F et al (2019) Lung nodule detection based on ensemble of hand crafted and deep features. J Med Syst 43:332. https://doi.org/10.1007/s10916-019-1455-6
Sarkar A (2020) Optimization assisted convolutional neural network for facial emotion recognition. Multimed Res 3(2):35
Srinivasa Rao TC, Tulasi Ram SS, Subrahmanyam JBV (2019) Enhanced deep convolutional neural network for fault signal recognition in the power distribution system. J Comput Mech Power Syst Control 2(3):39–46
Susan S, Aju D (2022) Optimised CNN based brain tumour detection and 3D reconstruction. Comput Methods Biomech Biomed Eng Imaging Vis 10:2–16. https://doi.org/10.1080/21681163.2022.2113436
Swamy SM, Rajakumar BR, Valarmathi IR (2013) Design of hybrid wind and photovoltaic power system using opposition-based genetic algorithm with Cauchy mutation. In: IET Chennai fourth international conference on sustainable energy and intelligent systems (SEISCON 2013), Chennai. https://doi.org/10.1049/ic.2013.0361.
Veronica BKJ (2020) An effective neural network model for lung nodule detection in CT images with optimal fuzzy model. Multimed Tools Appl 79:14291–14311. https://doi.org/10.1007/s11042-020-08618
Vinolin V, Vinusha S (2018) Enhancement in biodiesel blend with the aid of neural network and SAPSO. J Comput Mech Power Syst Control 1(1):11–17
Wang HZ, Wang GB, Li GQ, Peng JC, Liu YT (2016) Deep belief network based deterministic and probabilistic wind speed forecasting approach. Appl Energy 182:80–93
Wang Q, Shen F, Shen L et al (2019) Lung nodule detection in CT images using a raw patch-based convolutional neural network. J Digit Imaging 32:971–979. https://doi.org/10.1007/s10278-019-00221-3
Wang B et al (2020) A fast and efficient CAD system for improving the performance of malignancy level classification on lung nodules. IEEE Access 8:40151–40170. https://doi.org/10.1109/ACCESS.2020.2976575
Xie Y et al (2019) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans Med Imaging 38(4):991–1004. https://doi.org/10.1109/TMI.2018.2876510
Xu X, Wang C, Guo J, Yang L, Bai H, Li W, Yi Z (2019) DeepLN: a framework for automatic lung nodule detection using multi-resolution CT screening images. Knowl Based Syst 189:105128
Zhang Q, Kong X (2020) Design of automatic lung nodule detection system based on multi-scene deep learning framework. IEEE Access 8:90380–90389. https://doi.org/10.1109/ACCESS.2020.2993872
Zuo W, Zhou F, Li Z, Wang L (2019) Multi-resolution CNN and knowledge transfer for candidate classification in lung nodule detection. IEEE Access 7:32510–32521. https://doi.org/10.1109/ACCESS.2019.2903587
Zuo W, Zhou F, He Y (2020) An embedded multi-branch 3D convolution neural network for false positive reduction in lung nodule detection. J Digit Imaging 33:846–857. https://doi.org/10.1007/s10278-020-00326-0
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
Not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s00500-024-10085-7
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, M.K., Amalanathan, A. RETRACTED ARTICLE: Optimized convolutional neural network for automatic lung nodule detection with a new active contour segmentation. Soft Comput 27, 15365–15381 (2023). https://doi.org/10.1007/s00500-023-09000-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-023-09000-3