Abstract
Numerous lung nodule candidates can be produced through an automated lung nodule detection system. Classifying these candidates to reduce false positives is an important step in the detection process. The objective during this paper is to predict real nodules from a large number of pulmonary nodule candidates. Facing the challenge of the classification task, we propose a novel 3D convolution neural network (CNN) to reduce false positives in lung nodule detection. The novel 3D CNN includes embedded multiple branches in its structure. Each branch processes a feature map from a layer with different depths. All of these branches are cascaded at their ends; thus, features from different depth layers are combined to predict the categories of candidates. The proposed method obtains a competitive score in lung nodule candidate classification on LUNA16 dataset with an accuracy of 0.9783, a sensitivity of 0.8771, a precision of 0.9426, and a specificity of 0.9925. Moreover, a good performance on the competition performance metric (CPM) is also obtained with a score of 0.830. As a 3D CNN, the proposed model can learn complete and three-dimensional discriminative information about nodules and non-nodules to avoid some misidentification problems caused due to lack of spatial correlation information extracted from traditional methods or 2D networks. As an embedded multi-branch structure, the model is also more effective in recognizing the nodules of various shapes and sizes. As a result, the proposed method gains a competitive score on the false positive reduction in lung nodule detection and can be used as a reference for classifying nodule candidates.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung nodule detection is an effective means for early screening and diagnosis of lung cancer. However, to carry out this task, various images are provided by different screening methods [1, 2]; among which, the lung spiral scan is commonly used and a large number of computed tomography (CT) images can be obtained through this screening method. Thus, the lung nodule detection discussed in this paper is to find and mark the location of nodules from such multiple CT images.
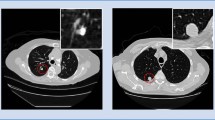
However, for a patient with lung nodules, there is generally only one or a few nodules in his (or her) lung, which often occupies few pixels among 100 to 400 lung slices, making manual detection more difficult. Figure 1 shows a nodule in a lung. The nodule is found to occupy only a few dozen pixels even in its center section, which is an area usually with the most nodules. On the contrary, it is also shown in Fig. 1 that the pixel size of a slice is 512 × 512, which is hundreds of times the size of a nodule.
In view of this status, the automated lung nodule detection system is proposed to reduce the burden on radiologists for manually recognizing a large number of CT images. Given that the nodules occupy a much smaller area than the entire lung slice does, the automated lung nodule detection system is generally divided into two steps [3]:
-
1.
Candidate detection
-
2.
False positive reduction
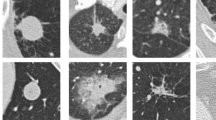
The candidate detection refers to the detection of candidates suspected lung nodules from a large number of CT slices. The purpose of candidate detection is to include as many real nodules as possible in the detection results, but allow for a large false positive rate. False positive refers to those pseudo-nodules that resemble true nodules. Figure 2 shows some of these false positives and their contrast with true nodules. It can be seen that false positives are similar to true nodules in appearance, but are often composed of blood vessels, lymph nodes, or other lesions. For this reason, the false positive reduction, as the second step of a complete nodule detection, aims to distinguish true nodules from false nodules as accurately as possible from candidates.
In these two steps, the task of false positive reduction faces more challenges, mainly attribute to the following two aspects:
-
1.
The lung nodules vary in size, shape, and even resolution, partly due to the different thicknesses of CT slices in the data set. Specifically, the CT slices in LUNA16 dataset were obtained from 7 different academic institutions which adopted different scanners with various parameters, resulting in the change in slice thickness and resolution.
-
2.
The surrounding texture environment of nodules is complex, including many tissues such as blood vessels, lymph nodes, and other pathological tissues which are highly similar to lung nodules.
In order to overcome these challenges, an embedded multi-branch 3D convolution neural network (CNN) is proposed to reduce the false positive. The main processes of this method include the extraction of 3D samples, the establishment and training of 3D model, the experimental results, and the evaluation of nodules’ classification performance. Different from other algorithms that are either 2D CNN methods or multi-3D CNN combination methods, the proposed method is based on a single 3D CNN network and easier to use. Moreover, the proposed method is more discriminative for pulmonary nodules of different resolution and sizes and can directly classify candidates without requiring some pretreatments such as voxel normalization. The main advantages of this method are as follows:
-
1.
A novel 3D CNN model is established. The model contains multiple embedded branches, and each branch comes from layers at different depths of the network. All of such branches can deal with feature maps in different levels, enabling the model to distinguish lung nodules in different resolution and sizes more easily.
-
2.
The model is based on 3D convolution calculation and employs 3D samples as input, so that it can extract the spatial correlation characteristics of the target more effectively than that of the traditional 2D method, thus improving the classification performance.
-
3.
Some effective tricks are proposed to make the network easier to converge in training. The network is complex and with a large number of parameters which make it hard to converge when trained. We adopt some tricks like adding batch normalization (BatchNorm) layer into the appropriate location of the network or adding Gaussian noise to the feature map of a particular layer to make the network more general and converge quickly and efficiently.
-
4.
A good performance achieved in false positive reduction makes the proposed method a valuable one for reference in this type of application.
Related Work
At present, more and more researchers are committed to solving the challenging problems in lung nodule detection. In their research, some traditional methods and deep learning methods are the most commonly used.
Traditional methods tend to make use of some low-level features such as the boundary character, shape character, and intensity character [4] as well as high-level features such as AM-FM [5] and artificial GLCM features [1] which are all man-made. Furthermore, these traditional features are combined for lung nodule detection or classification. For example, there are over 80 kinds of features that are used together in [6] for lung nodule classification, causing the traditional methods to require much pretreatment before detecting pulmonary nodules. Obviously, the traditional method is not an adaptive approach and complicated to use.
On the contrary, deep learning technology is widely used for it can adaptively extract target features and obtain better performance in lung nodule detection. Specifically, both the 2D CNN and 3D CNN methods are frequently used for this purpose [3, 7,8,9,10,11,12].
Take the example of 2D CNN used for the false positive reduction in lung nodule detection, Zagoruyko and Komodakis [7] proposed a 2D wide residual network to address the challenging problems of the false positive reduction. In the method, three patches are extracted from the axial, sagittal, and coronal views. Three wide residual networks process these three kinds of patches, respectively. As a result, the final prediction comes from the average value of outputs of the three networks. The method ranks fifth with a CPM score of 0.758, slightly lower than that of CADIMI method, which is also a 2D CNN method with a CPM of 0.783.
In fact, three teams among all of the five teams who won the top five places of LUNA16 challenge have chosen to use 2D CNN technology [7,8,9]. The reason is probably that the number of parameters of 2D CNN is small and easy to converge, which enables it to be complexly designed for complex tasks.
However, though the 2D CNN technology is mature and has been applied in many medical fields, it is still not as good as 3D CNN at sufficiently extracting the spatial information of the volumetric target. Therefore, some 3D CNN models as an alternative have been gradually used in pulmonary nodules detection [8, 10, 11].
As a typical example, Huang et al. [10] proposed a 5-layer 3D CNN which achieved a better classification effect than that of the corresponding 2D network while being trained only by 99 sets of CT scans through data enhancement in false positive reduction. The prior research by Anirudh [8] presented a 4-layer 3D CNN to classify lung nodule candidates and gained a sensitivity of 0.80 for 10 false positives per scan.
But, it is worth mentioning that the design of a single 3D CNN network cannot be too complex due to its various parameters and difficulty in converging, causing its performance limited in lung nodule detection. Therefore, Dou et al. successively proposed two schemes to combine three 3D CNNs for lung nodules’ prediction [11], which is also a common idea for multi-network combined method. Two schemes can be seen in Fig. 3, where the blue blocks with label X represent the input, the purple dots with label Y or {Yi, i = 1, 2, 3} represent the output, and other white blocks represent the layers in the middle of the network. It can be seen that in both schemes, all the three networks need to be trained separately to predict the results or extract features, which make these schemes not ideal “end to end” ones.
Two common schemes for the idea of multi-network combination. a The scheme obtains the final prediction result by weighted summation of the prediction results of the three separately trained models. b The scheme uses the three separately trained models to extract features, and then combines the features to train the classifier and predict
In accordance with the abovementioned, we come up with one kind of embedded multi-branch 3D CNN for false positive reduction in lung nodule detection. It is a single but complex 3D CNN network with a host of parameters. To make the proposed network easier to converge, we also apply some tricks to the scheme. The details are described in the next section.
Methodology
The Multi-branch 3D CNN Model
The proposed multi-branch 3D CNN model is shown in Fig. 4. The backbone of the network consists of nine main layers, namely seven 3D convolution layers and two 3D pooling layers. These nine layers can be divided into three parts. The first part is the shallow-layer part (SL), which includes two 3D convolution layers and one pooling layer. The second part is the middle-layer part (ML), including two 3D convolution layers and one pooling layer. The last part is the deep-layer part (DL) with three 3D convolution layers involved.
The embedded multi-branch 3D CNN model. The cubes with different colors represent the 3D convolutional layer, pooling layer, and cropping layer, respectively. The numbers beside the convolution layer represent the number and size of the convolution kernel, respectively, while the number beside the pooling layer represents the stride of the pooling operation
The network branches also can be divided into three parts. Each part is connected to the deepest layer of the corresponding part of the backbone, respectively. Thus, feature maps from the different depth layers of the backbone can be processed individually by each branch.
Behind each branch part, a 1 × 1 × 1 convolution layer is included to compress the multi-channel feature map into a single-channel one. After that, the 3D pooling layer or 3D crop layer is added into different branch parts, where the clipping range of the 3D crop layer is designed as the size of (1, 1, 1) to crop the edge of the feature mapping block by 1 pixel along each dimension. Thus, the feature map output from each branch will be in the same size. Finally, all the feature maps’ output from these three branches are merged and input to the full connection layer to train the classifier.
Through this kind of structure, each branch processes the feature maps with different receive field from the backbone (see Table 1). All of the feature maps also have different strides in backbone which are eventually changed into the same stride through pooling, indicating that the features of different scales from different levels can be extracted and synthesized by the model; thus, the model can work better in extracting the distinguishing characteristics of the target so as to address the challenging problems in false positive reduction.
These feature maps with different levels from branches are presented in Fig. 5, where the subfigure (a) is an original sample with size of 18 × 18 × 18 shown along the Z-axis. The subfigure (b) shows all 32 feature maps of the SL branch, in which each feature map is shown by a cross section located in the center of the feature map cube. In the same way, the subfigure (c) shows all 64 feature maps of the ML branch, and the subfigure (d) also displays 64 feature maps, only half of all the 128 feature maps of the DL branch.
Since these feature maps are also in different scales, that is, the size of each feature map cube of the ML branch is 9 × 9 × 9, and one of the DL branches is 5 × 5 × 5, both of which have few pixels for display. For the sake of clear presentation, the cross sections of the feature map in Fig. 5 c and d are uniformly converted into the size of 18 × 18 by double cubic interpolation.
It can be found that the feature maps from the SL branch show more basic features such as edge and direction information, whereas the feature maps from the ML and DL branches show more detailed features. By merging these features at different scales and levels, the proposed multi-branch 3D CNN model is more conducive to the analysis of lung nodules with different sizes and shapes than the general one, so as to improve the detection performance of lung nodules.
The Training Process
We denote the training sample set as (X, Y) = {(xm, ym), m = 1, 2, ⋯, M}, where xm represents the mth sample image, and ym ∈ {0, 1} is the corresponding ground truth label of xm. Since these samples need to be input into the network for analysis, we denote the collection of all the parameters of the network as W. Due to multiple branches contained in the proposed network, if each branch is regarded as a separate output from the network, the corresponding weights of the branch can be expressed as w = {w(1), ⋯, w(N)}. In addition, because the network contains a fusion part which fuses the output features of each branch, we denote the fusion weight as β = {β1, ⋯, βN}. Thus, our training program can be explained by a formula as follows.
The objective function of this model can be expressed as formula (1).
where Lfuse(W, w, β) is the loss function used for gradient iteration and is defined as the cross entropy function, as shown in formula (2).
In formula (2), \( {\overset{\wedge }{y}}_m \) is the prediction confidence that the image xm is judged as a nodule by the proposed network, defined as formula (3).
Since the proposed network is a multi-branch fusion network, the prediction \( \overset{\wedge }{y} \) of input sample x is actually obtained by the joint action of all branches; hence, it can be calculated according to formula (4).
where \( {a}_{branch}^{(n)} \) is the activation of the nth branch output, and σ(⋅) is the sigmoid function.
By combining the above formulas (1), (2), (3), and (4), the weights can be constantly updated until we get the optimized result (W, w, β)∗ and complete the training process.
Some Tricks for Network Generalization and Convergence
In order to make 3D CNN converge, a typical 3D network is usually not designed in a complicated manner. However, overly simple network structure may affect the performance of the network in classification tasks. Therefore, in order to improve the classification performance of lung nodule candidates, the network proposed in this paper is relatively complex, which requires tricks to facilitate convergence.
Batch normalization (BatchNorm) is an effective means to accelerate the convergence of the network [13, 14]. Besides, as the name implies, BatchNorm converts input feature maps to output by re-parameterizing them on batch. This kind of re-parameterization makes gradients of the loss more Lipschitz. This, in turn, enables us to use a larger learning rate and, in general, makes the convergence speed of the network accelerate significantly in the process of training.
We tried to add the BatchNorm layer to different locations in the network to accelerate convergence. The experiment results are shown in Table 2, in which the BatchNorm layer is added behind the input layer or behind the deepest convolutional layer of the three different parts (SL, ML, and DL) of the backbone, respectively; thus, four networks with different structures are obtained, and their loss functions are named loss1, loss2, loss3, and loss4, respectively. According to Table 2, in the four networks, only loss1 and loss2 are convergent in the training process, while loss3 and loss4 fall into local convergence and cannot reach the optimal state, which is considered as non-convergence.
The training results of these convergent networks are shown in Fig. 6, where loss2 performs better than loss1. Therefore, the BatchNorm layer is finally added behind the SL of the backbone.
However, in addition to adding the BatchNorm layer, we also add Gaussian noise to the network for training to generalize the proposed model. Through experiments, the factor of Gaussian noise is finally selected as 0.125. And this component is added to the network behind the input layer.
The Experiments
The Dataset for Experiments
The experiments are carried out on LUNA16 dataset [3], which is provided by the organizer of the LUNA16 challenge. The LUNA16 dataset is obtained by filtering the largest publicly available database of chest CT scans, the LIDC-IDRI dataset [15, 16]. Through screening, the LUNA16 dataset finally retained the location information of 1186 lung nodules from 888 patients. This information will be used as comparison information to evaluate the prediction results.
The LUNA16 dataset also contains the location and category information of 551,065 lung nodule candidates, detected by existing algorithm [6, 17, 18]. This information is used to conduct classification experiments on candidates and reduce the false positive rate of lung nodules detection.
The Evaluation Index for Experiments
Our task is to classify lung nodule candidates and reduce false positives. Thus, the metrics commonly used for classification and the competition performance metric (CPM) for the false positive reduction track in LUNA16 challenge are used to evaluate the work of this paper.
The metrics commonly used for classification include accuracy, precision, sensitivity, and specificity, which can be expressed by the following formulas (5), (6), (7), and (8), respectively.
where TP, TN, FP, and FN represent the true positive, true negative, false positive, and false negative, respectively; all of which are indicators generated by the classification task.
Among the commonly used classification indexes mentioned above, the sensitivity at seven predefined false positive rates is as follows: 1/8, 1/4, 1/2, 1, 2, 4, and 8 FPs per scan are taken as the main metric of the LUNA16 challenge. The average of these seven specific sensitivities is defined as CPM [3], which is another important metric provided by the competition.
The Experimental Results
-
1.
Parameters setting
The experiments are carried out on one GTX1080 Ti GPU. The deep learning framework of Keras is taken as the software platform. In order to make the network converge to the optimal value during training, we set the super-parameters of the network in the training process as follows:
We set the training epochs to 15, the batch-size to 128, and the learning rate to 0.01. For each convolution layer, we set the initial bias to 0.2 and adopt the “Relu” as activation way. As for the pooling layer, we set all the strides to 2 except the one in the shallow-layer branch, which is set to 4. In addition, in order to balance the huge difference in the number of positive samples and negative samples that may affect the network recognition ability, we set the mode for the parameter “class_weight” in the function “fit( )” in the Keras framework to “auto.”
-
2.
Data preprocessing
The proposed approach is an “end-to-end” one, which indicates that no pretreatments such as lung segmentation are needed. Instead, all of our samples were extracted based on candidate location information provided by the dataset. Moreover, as can be seen from the above, the proposed method is not likely to be affected by variations in the size and resolution of lung nodules when identifying the nodules. Therefore, the sample does not need any preprocessing such as voxel normalization either. Thus, all samples are directly obtained from those CT volumes which are stacked by original CT slices.
There are two steps to make the samples: the determination of the sample center points and the segmentation of the sample blocks. When making negative samples, we extracted the coordinate information of candidates marked as non-nodules from the data set and took these coordinates as the locations of the center points of the negative samples. Since the sample size is designed to be 18 × 18 × 18 and obtained through experiments, we get the sample block by extending 8 or 9 pixel distances from the center along either side of the X-axis, Y-axis, and Z-axis, respectively.
Whereas, when making positive samples, after we extracted the coordinate information of candidates marked as real nodules from the data set, we needed to expand these coordinate points to get more positive sample centers. This is because the number of positive candidates in the data set is much smaller than that of the negative candidates, and this treatment can further reduce the impact of sample imbalance. We move the obtained coordinate points along the X-axis, Y-axis, and Z-axis by corresponding pixel distances. Each move gets a new coordinate point, thus getting multiple coordinate points. Each of these coordinate points is taken as the center of the sample. Then, the expanded positive sample block can be obtained by using the method similar to the method of making negative sample block.
-
3.
Training performance
The model is trained from scratch and takes the gradient descent method as the optimization algorithm. The changing process of its loss and accuracy is shown in Figs. 6 and 7, respectively.
It can be seen from Figs. 6 and 7 that although the network is complexly designed and has many parameters, the training process converges quickly due to the tricks proposed in the previous section. It also can be seen that the validation loss and accuracy are slightly shaken in the last few epochs, which is caused by employing a larger learning rate, but this does not affect the overall convergence trend and final accuracy of the network.
-
4.
Performance on Acc., Pre., Sen., and Spe.
We used the trained model to predict the class of lung nodule candidates, and the probability that candidate was judged to be a nodule is known as prediction confidence.
Table 3 shows the average prediction confidence that real nodules or non-nodules are predicted to be nodules. It can be seen that among 543,381 candidates that can be cropped out in the size of 18 × 18 × 18 from the LUNA16 dataset, 1320 real nodules are predicted as nodules with an average confidence of 0.9673, whereas 542,061 non-nodules are predicted as nodules with an average confidence of 0.0161. The huge gap in prediction confidence indicates that the proposed model is well distinguishable for different classes.
We also conducted a separate classification experiment on 1186 nodules in the LUNA16 dataset, which had been marked with size information. And because 1162 of these 1186 real nodules could be completely reduced to the size of 18 × 18 × 18, excluding those too close to the edge, we figured out the diameter distribution of these 1162 nodules and classified the nodules within each diameter range. The experiment results are shown in Table 4.
In Table 4, the number of nodules correctly predicted within each diameter range is actually the number of TP, while the number of true nodules is equivalent to that of TP plus NF. Thus, the true positive rates (TPR) of nodules with different diameters can be obtained according to formula (7). As can be seen from Table 4, the TPR value of nodules with a diameter of 6–12 mm or greater than 15 mm is higher, while the TPR value of nodules with other diameters is relatively small. However, in general, the TPR values of nodules within all diameter ranges exceeded 96%, indicating that the proposed method has a good classification performance for nodules with various sizes.
What is more, in order to evaluate the classification performance more comprehensively, we compare our method with other classification methods on four commonly used classification evaluation indexes. The results are shown in Fig. 8 and Table 5, where our method is named as EMU_3D_CNN. It can be found from Fig. 8 that the EMU_3D_CNN method gains higher scores than those of the majority voting method [23] and multi-crop [19] on four classification metrics. In terms of the acc. index, the EMU_3D_CNN method also gains higher score than other methods mentioned in Table 5.
-
5.
Performance on CPM
As mentioned earlier, the sensitivity at seven predefined false positive rates and the CMP are taken as the main metrics of the LUNA16 challenge to evaluate the “the false positive reduction” work. Thus, we submitted the classification results of 551,065 candidates in the LUNA16 dataset to the evaluation software and obtained the evaluation results. Table 6 and Fig. 9 show these experimental results.
In Fig. 9, the FROC performance is shown in the form of three curves, where the solid curve represents the average sensitivity at different false positive rates, while the dotted curves represent the upper and lower sensitivity at different false positive rates, respectively. All the curves show that the sensitivity increases with the rise of the average number of false positives per scan, particularly at 1/8, 1/4, 1/2, 1, 2, 4, and 8 FPs per scan; the sensitivity scores are competitive.
Table 6 compares the performance of our EMU_3D_CNN method with that of the other two. The other two methods used the same version of the competition dataset as the method in this paper and won the first and third place of LUNA16 competition, respectively.
As can be seen from Table 6, the CPM score of our EMU_3D_CNN method is 0.830, close to but slightly higher than that of CUMedVis method (the top-ranked method) [11]. Maybe, it is because three single 3D networks are combined for prediction (as shown in Fig. 3) in the structure of CUMedVis, which is similar to the idea of a three-branch network structure in this paper. However, our model is a single network and only needs to be trained once, which is more convenient for training and using than the CUMedVis method.
Table 6 also shows that the CUMedVis method performs better than the DIAG CONVNET method [8]. That is because the DIAG CONVNET method adopts 2D CNN, while our EMU_3D_CNN method adopts 3D CNN, which once again proves that 3D CNN is better at processing spatial information of 3D targets than 2D CNN.
Fig. 10 shows some correctly classified nodules and their classification confidence. These nodules vary in shapes, sizes, and peripheral textural environment, but all can be classified correctly with a confidence greater than 0.5, indicating that the proposed method can correctly classify nodules of different sizes and shapes from complex peripheral environments with a low false positive rate.
Discussion
-
1.
Advantages and causes
An embedded multi-branch 3D CNN is proposed in this paper to reduce false positives in lung nodule detection. The model can learn more subtle features of the target, including features of different scales and levels, and process spatial correlation information of 3D samples more effectively, thus avoiding some identification errors while classifying lung nodule candidates.
The proposed approach is an “end-to-end” approach, which indicates that no manual feature extraction is required. Moreover, since the samples used in this method are all taken from the original CT images, there is no need to carry out image preprocessing such as grayscale transformation, morphological transformation, and frequency domain transformation.
It should be noted that we adopt a ten-fold cross-validation method, so the test set and training set are independent. In other words, the total sample set is randomly divided into approximately equal 10%, one of which is selected as the test set and the remaining 9 as the verification set. Therefore, the experimental results in this paper are scientific.
A large number of previous studies [3, 24] have shown that, in order to reduce false positives in lung nodules detection, the algorithms of combining multiple CNNs for classification of lung nodule candidates are worthy of recommendation. But the experiment results show that, as a single CNN algorithm, the embedded multi-branch 3D CNN model can get similar and slightly higher scores than multi-CNN combined method such as CUMedVis and DIAG CONVNET method for false positive reduction in lung nodule detection, which greatly simplifies the detection process of lung nodules.
The improvement of classification performance of the proposed single CNN method may be mainly due to its multi-branch structure. However, this kind of structure is rarely seen in previous 3D networks, for it will make the 3D network with a large number of parameters more complex and difficult to converge. In order to solve this problem, batch normalization is used in this paper to normalize the output of the corresponding layer of the network so as to speed up the convergence of the networks. This training strategy can be used as reference for the rapid convergence of networks with similar complex structures in the future.
-
2.
Limits and future work
As mentioned above, the proposed embedded multi-branch 3D CNN can identify nodules of different sizes and shapes with a low false positive rate. Even so, for nodules in different sizes, the prediction confidence is often different. As shown in Fig. 11, the average prediction confidence for nodules within each diameter range is displayed, where, for nodules with a diameter of 6–12 mm, the prediction confidence is higher, with a score of over 97%. However, for nodules larger than 12 mm in diameter, the prediction confidence is lower, with a score of about 93%. The reason may be that there are more cases of nodules with a diameter of 6–12 mm in the LUNA16 data set, which enables them to be fully represented in the training process. In contrast, the number of nodules with other sizes is relatively small and less well represented in the training set. We believe that this is a problem that will be improved as larger data sets become available in the future.
Finally, it is worth mentioning that in the above experiment, only 543,381 out of the 551,065 candidates in LUNA16 dataset were successfully cut out in size of 18 × 18 × 18, which indicates that some other candidates are too close to the edge to be cut out. This kind of inadequate cut may affect the identification accuracy. In order to solve this problem, we may also cut out targets of other sizes for auxiliary identification in the future, or only use one or several parts of the target for joint identification, or propose other algorithms specifically targeted at those marginal nodules for recognition, which will all be the work that we might try in the future.
Conclusion
The false positive reduction in lung nodule detection is often challenged by the fact that the lung nodules are three-dimensional and have a wide variation in size and shape. Therefore, an embedded multi-branch 3D CNN model is proposed to resolve this challenging problem. This model takes 3D volumes as the input and carries out 3D convolution calculation, which can more effectively deal with the spatial information of nodules. Moreover, as an embedded multi-branch structure, the model identifies lung nodules of different sizes and shapes more effectively. The experimental results on the LUNA16 dataset show that the proposed method has obtained a competitive score on the evaluation metrics of classification performance and the LUNA16 challenge metrics. In addition, experiments show that the network converges rapidly despite its complex structure and large number of parameters. To sum up, our model is more convenient to be trained and used, and as a single network, can actually achieve the effect similar to that of multi-network combination approach.
References
Demir Ö, Yılmaz ÇA: Computer-aided detection of lung nodules using outer surface features. Biomed Mater Eng 26(s1):S1213–S1222, 2015
Teramoto A, Fujita H, Yamamuro O, Tamaki T: Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med Phys 43(6):2821–2827, 2016
Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel S, Wille MW, Naqibullah M, Sanchez C, van Ginneken B: Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging,2016. https://doi.org/10.1109/TMI.2016.2536809
Setio AAA et al.: Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 42:1–13, 2017
Le L, Devarakota P, Vikal S et al.: Computer Aided Diagnosis Using Multilevel Image Features on Large-Scale Evaluation. Lect Notes Comput Sci:161–174, 2013
Aggarwal P, Vig R, Sardana HK: Patient-Wise Versus Nodule-Wise Classification of Annotated Pulmonary Nodules using Pathologically Confirmed Cases. J Comput 8(9), 2013
Jacobs C, van Rikxoort EM, Twellmann T, Scholten ET, de Jong PA, Kuhnigk JM, Oudkerk M, de Koning HJ, Prokop M, Schaefer-Prokop C, van Ginneken B: Automatic detection of subsolid pulmonary nodules in thoracic computed tomography images. Med Image Anal 18:374–384, 2014
Setio AAA, Jacobs C, Gelderblom J, van Ginneken B: Automatic detection of large pulmonary solid nodules in thoracic CT images. Med Phys 42(10):5642–5653, 2015. https://doi.org/10.1118/1.4929562
Zagoruyko S, Komodakis N: Wide residual networks. arXiv preprint arXiv:arXiv:1605.07146, 2016
Anirudh R: Lung nodule detection using 3D convolutional neural networks trained on weakly labeled data[C]//. SPIE Medical Imaging:978532, 2016
Huang X, Shan J, Vaidya V: Lung nodule detection in CT using 3D convolutional neural networks[C]// IEEE, International Symposium on Biomedical Imaging. IEEE, 2017
Dou Q, Chen H, Yu L et al.: Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans Biomed Eng 64(7):1558–1567, 2016
Ioffe S, Szegedy C: Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. 2015
Santurkar S, Tsipras D, Ilyas A, et al: How Does Batch Normalization Help Optimization?. 2018
Armato III, Samuel G, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP et al.: Data From LIDC-IDRI. Cancer Imaging Arch, 2015. https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX
Armato, III SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, van Beek EJR, Yankelevitz D et al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med Phys 38:915–931, 2011
Eman M, Nourhan Z, Mahmoud F: Automatic Classification of Normal and Cancer Lung CT Images Using Multiscale AM-FM Features. Int J Biomed Imaging 2015:1–7, 2015
Murphy K, van Ginneken B, Schilham AMR, de Hoop BJ, Gietema HA, Prokop M: A large scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classification. Med Image Anal 13:757–770, 2009
Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C et al.: Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recogn 61:663–673, 2017
Zhu W, et al: Deep lung: 3D deep convolutional nets for automated pulmonary nodule detection and classification, Arxiv 2017 [online]. Avaiable: arXiv: 1709.5538
Shen W, Zhou M, Yang F, Yang C, Tian J: Multi-scale Convolutional Neural Networks for Lung Nodule Classification. Inf Process Med Imaging 24:588–599, 2015
Yan X, Pang J, Qi H, Zhu Y, Bai C, Geng X, Liu M, Terzopoulos D, Ding X: Classification of lung nodule malignancy risk on computed tomography images using convolutional neural network: A comparison between 2d and 3d strategies. In ACCV, 2016
Farahani FV, Ahmadi A, Zarandi MHF: Lung nodule diagnosis from CT images based on ensemble learning[C]// Computational Intelligence in Bioinformatics & Computational Biology. IEEE, 2015
van Ginneken B, Armato SG, de Hoop B, van de Vorst S, Duindam T, Niemeijer M, Murphy K, Schilham AMR, Retico A, Fantacci ME, Camarlinghi N, Bagagli F, Gori I, Hara T, Fujita H, Gargano G, Belloti R, Carlo FD, Megna R, Tangaro S, Bolanos L, Cerello P, Cheran SC, Torres EL, Prokop M: Comparing and combining algorithms for computeraided detection of pulmonary nodules in computed tomography scans: the ANODE09 study. Med Image, 2010
Acknowledgments
The authors would like to thank the LUNA16 challenge organizers for providing the dataset and evaluation software.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) under Project 61471123 and the Research Foundation of Education Bureau of Hunan Province of China under Project 13C829.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zuo, W., Zhou, F. & He, Y. An Embedded Multi-branch 3D Convolution Neural Network for False Positive Reduction in Lung Nodule Detection. J Digit Imaging 33, 846–857 (2020). https://doi.org/10.1007/s10278-020-00326-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00326-0