Abstract
Remarkable progress has been made in image classification and segmentation, due to the recent study of deep convolutional neural networks (CNNs). To solve the similar problem of diagnostic lung nodule detection in low-dose computed tomography (CT) scans, we propose a new Computer-Aided Detection (CAD) system using CNNs and CT image segmentation techniques. Unlike former studies focusing on the classification of malignant nodule types or relying on prior image processing, in this work, we put raw CT image patches directly in CNNs to reduce the complexity of the system. Specifically, we split each CT image into several patches, which are divided into 6 types consisting of 3 nodule types and 3 non-nodule types. We compare the performance of ResNet with different CNNs architectures on CT images from a publicly available dataset named the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI). Results show that our best model reaches a high detection sensitivity of 92.8% with 8 false positives per scan (FPs/scan). Compared with related work, our work obtains a state-of-the-art effect.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the most aggressive disease among all the cancer-related diseases due to its significant morbidity and mortality. According to a World Health Organization survey, lung cancer has been the largest cause of cancer deaths among male patients, while in female patients, lung cancer deaths rank second only to breast cancer [1, 2]. Therefore, early detection is significant for lung cancer diagnosis, which can greatly increase a lung cancer patient’s chance of survival.
Low-dose computed tomography (CT) is a powerful method for detecting lung cancer early, by identifying the malignant primary lung nodules [3]. However, it is a quite tiring task for radiologists to carefully examine each CT image, of which there can be a large amount. Meanwhile, the diagnosis is very likely to be affected by the limitation of the radiologists’ experience and knowledge, which increases the possibility of misdiagnosis and failure to diagnose. To avoid the radiologists’ subjectivity factor and reduce their burden, computer-aided detection, which can find the location of lung nodules and predict the risk of lung cancer, can be used to help radiologists with the speed and accuracy of diagnosis [4, 5].
Before deep learning methods came out, image-based lung nodule detection was usually associated with two steps: first, image analysis did feature extraction [6] or hand-crafted features work [7, 8]. Second, they used a support vector machine (SVM) classifier [9] or random forest [10] method to classify these featured nodules. Descriptors of the histogram of oriented gradients (HOG) [11], local binary patterns (LBP) [12], and wavelet feature descriptors [13] were used for feature extraction. Without these automatic feature extraction methods, they can also extract hand-crafted features in terms of geometry [14], appearance [15], or texture [16]. For example, Gurcan et al. [6] used weighted k-means clustering to segment the lung structures from the lung region and classified them by designing a rule-based classifier to reduce the number of false positive (FP) objects per slice. Their work can achieve 84% sensitivity with 5.48 FP objects per slice. Elmar et al. [9] calculated several masks to eliminate the background and surrounding tissue and used SVM to categorize the regions of interest with a sensitivity of 84.93%, specificity of 80.92%.
In 2006, Geoffrey and his team [17] proposed a fast learning algorithm for deep belief nets. After that, deep learning methods developed quickly. Thanks to a large number of available datasets such as ImageNet and the computational power of the GPUs, convolutional neural networks (CNNs) have amazingly advanced the landscape of image classification [18], object detection [19], and semantic segmentation [20].
Recently, with the development of CNNs’ architectures such as AlexNet [21], VggNet [22], GoogLeNet [18], and ResNet [23], more and more teams use CNNs to do medical image diagnosis tasks. Rotem et al. [24] used AlexNet to extract valuable volumetric features from the input data and achieved a sensitivity of 78.9% with 20 false positives per scan (FPs/scan). Shuo et al. [25] present multi-view convolutional neural networks for lung nodule segmentation. The architecture can capture a set of nodule features from axial, coronal, and sagittal views in CT scans, so it has obtained a sensitivity of 83.72% with 20.71 FPs/scan. Kui et al. [26] also designed multi-view CNNs, but they used an MV-CNN for lung nodule classification, which consisted of benign, malignant primary, and metastatic malignant categories. Huang et al. [27] made full use of the spatial 3D context of lung nodules and proposed 3D CNNs. The results showed that 3D CNNs perform better than 2D CNNs, which can achieve a sensitivity of 90% at 5 FPs/scan. Yu et al. [28] also used 3D CNNs. However, their team used a multi-scale prediction strategy including multi-scale cube prediction and cube clustering which can detect extremely small nodules. The sensitivity of their scheme is 92.93% with 4 FPs/s. Recently, more teams apply object detection methods to lung nodule detection. Zhu et al. [29] used 3D Faster-RCNN method on a basis of dual-path network and a U-Net like encoder-decoder structure to obtain the features which achieve a sensitivity of 93.3% with 8 FPs/scan.
In this work, we propose a CAD system using the ResNet-like CNNs based on a prior CT image split, or what we called CT image segmentation. The architecture of a CAD system usually has two parts: (1) generating nodule candidates and (2) classifying candidates. However, we do not need to generate nodule candidates in this work. We split the raw CT images into several patches that were numbered in a certain order and divided into six types instead of the first part of former CAD systems. Then we put these six types of patches into CNNs to extract their features. We can predict if the patch has nodules and find the location of the patch in the CT images according to their numbers. The radiologists can use the information from the CAD system to make a correct diagnosis. We also evaluate the performance of different CNN models like AlexNet and GoogLeNet. Our contributions are summarized as follows:
1. We use an image split technique to reduce the complexity of the CAD system. In contrast with former work in this area, we do not need to do hand-crafted feature extraction or image pre-processing to generate nodule candidates, which can reduce the subjectivity factor.
2. The image split technique can easily find the location of nodules thanks to a certain number each patch has.
3. We use ResNet-like CNNs to classify six types of patches, which is more accurate than the two types of patches used in former work. This can increase the sensitivity of the CAD system and decrease the false positive rate at the same time.
Methods
Dataset
We use the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) dataset [30] to do training and testing experiments in this work. The LIDC-IDRI contains 1018 cases consisting of marked-up annotated lesions in lung cancer screening thoracic CT scans. Four experienced radiologists independently divided these lesions into three categories (“nodule ≥ 3 mm,” “nodule < 3 mm,” and “non-nodule ≥ 3 mm”). The XML file attached to each patient indicates the region of interest (ROI) of each nodule and its characteristics, such as texture, malignancy, and calcification, labeled by each of the four radiologists. The images of each patient comply with the Digital Imaging and Communications in Medicine (DICOM) standard and have a resolution of 512 × 512 pixels, where the number of slices may range from 65 to 764.
To rigorously evaluate the performance of our CNNs’ architecture, we partitioned the 600 scans into three subsets: training, validation, and testing sets. The training set was used to update the weights of the convolutional layers, fully connected layers, and softmax layers. The validation set was used to tune the values of hyper-parameters such as the learning rate, kernel size of each unit, and number of kernels. The testing set was used to evaluate the performance of the CNNs’ architecture and the whole CAD system.
CT Image Split
Due to the high resolution of raw CT images, it is quite difficult for the CNN model to directly determine whether the image has nodules. We came up with an effective way to solve this problem.
Former work usually used two or three specific detectors to extract nodule from CT scan and then put them into CNN classifier which was computationally inefficient. Some teams just used CNN classifier to categorize candidates into two types: nodules and non-nodules. According to the Ginneken’s work [31], it is an arduous task to classify adhesion pulmonary nodules attached lung wall because lung wall usually occupies most space of one patch which is extremely different from solitary pulmonary nodules in morphology. If we put solitary pulmonary nodules and pulmonary parenchyma adhesion type into one category which we will talk about in the Discussion section, the sensitivity will decrease a lot. Inspired by his studies, we considered categorizing patches into six types.
Before training, we should make six types of patches and label them as our train set. We split each slice of CT scans into several small patches in a certain order and numbered the patches in accordance with their locations in the scan. With the help of the diagnoses in the XML files provided by LIDC-IDRI, we marked patches as belonging to one of the six types so that the CNNs could more effectively detect lung nodules and locate them by patch number.
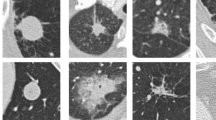
Figure 1 illustrates the six types of patches: (1) solitary solid nodules whose CT values are larger than – 450 HU, (2) large solitary solid nodules whose diameter is larger than 10 mm, (3) nodules attached to or surrounded by lung parenchyma, (4) lung parenchyma without nodules, (5) vessels and tissues, (6) no obvious tissues, vessels or lung parenchyma. Types 1, 2, and 3 are all nodules, and types 4, 5, and 6 are all non-nodules.
CNNs’ Architecture
In this work, we propose CNNs based on former researchers’ work [23]. Though the architecture of the CNNs is designed for classifying thousands of types of objects, it still has a remarkable performance in lung nodule detection task. As is shown in Fig. 2, the CNNs’ architecture consists of two parts. The first part consists of training CNNs to extract various features from the six types of patches we mentioned in section B and is composed of four residual blocks from RB1 to RB4. Every block has three different convolutional layers with leaky rectified liner units (Leaky ReLU) and batch normalization layer. The second part consists of the classifier that is composed of a global average pooling layer, followed by a softmax layer.
The system starts from one single CT scan. It gets one image from the whole scan and cuts it into 64 blocks, which we call patches. These patches are numbered in a certain order according to their location in this image. The classification part of the CNNs’ architecture will classify these patches into the six types we have talked above. With the help of these numbers, the CAD system can easily distinguish the nodules from the patches and find their locations in images.
Considering that the diameters of lung nodules usually range from 3 to 30 mm [32], we resize these patches to 64 × 64 pixels, which can keep the full feature information of nodules. However, this may lead to a failure involving the misdiagnosis of small nodules. The size of the first convolutional layer C1 is 3 × 3 with 32 kernels and the size of the second layer C2 is 3 × 3 with 64 kernels. And the strides of C2 is 2. The strides are set to decrease the size of feature maps and the number of the CNNs’ weights. Each kernel produces a 2D image output (e.g., 64 32 × 32 small patches after C2). Then the main body of the network is residual block. It consists of three different convolutional layers. The input of the residual block will be connected to the end of the convolutional block with a skip connection. In this way, the ResNet can access the earlier activations which were not modified in the convolutional block. The values of the kernels are initialized by the Xavier algorithm and can be updated during the training of CNNs using a back-propagation algorithm. The whole CNNs’ architecture is described in sequence as follows: C1:32 × 3 × 3, C2:64 × 3 × 3/2, RB1:× 1, C3:128 × 3 × 3/2, RB2:× 2, C4:256 × 3 × 3/2, RB3:× 4, C5: 512 × 3 × 3/2, RB4:× 2, AP1:4 × 4 where, e.g., RB2:× 2 means there are two the same residual blocks and AP1:4 × 4 means an average pooling layer with a kernel size of 4 × 4.
We used Leaky ReLU as the activation function in the convolutional layer. Unlike traditional activation functions such as sigmoid and tanh, Leaky ReLU is quite simple. The activation f(x) for an input x is obtained as f(x) = x when x > 0, f(x) = ax when x < 0. It is more in line with the transmission of a neural impulse of the human nervous system and can appropriately solve the problem of gradient dispersion in deep networks and introduce sparsity to the network to reduce the overfitting.
In the last of our network, we used a global average pooling layer instead of fully connected layer. We used a 4 × 4 size of kernels which meant every feature map will be followed by one output. It can reduce a large number of parameters compared with a fully connected layer. Furthermore, we used the batch normalization layer to control overfitting effect and speed up network’s convergence. Batch normalization layers are widely used in a convolutional network. The batch normalization layer’s work can be summarized as:
where μB is the mean of one mini-batch, and \( {\sigma}_{\mathrm{B}}^2 \) is the variance of one mini-batch. The subscript B means a mini-batch. γ and β are parameters which can be learned, output is yi.
The output layer global average pooling layer is followed by a softmax classifier that uses the cross-entropy cost function. The softmax classifier can give us a more intuitive output than another commonly seen classifier, SVM. Suppose that θi is the ith row weight of softmax layer. xi is the input vector, or we can call it the output of global average pooling layer (in this paper, the size of θi is 1 × 512 and xi is 512 × 1), and we have the probability assigned to the correct label yi as
the division performs the normalization to make the summation one. To maximize the probability of the correct class and improve the networks, we should minimize the cross-entropy loss. The loss function is defined as follows:
where {y(i) = j} means it is equal to 1 if the condition is met; otherwise, it is equal to 0. We add an L2-norm to penalize large values of the parameters.
Experiments
Data Augmentation
Optimization of the CNNs relies on the quality of the training dataset. If the training dataset is skewed, the weight of each layer will lead the deep learning algorithm to the local optimum, which means the normal balanced set cannot reach the same performance as the training set evaluated during the training period. Therefore, we use the data augmentation technique to prevent the overfitting of the training set. Meanwhile, the testing set does not have enough patches to evaluate the CNNs, so data augmentation is also applied to this set.
Since the number of non-nodules is much more than the number of nodules, we pay more attention to the nodules’ types. We use rotation, translation, and scaling techniques to enlarge the nodules set. It is noteworthy that a rotation technique usually cannot be used in data augmentation because of misleading image semantics. Here, however, the image semantics of the nodule patches do not change with rotation. Overall, there are 3286 patches with nodules and 4294 patches with non-nodules, as shown in Table 1. Finally, we resize each patch in all three sets to 64 × 64 to be put into the CNNs.
Evaluation Criteria
To evaluate the performance of the CAD systems and CNNs, some evaluation criteria need to be defined. The most intuitive performance metric is accuracy. However, it is not enough for medical diagnosis. We should pay more attention to missed diagnoses (regarding nodule patches as non-nodule patches) and misdiagnosis (regarding non-nodule patches as nodule patches) [33]. For this reason, sensitivity and misdiagnosis rates are applied to the evaluation criteria. Supposing that the number of true positives is TP, false positives is FP, true negatives is TN, and false negatives is FN, we have the formula for accuracy, sensitivity, and misdiagnosis as follows [9, 34]:
For the evaluation criteria of the CAD system, the evaluation is performed by measuring the sensitivity and the corresponding false positive rate per scan. We usually call these evaluation criteria the free receiver operating characteristic (FROC) analysis [34]. First, we set a threshold t; then the sensitivity and average number of false positives per scan are determined. The two values make coordinates in the FROC curve. If we change the threshold t by a value such as 0.001, we will get a new sensitivity and a new average number of false positives per scan in the FROC curve. In this way, we can draw a whole FROC curve. To be more easily compared with other work, we can get some certain sensitivity at the predefined false positive rate, such as 4 or 8 FPs/scan.
Training
The CNNs are implemented in a TensorFlow framework by using a GeForce RTX 2080 Ti GPU. We use the mini-batch gradient descent technique to update the parameters where the batch size is 32. As learning rate is a very important hyper-parameter during the training process, we should anneal it over time. With a high learning rate, the parameter vector may bounce around chaotically which can contribute to the system being unable to find a smaller loss of the results. However, if we set the learning rate too small, we will be wasting too much time on finding the best position for the system. So, we apply the step decay technique to the system, which means the learning rate will be reduced by some factor every few iterations. We also use the momentum technique to accelerate learning. The momentum algorithm accumulates an average of past gradients and continues to update in their direction. We set the momentum to 0.9. We have the annealing learning rate formula as follows:
where base_lr is 0.001, gamma is 0.9, stepsize is 100,000, and iter is the number of the current iteration.
We do one evaluation process of the validation set every few iterations. Finally, we train the CNNs for 30,000 batches and choose one CNNs’ architecture with the best parameters. In this work, we also try the image split technique in different CNN models like AlexNet and GoogLeNet.
The training curves of the three networks are shown in Fig. 3. As shown, the accuracy of AlexNet, GoogLeNet, and our ResNet became stable after 20 epochs.
Results and Comparison
When we begin to calculate the sensitivity of this CAD system, we regard the multi-classification task as the binary classification task which means if one patch is classified as type1, 2, or 3 patches, it will be regarded as nodules; otherwise, it will be regarded as non-nodules.
If we focus on the performance of the nodule classification task, the results of the binary classification for the networks are shown in Table 2. AlexNet can only achieve a 72.16% sensitivity along with 4 FPs/scan. There is no doubt that AlexNet networks are not deep enough to learn all the nodules’ features. GoogLeNet obtains a sensitivity of 75.25% with 4 FPs/scan because of the more convolutional layers, which is much better than AlexNet. Our ResNet can achieve a sensitivity of 89.60% with 4 FPs/scan, which is much higher than the sensitivity achieved by GoogLeNet and AlexNet. Within a certain range, increasing the depth of the network can improve accuracy. However, when the depth of the networks comes to a certain limit, the performance of the network cannot make good progress. That is why GoogLeNet can get better performance than AlexNet but still is not good enough. If we keep increasing the number of convolutional layers, the accuracy which we can see in Fig. 3 will start to become stable at one point and eventually degrade. It is obvious that shallower networks contain more feature information from patches than deeper network. Our ResNet-like network takes this advantage of shallower network. We connect the shallower network directly to deeper network to guide the whole CNNs to learn more features of nodules. The result of our ResNet-like network seems promising. ResNet also does better work at training speed compared with GoogLeNet and AlexNet.
To illustrate the effect of the networks, we inspect the FROC curves of AlexNet, GoogLeNet, and ResNet shown in Fig. 4 [35].
Table 3 shows the details of some CAD systems working with the LIDC-IDRI dataset. There are two types of CAD systems using CNNs. One is the type of CAD system that can work without any prior knowledge of nodules, like our work and the work of Rotem et al. This method can detect all types of nodules including ground-glass and juxta-pleural. Another type of CAD system, like that used by Setio et al., is a CAD system that generates all suspicious candidates according to the existing nodule detecting algorithms, which means it cannot detect those nodules that the algorithms do not cover, like ground-glass and juxta-pleural. So we can see that although these works are all done in recent years, the results are quite different, as shown in Table 4. The approaches which have a candidate generation part can have a small FPs/scan thanks to the small problem space, but they cannot detect all types of nodules. The approaches whose candidates include just the sub-space of the whole scan can detect all types of nodules with a substandard FPs/scan. Our work uses an image split to generate raw patches without any pre-processing. The system puts all types of nodules into the CNNs so that the networks can learn all the features of different nodules. Though the problem space is large, the result is still state-of-the-art. It can get a sensitivity of 92.8% with 8 FPs/scan.
To put our CAD systems in a broader context, the performances of existing CAD systems are reported in Table 4. All these schemes were implemented with deep learning methods. Setio’s scheme, Rotem’s scheme, Jiang’s scheme, Huang’s scheme, Khosravan’s scheme, Broyelle’s scheme, and Dou’s scheme all used CNNs to do classification task to reduce false positives. Setio’s scheme used 2D CNNs to implement lung nodule detection and others all used 3D CNNs. Except Rotem’s scheme and Khosravan’s scheme, these schemes all consisted of two steps whose first step was lung nodule detection and the second step was a FP reduction step. From the results, we can determine that those methods which had two steps usually had better performance in FPs/scan thanks to the little problem space which can be reduced in the first step.
Though Khosravan’s method just had one step, it still obtained much better performance even than those that had two steps. Khosravan’s scheme adopted dense network, which could keep the low-level information in shallower layers. Our ResNet is similar to its dense network in avoiding the loss of shallower layers’ information. Thus, both our scheme and their scheme generated few FPs with one step method.
Zhou’s scheme in Table 4 also obtained remarkable performance. They used Faster-RCNN deep learning method which is a quite popular method in object detection task. They adopted 3D Faster regions with 3D dual path blocks and a U-Net-like structure which could take advantage of both ResNet and DenseNet.
Compared with Setio et al. and Huang et al.’s approaches, the FPs/scan of our proposed system is little larger than theirs because of different pre-processing methods. They detect suspicious candidates in raw scans and remove the useless image data so that the number of non-nodules needing to be classified by networks is much smaller than that of our system. However, their methods may ignore some very small nodules and cannot detect certain types of nodules, which may lead to failure to diagnose. Additionally, in their pre-processing, they may ignore some nodules that their results do not notice. So, in application, the sensitivity of their work may be lower than their results. Our pre-processing method is simple but effective. It is convenient for users to immediately find the location of nodules in the scan of the patient. Our pre-processing method is similar to Rotem et al., but the performance of our network is much better than theirs both in sensitivity and FPs/scan. The reason why our scheme is better is that we divide the results into six types, which is of great help in improving the sensitivity. In the next section, we will study the influence of the number of types on our CAD systems.
Discussion
In this study, a lung nodule detection CAD system using an image spilt technique is proposed. Compared with other published CAD systems, our CAD system achieves a state-of-art performance whose detection sensitivity can obtain 92.8% with 8 FPs/scan. Furthermore, other published CAD systems need prior CT scan candidate generation steps, which is unnecessary for the application of our CAD system. We design an image split technique to generate several patches from raw CT scans so that the CNNs can classify each patch one by one. Once one patch from an image of a CT scan is classified by CNNs, the system can immediately find the location of the nodules in CT images. The classification work which is performed by CNNs has proven to be very remarkable, indicating that the CNNs can extract correct features of different types. So, CNNs are suitable for the problem of lung nodule detection.
We also tried different CNNs’ architectures based on our image split technique. Results are shown in Table 2. Compared with the simple AlexNet architecture, GoogLeNet and our ResNet perform better in detection accuracy and controlling overfitting. Compared with GoogLeNet, AlexNet has fewer parameters and layers, while it can achieve nearly the same accuracy as GoogLeNet. The ResNet we designed has more layers but fewer parameters. In application, the time of detection for ResNet is less than that of GoogLeNet and AlexNet. Above all, adopting more advanced networks can obtain faster speed of detection and higher accuracy of detection.
To figure out the effect of the number of patch types, we tried three different methods of classification. We set the number of patch types as 2, 4, and 6. At first, we categorized patches into two types which were nodules and non-nodules and get some results. But the results showed that the network could not classify non-nodules correctly because non-nodule patches were quite different in features. The rate of misdiagnosis was high. According to the results, we divided the non-nodules into three types. First one is patches of lung parenchyma without nodules; second one is patches of vessels and tissues; third one is patches of no obvious tissues, vessels, or lung parenchyma. In this way, the network can learn features of different non-nodule patches. The result of four types of patches showed that the accuracy of classification increased but it was still difficult for the CAD systems to distinguish between patches of nodules attached to or surrounded by lung parenchyma and patches of solitary nodules which was also mentioned in Ginneken’s work [31]. We also found that the size of nodules affected the accuracy of our CAD system. So we divided patches of nodules into three types on the basis of sizes and locations. The experiment showed that dividing patches into six types could increase the sensitivity of our CAD system and reduce the FPs/scan Table 5.
Conclusion
In this paper, we used deep learning methods for the problem of lung nodule detection and designed a new CAD system using CNNs based on an image split. The system consists of two parts: (1) obtaining six types of patches from the process of splitting images and (2) using ResNet to classify six types of patches to detect lung nodules. The first part of our proposed CAD system can reduce the complexity of the system compared with other existing systems. The promising results indicate that the CNN-based CAD system is suitable to be used to help radiologists with diagnosis work.
However, limited by the lack of large training sets of lung nodules, the detection sensitivity of our system is not good enough. We will explore more CNNs’ architectures using larger training sets and try to apply the deep learning methods to the diagnosis of other lung diseases.
References
Siegel RL, Miller KD, Jemal A: Cancer statistics, 2015. CA Cancer J Clin 65(1):5–29, 2015
Ma L, Wang DD, Zou B, Yan H: An eigen-binding site based method for the analysis of anti-EGFR drug resistance in lung cancer treatment. IEEE/ACM Trans Comput Biol Bioinform 14(5):1187–1194, 2017
Kaneko M, Eguchi K, Ohmatsu H, Kakinuma R, Naruke T, Suemasu K, Moriyama N: Peripheral lung cancer: screening and detection with low-dose spiral CT versus radiography. Radiology 201(3):798–802, 1996
Jiang H, Ma H, Qian W et al.: An automatic detection system of lung nodule based on multi-group patch-based deep learning network. IEEE J Biomed Health Inform 22:1227–1237, 2017
Messay T, Hardie RC, Rogers SK: A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal 14(3):390–406, 2010
Gurcan MN, Sahiner B, Petrick N, Chan HP, Kazerooni EA, Cascade PN, Hadjiiski L: Lung nodule detection on thoracic computed tomography images: Preliminary evaluation of a computer-aided diagnosis system. Med Phys 29(11):2552–2558, 2002
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006, 2014
Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, Kim J, Goldgof DB, Hall LO, Gatenby RA, Gillies RJ: Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol 7(1):72–87, 2014
Rendon-Gonzalez E, Ponomaryov V: Automatic lung nodule segmentation and classification in CT images based on SVM. International Kharkiv Symposium on Physics and Engineering of Microwaves, Millimeter and Submillimeter Waves. IEEE 1–4, 2016
Lee S L A, Kouzani A Z, Hu E J: A random forest for lung nodule identification. TENCON 2008 - 2008 IEEE Region 10 Conference. IEEE:1–5, 2008
Adetiba E, Olugbara OO: Lung cancer prediction using neural network ensemble with histogram of oriented gradient genomic features. ScientificWorldJourna 2015:1–17, 2015. https://doi.org/10.1155/2015/786013
Shan C: Learning local binary patterns for gender classification on real-world face images. Pattern Recognit Lett 33(4):431–437, 2012
Orozco HM, Villegas OOV, Sánchez VGC, Domínguez HJO, Alfaro MJN: Automated system for lung nodules classification based on wavelet feature descriptor and support vector machine. Biomed Eng Online 14(1):9, 2015
Tartar A, Akan A, Kilic N: A novel approach to malignant-benign classification of pulmonary nodules by using ensemble learning classifiers. Conf Proc IEEE Eng Med Biol Soc: 4651–4654, 2014
Kang G, Liu K, Hou B, Zhang N: 3D multi-view convolutional neural networks for lung nodule classification. PLoS One 12:e0188290, 2017. https://doi.org/10.1371/journal.pone.0188290
Han F, Wang H, Zhang G, Han H, Song B, Li L, Moore W, Lu H, Zhao H, Liang Z: Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging 28(1):99–115, 2015
Hinton GE, Osindero S, Teh YW: A fast learning algorithm for deep belief nets. Neural Comput, 2006. https://doi.org/10.1162/neco.2006.18.7.1527
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A: Going deeper with convolutions. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit, 2015.
Ren S, He K, Girshick R, Sun J: Faster r-cnn: towards real-time object detection with region proposal networks. Adv Neural Inf Process Syst: 91–99, 2015
Liang-Chieh C, Papandreou G, Kokkinos I, Murphy K, Yuille A: Semantic image segmentation with deep convolutional nets and fully connected crfs. International Conference on Learning Representations, 2015
Krizhevsky A, Sutskever I, Hinton GE: Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst:1097–1105, 2012
Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014
He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 770–778, 2016
Golan R, Jacob C, Denzinger J: Lung nodule detection in CT images using deep convolutional neural networks. International Joint Conference on Neural Networks: 243–250. 2016
Wang S, Zhou M, Gevaert O, Tang Z, Dong D, Liu Z, Tian J: A multi-view deep convolutional neural networks for lung nodule segmentation. Conf Proc IEEE Eng Med Biol Soc 2017, 1752–1755
Liu K, Kang G: Multiview convolutional neural networks for lung nodule classification. Int J Imaging Syst Technol, DOI:https://doi.org/10.1002/ima.22206, 2017
Huang X, Shan J, Vaidya V: Lung nodule detection in CT using 3D convolutional neural networks. 2017 IEEE 14th International Symposium on Biomedical Imaging 379–383, 2017
Gu Y, Lu X, Yang L, Zhang B, Yu D, Zhao Y, Gao L, Wu L, Zhou T: Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput. Biol. Med. 103:220–231, 2018
Zhu W, Liu C, Fan W, Xie X: Deeplung: Deep 3d dual path nets for automated pulmonary nodule detection and classification. 2018 IEEE Winter Conf. Appl. Comput. Vision, WACV: 673-681, 2018
Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA: The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931, 2011
Van Ginneken B, Setio AA, Jacobs C, Ciompi F: Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. IEEE Int. Symp. Biomed. Imaging:286–289, 2015
Han F, Zhang G, Wang H, et al: A texture feature analysis for diagnosis of pulmonary nodules using LIDC-IDRI database. IEEE International Conference on Medical Imaging Physics and Engineering, 2014, 14–18.
Htwe K Z, Yamamori K, Katayama T, Kyi T M: Automated lung nodule classification by artificial neural network and fuzzy inference system. Consumer Electronics, 2016 IEEE, Global Conference on IEEE, 2016 1–2.
Van Ginneken B, Armato SG, de Hoop B, van Amelsvoort-van de Vorst S, Duindam T, Niemeijer M, Murphy K, Schilham A, Retico A, Fantacci ME, Camarlinghi N: Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: the ANODE09 study. Med Image Anal 14(6):707–722, 2011
Fawcett T: An introduction to ROC analysis. Pattern Recognit Lett 27(8):861–874, 2006
Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MMW, Naqibullah M, Sanchez CI, van Ginneken B: Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 35(5):1160–1169, 2016
Jiang H, Ma H, Qian W, Gao M, Li Y: An automatic detection system of lung nodule based on multi-group patch-based deep learning network. IEEE J Biomed Health Inform (99):1–1, 2017
Khosravan N, Bagci U: S4ND: Single-shot single-scale lung nodule detection. Med Image Comput Comput Assist Interv:794–802, 2018
Broyelle A: Automated Pulmonary Nodule Detection on Computed Tomography Images with 3D Deep Convolutional Neural Network, School of Computer Science and Communication KTHRoyal Institute of Technology, Stockholm 2018.
Dou Q, Chen H, Jin Y, Lin H, Qin J, Heng PA: Automated pulmonary nodule detection via 3d convnets with online sample filtering and hybrid-loss residual learning. Med Image Comput Comput Assist Interv:630–638, 2017
Acknowledgments
This work is supported in part by National Natural Science Foundation of China (61772331). We would also like to thank the Shanghai Chest Hospital and department of micro/nanoelectronics at Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(HDF5 50,485 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Shen, F., Shen, L. et al. Lung Nodule Detection in CT Images Using a Raw Patch-Based Convolutional Neural Network. J Digit Imaging 32, 971–979 (2019). https://doi.org/10.1007/s10278-019-00221-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-019-00221-3