Abstract
Major innovations in deep neural networks have helped optimize the functionality of tasks such as detection, classification, segmentation, etc., in medical imaging. Although Computer-Aided Diagnosis (CAD) systems created using classic deep architectures have significantly improved performance, the pipeline operation remains unclear. In this work, in comparison to the state-of-the-art deep learning architectures, we developed a novel pipeline for performing lung nodule detection and classification, resulting in fewer parameters, better analysis, and improved performance. Histogram equalization, an image enhancement technique, is used as an initial preprocessing step to improve the contrast of the lung CT scans. A novel Elagha initialization-based Fuzzy C-Means clustering (EFCM) is introduced in this work to perform nodule segmentation from the preprocessed CT scan. Following this, Convolutional Neural Network (CNN) is used for feature extraction to perform nodule classification instead of customary classification. Another set of features considered in this work is Bag-of-Visual-Words (BoVW). These features are encoded representations of the detected nodule images. This work also examines a blend of intermediate features extracted from CNN and BoVW characteristics, which resulted in higher performance than individual feature representation. A Support Vector Machine (SVM) is used to distinguish detected nodules into benign and malignant nodules. Achieved results clearly show improvement in the nodule detection and classification task performance compared to the state-of-the-art architectures. The model is evaluated on the popular publicly available LUNA16 dataset and verified by an expert pulmonologist.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Lung cancer
- Nodule classification
- Nodule detection
- Fuzzy C-Means clustering
- Medical imaging
- Bag-of-Visual-Words
1 Introduction
One of the most lethal cancers amongst all the cancer is lung cancer. Considering the statistics throughout the world, it nearly affects up to 12.9% of the overall population [1]. The investigation of lung cancer is predominantly conducted using medical imaging techniques like Chest radiographs (also popularly known as X-rays) or Computed Tomography (CT) scanning. Lung cancer screening is a time-consuming skill that requires expert radiologists. Not to mention, it also consists of intra-observer and inter-observer variability among the decisions of different radiologists in identifying nodules or tumors, which makes the task even more tedious [2, 3]. Lung cancer is one of the highest mortality and morbidity rate, with no visible tumor presence symptoms until the patient has reached advanced stages. Therefore, early detection of cancer is one of the critical problems to be addressed. Computer-Aided Diagnosis (CAD) systems are developed to provide a second opinion to radiologists to overcome these issues. Automating this process will reduce the hassle for the doctors and radiologists and the quick and accurate diagnosis.
CAD systems work in two phases, and the initial phase involves the detection of pulmonary nodules in the CT scans. This is time-consuming with a high mistake rate since other anatomical structures are morphologically similar, leading to misunderstanding as a nodule. Therefore, this task requires careful examination and experienced radiologists to assign a candidate in the CT scan as a nodule. This task is named lung nodule detection. The final phase involves classifying the detected nodules into non-cancerous and cancerous. Not all nodules present in the thoracic region are cancerous nodules. The classification of the nodules is mainly based on the nodule size and malignancy type. Specific guidelines are provided by the Lung CT Reporting and Data Systems (Lung-RADS) that need to be followed in the follow-up strategy to categorize the lung nodules [3].
Related Works. In recent literature, Deep Convolutional Neural Networks (DCNN) are used to detect, segmentation, and classify lung nodules. Shen et al. [4] proposed a deep learning model based on crop operation, avoiding the typical segmentation of nodules. The multi-crop CNN model (MC-CNN) is used to identify the malignancy rate in the lung nodules. The model resulted in robust performance even after the exclusion of the segmentation step for lung malignancy analysis. The literature mentioned above used two-dimensional CT scans to perform any lung cancer diagnosis task. However, one crucial aspect overseen in two-dimensional CT scans is volumetric information extracted from three-dimensional CT scans. Few CAD systems have used three-dimensional CT scans to extract different attributes such as nodule shape, texture, type, etc., using volumetric information. This improved the CAD system’s performance and resulted in a more accurate diagnosis [3, 5, 6].
Our Contribution. In this work, a novel segmentation method named Elagha initialization based Fuzzy C-Means clustering (EFCM) is proposed to perform segmentation of nodule regions from a given CT scan. Followed by nodule detection, a hybrid blend of features extracted from segmented images is used to classify lung nodules. The deep architectures are being used for various medical imaging tasks such as detecting nodules, segmenting the nodule regions, and classifying them into cancerous and non-cancerous. In our work, deep architecture is used as a feature extraction technique. The features from different intermediate layers from a deep CNN are validated for the lung nodule classification’s best performance. Another set of features is also used to achieve better discriminative information in the nodule structure, Bag-of-Visual-Words (BoVW) features. These features build a visual dictionary for various input data, and using those dictionary values, the features of unseen test images are extracted. The combination of BoVW and CNN features outperformed the results of the individual feature representations and state-of-the-art nodule classification systems. The results achieved are verified by an expert pulmonologist.
2 Materials and Methods
2.1 Materials
The dataset utilized for the evaluation of the proposed method is Lung Nodule Analysis (LUNA-2016). This dataset is a curated version of the publicly available lung cancer CT dataset LIDC-IDRI. The number of CT scans in the dataset is 888, which includes 5,51,065 nodule candidates. There are a total of 1186 positive nodules in the dataset [7]. The LUNA16 dataset does not contain the malignancy rate of the nodules. However, the dataset includes the scans taken from the LIDC-IDRI dataset [8]. Each radiologist’s malignancy score is provided in a Comma Separated Value (CSV) file for all the nodule scans. Based on all four radiologists’ average malignancy score, the nodule is assigned as a benign nodule or a malignant nodule.
2.2 Methods
Architecture. Figure 1 demonstrates the architecture of the proposed method. The CT scans consist of candidates that need to be identified as nodules and non-nodules, which is performed using the EFCM method. Once the nodules are categorized, it needs to be further classified into benign and malignant nodules. The nodule can be assigned a malignant label based on the malignancy score provided by expert radiologists. The average score of all the radiologists is calculated, and the nodule is assigned with the corresponding label. Once we get the two classes, the classification is performed using two sets of feature representations, BoVW, deep features, and the combination of both features. The classifier used is the SVM for the final decision.
Image Enhancement. Enhancing the images draws more attention towards certain characteristics of an image, making the images more precise, sharp, and detailed. This, in turn, can be used for better analysis and information extraction from the images. In this work, the Histogram Equalization (HE) technique is used where the contrast is altered by adjusting the intensity of the image, which provides an enhanced CT scan image. The comparative frequency of occurrence of different gray levels in the image is represented in the histogram [9].
The histogram \(h(r_k)\) of an image consists of a L total intensity values \(r_k\) within a range of [0–255] (refer Eq. 1).
where, \(n_k\) is the number of pixels with an intensity value of \(r_k\) in the image.
The histogram can be obtained by plotting the \(p(r_k)\) which is shown in below Eq. 2:
The HE of an image is a transformation function i.e., Cumulative Distribution Function (CDF) is given in below Eqs. 3 and 4:
Image Segmentation. The proposed work uses a novel clustering approach named Elagha initialization based Fuzzy C-Means clustering (EFCM) to segment the nodule region from the given input CT scan. At first, FCM partitions the image into several clusters, and then the cluster centroids are selected randomly to compute the Euclidean distance. This random selection of initial centroids may lead to the local optimum solution. Thus, to overcome this drawback, Elagha initialization is used for the initialization of centroids. It generates the initial centroids based on the overall shape of the data. This modification in traditional FCM is termed EFCM.
EFCM method divides the input image (X) into M clusters such that \(x_j = x_1, x_2, ..., x_m\). Then, Elagha initialization calculates the initial cluster centroids by identifying the boundaries of data points and divides them into F rows and F columns to calculate the initial centroids. The width \(w_j\) and height \(h_j\) of the grid cell is computed as shown in Eqs. 5 and 6:
where, \(w_{j,max}\) and \(w_{j,min}\) represents the maximum and minimum widths, \(h_{j,max}\) and \(h_{j,min}\) signifies the maximum and minimum heights respectively. The N number of initial cluster centroids (\(c_i\)) is given by Eq. 7,
After initialization of centroids, the membership function calculation of each pixel is done using Eq. 8:
where, q indicates the power exponent, \(d_{ij}\) is the Euclidean distance between samples \(x_j\) and cluster centroid \(c_i\) and is given by, Eq. 9:
The objective function \(\xi \) used for the initialization of FCM algorithm is given by Eq. 10:
The clusters are formed for nodule and non-nodule regions into separate groups based on the Euclidean distance. The output of the EFCM algorithm is a segmented image consisting of lung nodule regions.
Bag-of-Visual Words (BoVW). Learning feature representations from images using the BoVW method is a two-tiered process. The information from the segmented images is extracted from a pre-generated codebook or dictionary consisting of low-level local features, also known as visual words. The image descriptors used in this work are SIFT features. A visual dictionary is represented using a histogram named “Bag of Visual Words (BoVW),” which is used as a mid-level feature representation [10]. The words in the image mean information in a patch of an image. The patch size must be larger than a few pixels to retrieve more and better information, as it should consider key parts like corners or edges.
SIFT operation is based on the local edge histogram technique. The SIFT technique is one of the popular methods that work very effectively for the BoVW method. Densely sampled SIFT features are extracted from the images. K-means algorithm is used to get cluster centers to generate a visual codebook or dictionary on these features. A histogram is built to the nearest code in the codebook based on the number of occurrences of a feature in each image. The image is then divided into sub-regions of size \(2 \times 2\), and histograms are built for each sub-region. Once all the histograms are generated, all the sub-region histograms are concatenated to form a single feature vector.
Deep Features. In medical imaging, deep architectures are mostly used for final decision-making. However, in this work, deep architecture is used as a feature representation. The deep learning models are well-known for learning hierarchical information from the input images. The higher the layers, the more information the network learns. This novel set of features are used for classifying cancerous and non-cancerous nodules. Images of both categories are trained separately using a deep CNN architecture, and intermediate features of both the classes are extracted. The deep features learn better representations as the network gets deeper.
Nodule Classification. Classification of detected nodules into cancerous and non-cancerous is performed using an SVM classifier. The model is trained using BoVW, deep features, and the combination of these features. The kernel used for SVM is linear. The hyperparameters set for the SVM model are cross-validation parameter set to 5 and the cost parameter set to 0. The model is tested using probability estimates generated from the trained model for the classification.
3 Results and Discussion
3.1 Nodule Segmentation
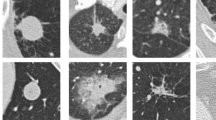
A novel segmentation EFCM approach is proposed in this study to get the region of interest from the input CT image, which is the nodule region. The results obtained from the proposed method are illustrated in Fig. 2. The figure depicts five input CT scans, along with their ground truths and predicted masks. The visual representation of the results clearly shows that the proposed model significantly predicts the mask of the nodule. This yields an exact nodule region extraction from the input CT scan eliminating the additional background noise in the image. This segmentation of the nodule helps in getting better nodule information.
Figure 3 shows the dominance of the proposed EFCM segmentation method. The proposed model obtains the Dice Score Coefficient (DSC) of 97.10%, whereas existing methods obtain lower values, such as U-Net of 80.36%, V-Net (92.86%), Fully Connected Network (FCN) U-Net (91.20%), and Mask Region-based CNN (Mask RCNN) (71.16%). Also, the Intersection-over-Union (IoU) of the proposed technique is 91.96%, but the existing methods show lesser values. Likewise, the Sensitivity (SEN) value of 95.35% makes the proposed model preferable to the current techniques. On the other hand, the proposed system attained the Positive Predictive Value (PPV) of 96.30%, which is higher than the existing methods. This higher DSC, IoU, SEN, and PPV show the improved performance of the proposed EFCM segmentation model.
The evaluation of the nodule detection system for the LUNA16 dataset is performed using a primary performance metric named False Positives per scan (FPs/scan). Figure 4 illustrates the Free-Response Receiver Operating Characteristic (FROC) curve achieved for the proposed EFCM method. The graph depicts that the proposed method resulted in low FPs/scans, proving it is a better performing system. The FPs/scan result for the proposed EFCM model is 2.7 FPs/Scan with a sensitivity of 95.35%.
The proposed EFCM segmentation method is compared with the existing lung nodule detection systems in Table 1. The methods considered for comparison are mostly deep learning architectures such as U-Net [15], dual branch residual network [17], convolution neural network [21], deep Fully Convolution Networks (FCN) [19], receptive field-regularized (RFR) V-Net [22], and so on. In recent trends, deep learning architectures have taken over image segmentation techniques. However, in the proposed method, a clustering approach for segmentation attained better results in both DSC and IoU.
3.2 Nodule Classification
The performance of the lung nodule classification system is evaluated on the publicly available LUNA16 dataset. The accuracy obtained for the lung nodule classification task is 96.87%. The performance metrics considered for the evaluation of the proposed method are accuracy, error rate, specificity, sensitivity, FPR, and F-score. The results are presented in Table 2.
A layer-wise feature comparison is performed for deep and BoVW + deep features. Figure 5 illustrates the accuracy values obtained for different intermediate layers for deep features. The number of layers considered is from 1 to 10. It can be noticed that the performance of the system increases with the increase in layers. After 7\(^{th}\) layer, there is a degradation of accuracy observed in the figure. This is due to overfitting the model for more layers as complexity increases, and less data is available to train the deep architecture. Therefore, the number of layers must be monitored to prevent the model from overfitting.
The proposed method is compared with state-of-the-art lung nodule classification systems. The systems previously proposed for performing lung nodule classification system utilizes deep learning architectures such as Artificial Neural Network (ANN), CNN, multi-scale CNN, Stacked Auto Encoder (SAE), etc. The results are presented in Table 3. It can be noted from the table that the proposed method achieved better performance as compared to the state-of-the-art lung nodule classification systems. Another major issue in training these networks is it is computationally expensive as it requires a lot of time to train a deep model. However, in our method, we used CNN as a feature extractor rather than a classifier. It does not require much time to extract intermediate features. It is also computationally less expensive as the classifier used does not require much time to learn BoVW and deep features. Because BoVW features are encoded, representations do not take up more time for calculation.
Figure 6 illustrates the quantitative analysis of the three feature representations using the Receiver-Operating Characteristics (ROC) curve. The ideal system provides an Area Under Curve (AUC) of 1. The AUC values attained for BoVW, deep, and BoVW + Deep features are 0.83, 0.88, and 0.92, respectively. The classification performance analysis is presented for the SVM classifier. It can be noted from the Figure that BoVW + Deep features resulted in the highest AUC. The feature combination worked effectively to improve the performance of the system.
4 Conclusion
Lung cancer is considered to be one of the deadliest diseases. In this work, a novel clustering-based segmentation method named EFCM is proposed to extract lung nodules from the given CT scan. A hybrid of two different types of feature representations for lung nodule classification is proposed in the work. The method glorifies that deep learning can be used as a classifier and as a suitable feature extractor. The segmentation method introduced in this work performs better than existing segmentation methods in terms of DSC, IoU, and PPV. The proposed method acknowledges that a combination of certain feature representations can enhance the system’s performance in terms of various evaluation metrics such as accuracy, sensitivity, etc. It also reduces the computational cost of the system by reducing the system’s learning parameters. The proposed system effectively combines the encoded feature representation method BoVW and deep features extracted from intermediate layers of a CNN. The performances obtained in the medical imaging tasks are rather critical and also require quicker output. The proposed method provided better and faster results than other CAD systems proposed for the lung nodule classification tasks. In future work, other sets of feature representations and also deeper networks with larger datasets can be explored for performing lung nodule classification.
References
Ferlay, J., et al.: Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136(5), E359–E386 (2015)
Shen, S., Han, S.X., Aberle, D.R., Bui, A.A.T., Hsu, W.: An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification. arXiv:1806.00712 (2018)
Abid, M.M.N., Zia, T., Ghafoor, M., Windridge, D.: Multi-view convolutional recurrent neural networks for lung cancer nodule identification. Neurocomputing 453, 299–311 (2021)
Shen, W., et al.: Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn. 61, 663–673 (2017)
Kang, G., Liu, K., Hou, B., Zhang, N.: 3D multi-view convolutional neural networks for lung nodule classification. PLOS One 12(11), 1–21 (2017)
Dou, Q., Chen, H., Yu, L., Qin, J., Heng, P.: Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans. Biomed. Eng. 64(7), 1558–1567 (2017)
Setio, A.A.A., et al.: Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med. Image Anal. 42, 1–13 (2017)
Armato, S., III., et al.: The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 38, 915–931 (2011)
Salem, N., Malik, H., Shams, A.: Medical image enhancement based on histogram algorithms. Procedia Comput. Sci. 163, 300–311 (2019)
Sundarambal, B., Subramanian, S., Muthukumar, B.: A hybrid encoding strategy for classification of medical imaging modalities. J. Ambient Intell. Humaniz. Comput. 12(6), 5853–5863 (2020). https://doi.org/10.1007/s12652-020-02129-1
da Silva, G.L.F., de Carvalho Filho, A.O., Silva, A.C., de Paiva, A.C., Gattass, M.: Taxonomic indexes for differentiating malignancy of lung nodules on CT images. Res. Biomed. Eng. 32(3), 263–272 (2016)
Song, Q., Zhao, L., Luo, X., Dou, X.: Using deep learning for classification of lung nodules on computed tomography images. J. Healthc. Eng. 2017, 1–7 (2017)
Shen, W., Zhou, M., Yang, F., Yang, C., Tian, J.: Multi-scale convolutional neural networks for lung nodule classification. In: Ourselin, S., Alexander, D.C., Westin, C.-F., Cardoso, M.J. (eds.) IPMI 2015. LNCS, vol. 9123, pp. 588–599. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-19992-4_46
Gupta, A., Das, S., Khurana, T., Suri, K.: Prediction of lung cancer from low-resolution nodules in CT-scan images by using deep features. In: International Conference on Advances in Computing, Communications and Informatics (ICACCI), pp. 531–537 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. arXiv:1505.04597, pp. 234–241 (2015)
Wu, B., Zhou, Z., Wang, J., Wang, Y.: Joint learning for pulmonary nodule segmentation, attributes and malignancy prediction. arXiv:1802.03584, pp. 1109–1113 (2018)
Cao, H., et al.: Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 86, 105934 (2020)
Aresta, G., et al.: iW-Net: an automatic and minimalistic interactive lung nodule segmentation deep network. Sci. Rep. 9(1), 1–9 (2019)
Roy, R., Chakraborti, T., Chowdhury, A.S.: A deep learning-shape driven level set synergism for pulmonary nodule segmentation. Pattern Recogn. Lett. 123, 31–38 (2019)
Messay, T., Hardie, R.C., Tuinstra, T.R.: Segmentation of pulmonary nodules in computed tomography using a regression neural network approach and its application to the lung image database consortium and image database resource initiative dataset. Med. Image Anal. 22, 48–62 (2015)
Wang, S., et al.: Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med. Image Anal. 40, 172–183 (2017)
Dodia, S., Basava, A., Mahesh, P.A.: A novel receptive field-regularized V-net and nodule classification network for lung nodule detection. Int. J. Imaging Syst. Technol. 32, 88–101 (2021)
Shaukat, F., Raja, G., Ashraf, R., Khalid, S., Ahmad, M., Ali, A.: Artificial neural network based classification of lung nodules in CT images using intensity, shape and texture features. J. Ambient Intell. Humaniz. Comput. 10, 4135–4149 (2019)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Dodia, S., Annappa, B., Padukudru, M.A. (2022). A Novel Artificial Intelligence-Based Lung Nodule Segmentation and Classification System on CT Scans. In: Raman, B., Murala, S., Chowdhury, A., Dhall, A., Goyal, P. (eds) Computer Vision and Image Processing. CVIP 2021. Communications in Computer and Information Science, vol 1568. Springer, Cham. https://doi.org/10.1007/978-3-031-11349-9_48
Download citation
DOI: https://doi.org/10.1007/978-3-031-11349-9_48
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-11348-2
Online ISBN: 978-3-031-11349-9
eBook Packages: Computer ScienceComputer Science (R0)