Abstract
Purpose
Lung cancer detection at its initial stages increases the survival chances of patients. Automatic detection of lung nodules facilitates radiologists during the diagnosis. However, there is a challenge of false positives in automated systems which may lead to wrong findings. Precise segmentation facilitates to accurately extract nodules from lung CT images in order to improve performance of the diagnostic method.
Methods
A multistage segmentation model is presented in this study. The lung region is extracted by applying corner-seeded region growing combined with differential evolution-based optimal thresholding. In addition to this, morphological operations are applied in boundary smoothing, hole filling and juxtavascular nodule extraction. Geometric properties along with 3D edge information are applied to extract nodule candidates. Geometric texture features descriptor (GTFD) followed by support vector machine-based ensemble classification is employed to distinguish actual nodules from the candidate set.
Results
A publicly available dataset, namely lung image database consortium and image database resource initiative, is used to evaluate performance of the proposed method. The classification is performed over GTFD feature vector and the results show 99% accuracy, 98.6% sensitivity and 98.2% specificity with 3.4 false positives per scan (FPs/scan).
Conclusion
A lung nodule detection method is presented to facilitate radiologists in accurately diagnosing cancer from CT images. Results indicate that the proposed method has not only reduced FPs/scan but also significantly improved sensitivity as compared to related studies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lung cancer is considered as a second most probable cancer in human beings. Moreover, the graph of survival rate is curved toward the very low end. Due to this fatal disease, around 1.69 million deaths have been reported worldwide during the year 2015 along with several new cases [1]. The human cells grow in a well-controlled manner. The uncontrolled growth of these cells leads to cancer. In lung cancer, these abnormal cells grow quickly to form a nodule [2]. As the nodule size increases, lungs functionality is severely disturbed and leads to death within a short time [3]. Therefore, it has become a compulsion to identify lung nodules at earlier stages. Medical imaging helps in early diagnosis of lung cancer. The most popular one is computed tomography (CT) [4]. During a CT scan, more than 100 images of a patient are generated. Radiologists have to examine all these images during the diagnosis. Handling such large number of images is error-prone and time-consuming. Computer-based systems help the radiologists in diagnosis process by providing the second opinion [5]. However, there is a significant challenge of sensitivity and number of FPs/scan. One of the major steps of a diagnosis system is segmentation. Accurate segmentation of lung region and pulmonary nodules is critical that helps the radiologists in decision making [6]. This paper presents a multistep segmentation method to improve the diagnosis procedure. In addition to segmentation, the selection of relevant features and a robust classifier helps to improve accuracy and reduce false positives. Considering aforementioned, GTFD is created for better representation of nodule candidates. Finally, SVM-ensemble classifier is applied, which is based on three base classifiers, namely SVM-linear, SVM-polynomial and SVM-RBF (radial-based function). The proposed method is evaluated over a widely used publicly available standard dataset: LIDC–IDRI. The results show improved performance of the proposed method in terms of accuracy, sensitivity and specificity with low FPs/scan as compared to earlier methods.
Related work
Automated diagnostic systems for lung cancer have remained a key area of research in the recent years. Several such systems have been developed based on image processing and machine learning techniques [7]. However, there is a primary issue of accuracy and false positive rate (FPR) in these diagnosis systems. Lung nodule detection methods are mainly comprised of segmentation, feature extraction and classification [8]. The segmentation is further divided into lung region extraction and nodule candidate detection. Several segmentation methods have been proposed including thresholding [9], template matching [10], 3D template matching [11] to name a few. Thresholding is a simple and popular technique used for lung volume segmentation and nodule candidate detection. Different variations of thresholding techniques are reported in literature including simple thresholding [9], optimal thresholding [12], Otsu thresholding [13], thresholding combined with morphological operations [14], fuzzy thresholding [15] and multiple thresholds [16,17,18,19]. Some researchers also used combinations of different segmentation methods to improve the segmentation quality. ..Tan et al. [20] proposed a segmentation method by combining watershed, active contours and Markov random field. ..Shen et al. [21] used bidirectional chain code for nodule segmentation and then SVM for classification. The use of classifier is also a very significant step in the reduction in false positives. It distinguishes actual nodules from the set of nodule candidates on the basis of features provided for training and testing. Artificial neural networks (ANN) [22, 23] and SVM are popular techniques used for nodule classification. ..Nibali et al. [24] proposed deep residual network-based nodule classification method. Their major focus was on the performance evaluation of deep learning in the field of lung nodule detection. ..de Carvalho Filho et al. [25] applied thresholding combined with a genetic algorithm for nodule segmentation and finally SVM for classification. Taşcı, Uğur [13] presented a segmentation method based on Otsu thresholding and morphological operations. Shape and texture features were used as input to a regression-based classifier. .Teramoto, Fujita [26] proposed the cylindrical nodule enhancement filter to improve FPR. They used thresholding for segmentation of region of interest (ROI) and SVM for nodule classification.
Segmentation of lung nodules is a challenging phase in the detection process due to complex structure of lung. Furthermore, handling of small, boundary attached (juxtapleural) and vessel attached (juxtavascular) nodules is a big challenge. Precise feature extraction and powerful classification algorithms assert their impact on the performance of a diagnosis system. The proposed technique addresses these issues in order to give a better detection method for lung cancer.
Proposed method
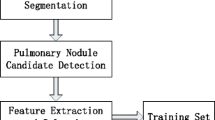
An automated diagnosis method to detect lung nodules on the basis of various image processing and machine learning techniques is presented in this paper. Figure 1 shows working model of the proposed method. Segmentation comes first as a vital step in the presented technique which deals with two major parts, i.e., lung region extraction and nodule candidate detection. These two stages are further divided into substeps. Other two stages are the creation of GTFD and SVM-ensemble-based nodule classification, respectively.
Nodule segmentation
Pulmonary nodule segmentation is an escalating phase in the performance of an automatic diagnostic system. During the diagnosis procedure, precise and accurate segmentation of nodules impacts the performance. Segmentation of smaller nodules, ranging from 3–30 mm, juxtapleural nodules and vascular attached nodules is a significant test for the segmentation method.
Lung region extraction
In the first step, the image background is removed by using corner-seeded region growing combined with thresholding approach. The thresholding is a straightforward and efficient method for image segmentation. It performs segmentation on the basis of pixel intensities. It is observed that simple thresholding is not useful for lung region extraction due to overlapping between the intensities of background and some sections of ROI. To overcome this issue, the background of input images is removed. Furthermore, the threshold generated for one slice is not useful to other slices because of its variation in the gray level for each slice as shown in Fig. 2. Therefore, the suggested segmentation technique used a combination of differential evolution-based optimal thresholding [27] and corner-seeded region growing.
After the removal of image background, differential evolution-based optimal thresholding is used to identify the lung boundary and extraction of lung region. An initial threshold of − 950 HU is applied because most of the lung region lies in the range of − 950 HU to − 500 HU. It is an iterative step in which the threshold is recalculated in each iteration.
In an image, a histogram gives the probability distribution of gray levels. This probability distribution is calculated by Eq. (1).
The total number of categories within the image is represented by K. \(P_i\) and \(p_i(x)\) are the probability and probability distribution functions within the category i, respectively. \(\mu _i\) and \(\sigma _i\) are the mean and standard deviation. To compute optimal threshold, the overall probability error in two different categories is minimized by Eq. (2).
This error is with respect to the threshold \(T_i \). Then the overall error is calculated as given by Eq. (3).
Henceforth, a threshold image is generated that contains the lung mask. To refine the lung boundary and extraction of juxtapleural nodules, morphological operations are applied. Morphological closing is performed on the binary mask, where a disk of size 25 is taken as the structuring element. The process of closing also includes some extra regions which can affect the accuracy of segmentation. To solve this problem, morphological opening with the same structuring element, a disk of size 25, is applied. Figure 3 highlights the process of juxtapleural nodule extraction. Finally, the lung region is extracted by using the resultant mask and input CT image. The whole procedure is repeated for all the slices of each scan. Figure 4 shows the step-by-step results of segmentation.
Nodule candidate detection
After ROI extraction, the next step is to extract nodule candidates by eliminating vessels and other unwanted objects. For this purpose, a combination of morphological operations, edge detection, bounding box and shape information are applied to the lung ROI for eliminating vessels and noise objects. The major issue in nodule candidate detection is juxtavascular nodules due to their attachment to the vessels [28]. In most of the previously known techniques, these nodules are eliminated with the removal of vessels. To avoid nodule elimination, we use 3D connectivity information. Since nodule is a 3D object, therefore, it must show its existence in consecutive slices of a scan. Considering this property of nodule, the presence of each object with its predecessor and successor slices is investigated. Let p(i, j, k) is a candidate for a nodule pixel, where k is the slice number and i, j represent the location of pixel within that slice. Then 3D neighborhood is checked by comparing the intensity of that pixel. Each pixel p(i, j) of slice k has nine neighborhoods in the \(k-1\) slice and nine in the \(k+1\) slice as shown in Fig. 5. The similar intensity pixels must be there within the eighteen neighborhood pixels, either in predecessor or in successor slices or in both.
The morphological opening is used to separate juxtavascular nodules from vessels. The structuring element used for this purpose is a ‘sphere’ of size 2 which is effective in 3D lung structure. The nodule is a round-shaped object and the use of a ‘sphere’ as structuring element preserves the nodules and breaks the connection of nodules and vessels. Afterward, 3D connected component analysis is performed and regions with centers within 5 mm are merged. Canny edge detection method is used to identify the boundaries of each object. It results in a large number of candidate objects. To refine this set of objects, the nodule size and shape information are taken into account. The nodule is round-shaped object ranges from 3 to 30 mm in size within ROI. Considering these properties, the nodule candidate set is refined on the basis of diameter, elongation and circularity. The objects with diameter less than 3 mm and greater than 30 mm are removed and considered to be unwanted objects as their sizes do not match with the size specifications of nodules. In addition to diameter, those objects are excluded which are too elongated i.e., their major axis length is 2.25 times more than the minor axis length. The third parameter is circularity, which defines roundness of the object. The roundness of an object ranges from 0 to 1, where 1 represents perfect round. All those objects which have circularity less than 0.65 are also excluded from the candidate set. The elongation and circularity threshold values are empirically selected based on the observation that if circularity value is increased more than 0.65 and elongation value is decreased less than 2.25, this causes the removal of some of the actual nodules. On these parameters, nodule candidate set is refined and nodules are distinguished from other objects like vessels and noise objects as presented in Fig. 6. Some of the extracted nodule candidates are illustrated in Fig. 7.
Geometric texture feature descriptor (GTFD)
In object detection and classification, precise and relevant features play a significant role. In recent studies, a variety of features including geometric, texture, local binary patterns (LBP), wavelet, intensity and gradient are used for the detection and classification of nodules because a single type of feature vector cannot give an accurate representation of objects resulting in unsatisfactory performance. To overcome these issues, researchers also used hybrid feature vectors by encapsulating more types of features in a single feature vector. In this study, a GTFD by combining texture and shape features in 2D and 3D is devised that successfully delineates the nodules and non-nodules.
Geometric features
Geometric features like circularity, area and volume are the shape descriptors of an object [29]. Therefore, these features are helpful to distinguish nodules from other objects within the lung region. The median slice of a scan contains maximum information of objects present in the scan. Therefore, 2D geometric features are extracted from the median slices. However, 3D geometric features are extracted from 3D segmented objects. The features include area, radius, circularity, volume, compactness and elongation. Equations (4)–(9) give the mathematical representation of these features.
where p is pixel within the segmented object O of slice S of a 3D scan, D represents diameter of the nodule candidate and V accounts for the voxels of a candidate object.
Texture features
Texture features characterize the regions of images on the basis of their pixel intensities [30]. Ten texture features, four 2D and six 3D features, are used in this study. The 2D texture features are given by Eqs. (10)–(13) and 3D texture features are specified from Eqs. (14)–(18).
where p(i) is ith pixel in a 2D slice, p(i, j) is ith voxel in 3D volume at slice j and I represents maximum intensity within the slice.
Ensemble classification
SVM-ensemble classification is used to identify the actual nodules from a set of candidate nodules. An ensemble classifier makes final decision on the test dataset derived from the output of individually trained base classifiers [31]. Let there are n base classifiers as: \(\{ {C_1 ,C_2 ,C_3 \ldots C_n}\}\) over a dataset X and target T. All classifiers are trained and then evaluated over the same test examples. The decisions of individual classifiers are then combined to make a final decision. Majority voting is a useful combiner in which decision about the class of a test example is made through the number of votes, determined by Eq. (19).
where \(f_e(x)\) assigns class labels to the test feature vector x on the basis of maximum votes given to a class and n represents the total number of base classifiers. Let if there are three base classifiers, two predict that the test example is of class A and one predicts it in class B, then majority voting predicts it as class A. In this study, SVM is used as a base classifier with three different kernels including linear, polynomial and Gaussian RBF. These kernels are represented mathematically in Eqs. (20)–(22).
where \(x_i \) and \(x_j \) are vectors in input space, d represents the degree of polynomial, \(\Vert x_i -x_j \Vert ^{2}\) is the Euclidean distance and \(\upsigma \) is standard deviation, defined as Gaussian distribution. SVM is known to be a good classifier that learns easily and produces good classification results. However, a single classifier may not learn all the parameters to produce good global optimum result. To meet this limitation, an SVM-based ensemble classifier is developed for nodule classification. Figure 8 shows the workflow of SVM-ensemble classifier for the proposed method to distinguish between nodules and non-nodules. During the training phase, each individual SVM-based classifier is trained over the labeled data provided for training. SVM-linear, SVM-polynomial and SVM-RBF are trained as base classifiers. The individual classifiers are aggregated on the basis of voting. In the testing phase, an unlabeled test example is supplied as input to all the individual classifiers simultaneously and final decision is made on the basis of Eq. (19).
Results and discussions
In this study, an authentic publicly available dataset LIDC–IDRI [32] is used to evaluate the performance of proposed method. The tube current range, from 40 to 627 mAs, is used for image acquisition. The tube potential energies have a range from 120 to 140 kV. The low-dose scans are more effective to identify small size lung nodules [33]. The dataset contains 1018 CT scans of 1010 patients collected from 7 academic centers. Each scan contains approximately 100 to 400 slices with slice thickness between 0.6 to 5 mm and dimension of \(512\times 512\) pixels. Each image has a 12-bit grayscale resolution with the pixel size ranging from 0.5 mm to 0.8 mm. Out of 1018 cases, 121 cases have slice thickness \(\ge \) 3 mm which is not suitable for the computer-based diagnosis [34]. In addition to this, there are nine cases which have inconsistent slice spacing. By excluding these cases, a total of 888 scans are left for the automated diagnosis. Every scan of dataset is reviewed by four experienced radiologists for nodule annotation. The annotation information for each scan is provided in XML format. All the radiologists have a consensus on 777 nodules. In this study, those nodules are considered for experimentations which are marked by all the four radiologists. Around 45% scans of dataset contain no nodules which are annotated by all the four radiologists and some scans have multiple nodules. Considering these points, those scans are excluded which have zero nodules and those scans are selected which contain number of nodules in the range of 1–8. As a result, 250 scans are finally selected with 567 nodules. Candidate nodules are classified according to annotations provided by the expert radiologists. If a nodule candidate meets the criteria, it is considered as hit and classified as a nodule. On the other hand, if it does not meet the criteria, it is taken as miss and considered as a non-nodule. A candidate is considered hit if its radius is 0.8 to 1.5 times of the actual nodule or its distance with reference to any corresponding annotated nodule is less than 1.5 times the radius of that nodule .[12]. The performance of classifiers is checked over different distributions of data into training and testing ratio. Following evaluation parameters, as mentioned from Equations (23) to (26), are used to analyze the performance.
MATLAB 2016a is used as an implementation tool. Three different distributions of data are used as 80–20, 70–30 and 50–50% for training and testing purpose, respectively. The hold-out validation is used for training and testing the classifiers. The process is repeated 10 times to avoid the biasness and average score is computed. Various experiments are executed with different combinations of feature sets. The results are shown in Tables 1, 2 and 3 with geometric, texture and GTFD features, respectively. SVM is trained with three different kernels, namely SVM-linear, SVM-polynomial and SVM-RBF. The output of individual classifiers over the test examples is evaluated. Majority voting is applied to make a final decision by SVM-ensemble. From Tables 1, 2 and 3, it is clear that SVM-ensemble has produced much better results as compared to individual classifiers.
Performance of the proposed method is evaluated with the individual as well as combined feature sets. The results show that combined feature vectors produced a better performance as compared to single feature type. The performance with texture features is lower than the geometric features. However, when GTFD is used by encapsulating both geometric and texture features, much better results are obtained as compared to single type of feature vector. Furthermore, best results are shown when experimentation is done over the combination of 70–30 i.e., 70% training and 30% testing. Table 3 depicts that GTFD presented better results than individual features. In comparison with Tables 1 and 2, it gives a clear picture that geometric features are better descriptors of nodules as compared to texture features. However, on an individual basis, they cannot produce best results. Figure 9 illustrates a receiver operating characteristic (ROC) curve for the results of classification. It is evident that ensemble-based SVM shows better results as compared to RBF, polynomial and linear kernels.
The nodule candidate detection process comprises of two stages. In the first stage, 66021 nodule candidates are detected with 261.8 FPs/scan. At this step, all 567 actual nodules are also recognized with 100% sensitivity. The second stage is related to candidate refinement on the basis of nodule size and shape information. In this stage, the number of candidates is reduced to 46176 and FPs/Scan is decreased to 182.5 with 45614 total false positives. However, this refinement phase has missed 5 actual nodules and sensitivity is reduced to 99.1%. The classification phase, based on SVM-ensemble, reduces the number of false positives to 838 which is 3.4/scan. The free-response receiver operating characteristic (FROC) curve in Fig. 10a illustrates the number for FPs/scan produced by four classifiers. The SVM with linear kernel delivers 9.6 FPs/scan with 89.1% sensitivity, SVM-polynomial produces 6.4 FPs/scan with 93.6% sensitivity and SVM-RBF shows comparatively good results with 4.2 FPs/scan and sensitivity of 97.7%. However, SVM-ensemble reduces the FPs/scan to 3.4% with the highest sensitivity of 98.6%. In Fig. 10b, another FROC curve is given which illustrates the comparison of classifiers’ sensitivity at 3.4 FPs/scan. It depicts that SVM-ensemble has highest sensitivity i.e., 98.6% and SVM-linear has the lowest. SVM-RBF has better sensitivity than SVM-polynomial at 3.4 FPs/scan. It is evident from FROC curves that minimum FPs/scan with SVM-ensemble is reduced to 1 but at the cost of sensitivity degradation with 92.1%.
For estimating the significance of proposed method in relation to reported techniques in the literature, a comparison of datasets and their results is presented in Tables 4 and 5, respectively.
Ye et al. [15] used clinical data of 108 scans containing 220 nodules of size 3–20 mm with a slice thickness of 0.5–2 mm. Cascio et al. [22] and Choi, Choi [12] used 84 CT scans of LIDC dataset consisting of 148 nodules marked by any of the radiologists. Dehmeshki et al. [10] made use of 70 CT scans of a clinical dataset containing 179 nodules of size 3–20 mm with a slice thickness of 0.5–1.25 mm. Kuruvilla, Gunavathi [23] utilized a clinical dataset of 155 scans with a slice thickness of 0.75–1.25 mm including 110 nodules. In addition, they have also used some scans of LIDC dataset without LIDC annotations. Han et al. [19] used 205 CT scans of LIDC–IDRI dataset containing 490 nodules marked by any of the radiologists. The nodules included in their study were of 3–30 mm size. Teramoto, Fujita [26] made use of 84 CT scans of LIDC dataset with 103 nodules of size 5–20 mm. The nodules included in the work were marked by any of the radiologists. Nibali et al. [24] utilized LIDC–IDRI dataset for experimentations but details are not provided. Tan et al. [9] made use of 125 CT scans of LIDC dataset containing 80 nodules of size 3–30 mm marked by all the four radiologists. Riccardi et al. [14] used 154 CT scans of LIDC dataset consisting of 117 nodules with size 3–30 mm marked by all the four radiologists. Proposed method is evaluated over 250 scans of LIDC–IDRI dataset containing 567 nodules with size 3–30 mm annotated by all the four radiologists. The statistics in Table 4 highlight the fact that the proposed method is tested over a more significant number of scans and nodules as compared to other research works reported in the literature.
Table 5 compares the results of the proposed and existing methods. Kuruvilla, Gunavathi [23] produced 91.4% sensitivity but with a very high FPs/scan of 30. Moreover, they did not provide specificity. Dehmeshkiet.al [10] also produced very high FPs/scan of 14.6 with 90% specificity. These results were taken on clinical datasets instead of some standard dataset. Choi, Choi [12] claimed 97.5% sensitivity as well as specificity with 6.76 FPs/scan. Ye et al. [15] produced 8.2 FPs/scan with 90.2% sensitivity. Cascio et.al [22] claimed 88% sensitivity and 97% accuracy with 6.1 FPs/scan. Teramoto, Fujita [26] achieved a low FPs/scan of 4.2 but at the cost of accuracy which is as low as 80%. Han et al. [19] claimed a low FPs/scan of 4.14 but their sensitivity was low as 89.2%. Nibali et al. [24] showed 89.9, 91.07 and 88.64% accuracy, sensitivity and specificity, respectively. Tan et al. [9] produced 87.5% sensitivity with 4.0 FPs/scan and Riccardi et al. [14] claimed 6.5 FPs/scan with a low sensitivity of 71.0%. By concluding the whole discussion, it is evident that the proposed method has shown better results i.e., 99.0% accuracy, 98.6% sensitivity and 98.2% specificity with a 3.4 FPs/scan which is a clear proof of the effectiveness of the presented method.
Conclusion
A critical issue with the nodule diagnosis methods is handling small, boundary attached and vessel attached nodules. If a detection method misses any nodule, it leads to inaccurate diagnosis which might be life threatening. To handle such problems, a nodule segmentation method from 3D CT scan is presented. The first step of proposed method is based on hybrid multistep segmentation technique to extract lung ROI and nodule candidates. SVM-ensemble is used to distinguish nodules from the set of selected candidates. The classification is based on a GTFD feature vector encapsulating texture and geometric features. The proposed method is tested on a standard publicly available dataset LIDC–IDRI. The presented method achieved 99.0% accuracy with 98.6% sensitivity and 98.2% specificity. The number of FPs/scan is also low as 3.4/scan. These results show the significance of presented method that it will facilitate radiologists during the diagnosis process of lung cancer.
References
Organization WH (2017) Cancer fact sheet. http://www.who.int/mediacentre/factsheets/fs297/en/. Accessed 09 June 2017
Gould MK, Fletcher J, Iannettoni MD, Lynch WR, Midthun DE, Naidich DP, Ost DE (2007) Evaluation of patients with pulmonary nodules: when is it lung cancer? ACCP evidence-based clinical practice guidelines. Chest J 132(3–suppl):108S–130S
Petkovska I, Brown MS, Goldin JG, Kim HJ, McNitt-Gray MF, Abtin FG, Ghurabi RJ, Aberle DR (2007) The effect of lung volume on nodule size on CT. Acad Radiol 14(4):476–485
Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR (2012) Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 307(22):2418–2429
Endo M, Aramaki T, Asakura K, Moriguchi M, Akimaru M, Osawa A, Hisanaga R, Moriya Y, Shimura K, Furukawa H (2012) Content-based image-retrieval system in chest computed tomography for a solitary pulmonary nodule: method and preliminary experiments. Int J Comput Assist Radiol Surg 7(2):331–338
Yim Y, Hong H (2008) Correction of segmented lung boundary for inclusion of pleural nodules and pulmonary vessels in chest CT images. Comput Biol Med 38(8):845–857
Naqi SM, Sharif M (2017) Recent developments in computer aided diagnosis for lung nodule detection from CT images: a review. Curr Med Imaging Rev 13(1):3–19
van Ginneken B, Armato SG III, de Hoop B, van Amelsvoort-van de Vorst S, Duindam T, Niemeijer M, Murphy K, Schilham A, Retico A, Fantacci ME (2010) Comparing and combining algorithms for computer-aided detection of pulmonary nodules in computed tomography scans: the ANODE09 study. Med Image Anal 14(6):707–722
Tan M, Deklerck R, Jansen B, Bister M, Cornelis J (2011) A novel computer-aided lung nodule detection system for CT images. Med Phys 38(10):5630–5645
Dehmeshki J, Ye X, Lin X, Valdivieso M, Amin H (2007) Automated detection of lung nodules in CT images using shape-based genetic algorithm. Comput Med Imaging Graph 31(6):408–417
Ozekes S, Osman O, Ucan ON (2008) Nodule detection in a lung region that’s segmented with using genetic cellular neural networks and 3D template matching with fuzzy rule based thresholding. Korean J Radiol 9(1):1–9
Choi W-J, Choi T-S (2014) Automated pulmonary nodule detection based on three-dimensional shape-based feature descriptor. Comput Methods Programs Biomed 113(1):37–54
Taşcı E, Uğur A (2015) Shape and texture based novel features for automated juxtapleural nodule detection in lung CTs. J Med Syst 39(5):46
Riccardi A, Petkov TS, Ferri G, Masotti M, Campanini R (2011) Computer-aided detection of lung nodules via 3D fast radial transform, scale space representation, and Zernike MIP classification. Med Phys 38(4):1962–1971
Ye X, Lin X, Dehmeshki J, Slabaugh G, Beddoe G (2009) Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE Trans Biomed Eng 56(7):1810–1820
Mukhopadhyay S (2016) A segmentation framework of pulmonary nodules in lung CT images. J Digit Imaging 29(1):86–103
Armato SG III, Giger ML, MacMahon H (2001) Automated detection of lung nodules in CT scans: preliminary results. Med Phys 28(8):1552–1561
Elizabeth DS, Nehemiah HK, Raj C, Kannan A (2012) A novel segmentation approach for improving diagnostic accuracy of CAD systems for detecting lung cancer from chest computed tomography images. J Data Inf Qual 3(2):4
Han H, Li L, Han F, Song B, Moore W, Liang Z (2015) Fast and adaptive detection of pulmonary nodules in thoracic CT images using a hierarchical vector quantization scheme. IEEE J Biomed Health Inform 19(2):648–659
Tan Y, Schwartz LH, Zhao B (2013) Segmentation of lung lesions on CT scans using watershed, active contours, and Markov random field. Med Phys 40(4):4350201–4350210
Shen S, Bui AA, Cong J, Hsu W (2015) An automated lung segmentation approach using bidirectional chain codes to improve nodule detection accuracy. Comput Biol Med 57:139–149
Cascio D, Magro R, Fauci F, Iacomi M, Raso G (2012) Automatic detection of lung nodules in CT datasets based on stable 3D mass-spring models. Comput Biol Med 42(11):1098–1109
Kuruvilla J, Gunavathi K (2014) Lung cancer classification using neural networks for CT images. Comput Methods Programs Biomed 113(1):202–209
Nibali A, He Z, Wollersheim D (2017) Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg 12(10):1799–1808
de Carvalho Filho AO, de Sampaio WB, Silva AC, de Paiva AC, Nunes RA, Gattass M (2014) Automatic detection of solitary lung nodules using quality threshold clustering, genetic algorithm and diversity index. Artif Intell Med 60(3):165–177
Teramoto A, Fujita H (2013) Fast lung nodule detection in chest CT images using cylindrical nodule-enhancement filter. Int J Comput Assist Radiol Surg 8(2):193–205
Cuevas E, Zaldivar D, Pérez-Cisneros M (2010) A novel multi-threshold segmentation approach based on differential evolution optimization. Expert Syst Appl 37(7):5265–5271
Dhara AK, Mukhopadhyay S, Mehre SA, Khandelwal N, Prabhakar N, Garg M, Kalra N (2017) A study of retrieval accuracy of pulmonary nodules based on external attachment. In: SPIE medical imaging. International Society for Optics and Photonics, pp 101343T–101346
Lafarge F, Descombes X (2010) Geometric feature extraction by a multimarked point process. IEEE Trans Pattern Anal Mach Intell 32(9):1597–1609
Haralick RM, Shanmugam K (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 6:610–621
Yang F, Xu Y-Y, Wang S-T, Shen H-B (2014) Image-based classification of protein subcellular location patterns in human reproductive tissue by ensemble learning global and local features. Neurocomputing 131:113–123
Armato SG III, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA (2011) The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931
Moyer VA (2014) Screening for lung cancer: US Preventive Services Task Force recommendation statement. Ann Intern Med 160(5):330–338
Jacobs C, Rikxoort EM, Murphy K, Prokop M, Schaefer-Prokop CM, Ginneken B (2016) Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database. Eur Radiol 26(7):2139–2147
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies performed with human participants or animals by any of the authors.
Informed consent
This article does not contain patient data.
Rights and permissions
About this article
Cite this article
Naqi, S.M., Sharif, M. & Yasmin, M. Multistage segmentation model and SVM-ensemble for precise lung nodule detection. Int J CARS 13, 1083–1095 (2018). https://doi.org/10.1007/s11548-018-1715-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1715-9