Abstract
Using lignin to produce high-value-added aromatic fine chemicals and high-grade biofuels such as aromatics, cycloalkanes, and alkanes can reduce the dependence on fossil resources and highly improve the competitiveness of biorefining industry. The biological valorization of lignin includes the biological depolymerization and bioconversion of lignin. With the development of bioprospecting and systems biology technology, more and more lignin-degrading microorganisms have been discovered and separated from the natural habitat of lignin decomposition. The physiological and biochemical characteristics of microorganisms and the molecular- and systematic-level degradation mechanism on lignin and lignin-derived aromatic compounds have also been deeply recognized. All of these have laid a theoretical foundation for precisely controlling the depolymerization and metabolism of lignin and establishing the biological processing pathway of lignin. This chapter will introduce the research progress of lignin valorization from the aspects of lignin-degrading microorganisms and enzymes, lignin degradation metabolic pathways, and the application of biosynthesis in lignin conversion.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Lignin biodegradation
- Lignin-degrading microbes
- Lignin-degrading enzymes
- Metabolic pathway
- Lignin valorization
- Bioprospecting
- Biorefining industry
- Biosynthesis
- Biofuels
- Biocatalytic conversion
12.1 Introduction
Although more complex in structure, lignin has a higher carbon content and lower oxygen content than either polysaccharides or whole-cellulose raw materials, which makes it an attractive raw material for the production of biofuels and chemicals. More importantly, the highly functional and aromatic nature of lignin offers potentiality for the direct preparation of special and fine aromatic chemicals [103]. As a result, the production of chemicals from lignin has attracted worldwide attention [73].
Researchers around the world devote increasing attention to the valorization of lignin by biological methods. In contrast to thermochemical lignin depolymerization methods which consume large amounts of energy, chemicals, and expensive catalysts, lignin biodepolymerization methods mainly use microorganisms and enzymes to break down the bonds between lignin units or bonds between lignin and carbohydrate [115]. Generally speaking, the bioprospecting of lignin-degrading microbes and enzymatic systems and the understanding of molecular and systematic degradation mechanisms and metabolic pathways of lignin and aromatics by means of systems biology analyses are two major focuses of the lignin value-added utilization [74]. At present, a large number of bacteria and fungi have been screened from environment, which contain various enzymes and have the characteristics of lignin degradation and transformation. It is very important for the cognition and accurate control of lignin biodegradation and the establishment of biological processing for lignin valorization.
12.2 Lignin-Degradable Microorganisms
12.2.1 Fungi
The degradation of lignocellulose by fungi , including white-rot fungi, brown-rot fungi, and soft-rot fungi, mainly attributes to the extracellular enzymes, including polysaccharide hydrolases and lignin-degrading enzymes [81]. Fungi can degrade lignocellulose rapidly, which makes it widely used in the fields of crop straw decay, wastewater treatment, bioconversion of lignocellulose, and so on [110]. Lignin-degrading fungi can be divided into three categories, white-rot fungi, brown-rot fungi, and soft-rot fungi. The lignin degradability is different significantly. White-rot fungi mainly degrade lignin and polysaccharides. Brown-rot fungi and soft-rot fungi mainly degrade cellulose, hemicellulose, and some polysaccharides. However, brown-rot fungi hardly degrade lignin; soft-rot fungi just degrade lignin slightly and slowly. Therefore, the degradation of lignin mainly relays on white-rot fungi.
White-rot fungi are the general name for a group of filamentous fungi that make wood white and rotten. It is the only microorganism in nature that can completely mineralize lignin. There are many white-rot fungi that can degrade lignin, mainly Basidiomycota and some Ascomycetes. The most studied white-rot fungi include Phanerochete chrysosporium, Coriolus versicolor, Trametes versicolor, Phlebia radiata, Panus conchatus, Pleurotus pulmonarius, Pycnoporus cinnabarinus, etc. [52]. Nowadays, P. chrysosporium is always used as the type strain of white-rot fungi. White-rot fungi have established unique lignocellulose degradation system in the long-term biological evolution, and they are the only known microorganism that can effectively degrade lignin into CO2 and H2O by pure culture. Actually, the degradation of lignin by white-rot fungi is a co-metabolic process between lignin and another extra nutrient used as carbon source. During the cultivation, these nutrients, as co-substrates, produce water-soluble extracts (such as ammonia peroxide, VA, organic acid, etc.) which could induce the aromatic oxidation and ring cleavage of lignin [61].
Therefore, the degradation of lignocellulose by white-rot fungi can be divided into two modes. The first mode is nonselective delignification, which means degrading lignin, cellulose, and hemicellulose simultaneously. The reported fungi that possessed this mode mainly included Trametes versicolor, Irpex lacteus, Phanerochaete chrysosporium, Heterobasidion annosum, Phlebia radiate, and some ascomycetes such as Xylaria hypoxylon. However, this type of degradation is limited around the hypha and degrades only a small amount of lignocellulose. The second mode is selective delignification which means lignin and hemicelluloses are attacked before cellulose. This mode mainly exists in Ganoderma australe, Ceriporiopsis subvermispora, Phellinus pini, Phlebia tremellosa, and some Pleurotus. In most white-rot fungi, both modes are available and are not mutually exclusive. The lignin degradability of white-rot fungus depends on its lignin-degrading enzymes, which initiate the free radical chain reaction and result in the C–C and C–O cleavage, demethylation, hydroxylation, benzyl alcohol oxidation, aromatic ring opening, etc. The products would be further degraded into CO2 and H2O through different pathways. In addition to white-rot fungi, other lignin degradability fungi, such as Aspergillus oryzae and Aspergillus niger, can also catalyze the degradation of lignin by ligninolytic enzymes [31, 136].
Although there had a lot of researches on the fungi degradation of lignin since 1980, the commercial application is still limited owing to the strict growth conditions and additional energy and chemical inputs [114].
12.2.2 Bacteria
Bacteria have many advantages over fungi on the lignin degradation, since they can tolerate a wider range of pH, temperature, and oxygen concentration, grow faster, have more biochemical functions, and have better environmental adaptability. They have attracted increasing attentions in the value-added utilization of lignin. However, the current understanding is that bacteria can only degrade low molecular weight of lignin-derived compounds. Therefore, they may play an important role in the final stage of lignin degradation [131]. In recent years, a large number of bacteria that can depolymerize lignin have been found in composted soil, rainforest, eroded bamboo slips, sludge from pulp mills, and intestines of wood-eating insects [132]. There are many types of bacteria that can degrade lignin, which are mainly Actinobacteria, Proteobacteria (mainly α-Proteobacteria and γ-Proteobacteria), and Firmicutes. Actinomycota includes Streptomyces, Microbacterium, and Rhodococcus. Proteobacteria includes Pseudomonas, Burkholderia, and Enterobacter [44].
12.2.2.1 Actinomycetes
Actinomycetes can penetrate insoluble substrates and increase the water solubility of lignocellulose. It is mainly involved in the initial degradation and humification of organic matters. They secrete a series of enzymes and convert lignin into low-molecular-weight lignin-derived compounds [43]. With the development in isolation and identification technology, more and more lignin-degrading Actinomycetes have been found, including Streptomyces sp., Thermoactinomyces sp., Arthrobacter sp., Micromonospora sp., Nocardia sp., Thermomonospora sp., etc. They can produce a variety of enzymes, such as lignin peroxidase, laccase, xylanase, carboxymethyl cellulase, p-nitrophenyl-β-D-glucosidase, etc., responsible for the degrading of lignocellulose [4]. With these enzymes, they can change the cell wall structure of lignocellulose effectively through selective cleavage and structure modification of lignin, thereby promoting the enzymatic hydrolysis of cellulose to release sugar [140]. It obviously modified the carbonyl and methoxy groups in the structure of lignin, especially resulting in the significant reduction of guaiacol units [141].
The advantages of Streptomyces in lignin degradation are becoming increasingly apparent. For example, Streptomyces griseorubens can degrade more than 60% of lignin by secreting extracellular laccase and lignin peroxidase, as well as exo-1,4-β-glucanase, endo-1,4-β-glucanase, and β-xylosidase [34, 130]. Streptomyces viridosporus T7A and S. setonii 75Vi2 can degrade lignin and carbohydrate in softwood, hardwood, and herbal, especially in herbs, with the generation of lignin-carbohydrate complex named acid-precipitable polymeric lignin (APPL) by cleaving p-hydroxy ether bonds in lignin [21]. The degradation rate of lignin by S. viridosporus T7A can reach 19.7% within 8 weeks [135]. Streptomyces can also degrade alkaline lignin into phenolic compounds. Meanwhile, the coculture with white-rot fungi can effectively enhance the degradation rate of alkaline lignin [137]. In particular, the recognition of some thermophilic Actinomycetes, such as Thermomonospora fusca, T. alba, Micromonospora sp., etc., makes the industrial application of Actinomycetes in lignin valorization possible [18, 67].
12.2.2.2 Proteobacteria
The major lignin-degrading Proteobacteria is α-Proteobacteria and γ-Proteobacteria. As the most important genus of α-Proteobacteria, Sphingobacterium sp. has been widely used in lignin degradation, which can use Klason lignin as the sole carbon source with the secretion of lignin peroxidase and laccase [14]. Sphingobacterium sp. can oxidize guaiacyl (G) and syringyl (S) units in Klason lignin to produce small molecule aromatic compounds and ketone compounds, such as guaiacol, p-hydroxybenzoic acid, vanillic acid, vanillin, 4-hydroxy-2-butanone, methyl vinyl ketone, etc. [27].
The best known lignin-degrading γ-Proteobacteria is Pseudomonas fluorescens, Pseudomonas putida, Enterobacter lignolyticus, and E. coli. Their strong lignin degradability could be attributed to the plentiful lignin-degrading enzymes including laccase, lignin peroxidase (LiP), and manganese-dependent peroxidase (MnP), as well as some kind of lignin-degrading auxiliary enzymes [132]. For example, Pseudomonas fluorescens mainly secretes LiP; dye-decolorizing peroxidase (DyP) and MnP are the major enzymes in Pseudomonas putida; catalase, peroxidase, and DyP are the major enzymes in Enterobacter lignolyticus; and laccase is the major enzymes responsible for lignin degradation of Escherichia coli [44].
12.2.2.3 Firmicutes
The lignin-degrading Firmicutes are mainly belonging to Bacillus, such as Basic Bacillus, B. ligniniphilus, B. firmus, thermophilic B. subtilis, and B. licheniformis. It was suggested that the lignin-degrading pathway in B. ligniniphilus included gentisate pathway, benzoic acid pathway, and β-ketoacidic acid pathway, among which β-ketoacidic acid pathway included catechuic acid branches and protocatechuic acid branches. Zhu et al. [147] reported a B. ligniniphilus strain with high tolerance to the extreme environments, which can grow on alkaline lignin by using it as a sole carbon or energy source. And up to 42% of monophenol aromatic compounds were identified in the fermented residues, including phenylacetic acid, 4-hydroxybenzoic acid, and vanillic acid. Thermophilic B. subtilis and B. licheniformis have also been found to degrade KL lignin significantly [29].
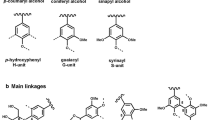
Other lignin-degrading Firmicutes include Acetoanaerobium, Trabulsiella, Paenibacillus glucanolyticus, etc. Duan et al. [26] reported an Acetoanaerobium strain that could oxidize the p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) unit of KL structure with the formation of benzene-propanoic acid, ferulic acid, syringic acid, and some low-molecular-weight acid compounds, such as adipic acid, hexanoic acid, and 2-hydroxybutyric acid. Trabulsiella guamensis IIPTG13 reported by Suman et al. [119] caused the degradation of over 60% of guaiacylglycerol-β-guaiacyl ether (GGE). And a variety of compounds containing polar functional groups was detected in the fermented residues, mainly including organic acids, fatty acids, and other phenolic derivatives. In particular, some compounds such as 4-hydroxy-3-methoxybenzoic acid, 3-methoxybenzoic acid, and 2-(4-hydroxy-3-methoxyphenyl) acetic acid were similar to coniferyl alcohol and sinapyl alcohol (lignin precursor).
12.3 Lignin-Degrading Enzymes
The irregularity, complexity, and specificity of the three-dimensional structure of lignin polymers determine that the lignin-degrading enzyme system is a nonspecific enzyme system. Compared with the enzymatic hydrolysis of cellulose and hemicellulose, the lignin-degrading process was oxidation rather than hydrolysis [52]. The high-molecular-weight lignin polymers generate unstable free radicals under the catalysis of enzymes, which triggers the spontaneously nonenzymatic cleavage reactions of lignin.
Generally, the lignin-degrading enzymes are divided into three categories: peroxidase, laccase, and lignin degradation auxiliary enzyme (LDA) [96]. Peroxidase mainly includes lignin peroxidase (LiP), manganese peroxidase (MnP), versatile peroxidase (VP), and dye-decolorizing peroxidase (DyP). Laccase is a type of copper-containing polyphenol oxidase which can directly use oxygen to catalyze phenolic hydroxyl groups with phenoxy radicals and water as products. LDA enzymes cannot degrade lignin directly, but it is necessary for the complete degradation of lignin. It mainly includes glyoxal oxidase, aryl alcohol dehydrogenase, heme-thiolate haloperoxidases, flavin adenine dinucleotide-dependent glucose dehydrogenase (FAD-GDH), pyranose 2-oxidase, and other enzymes like quinone reductase, superoxide dismutase, methanol oxidase, etc. Different lignin-degrading microbes usually secrete one or more lignin-degrading enzymes in different combinations, which would degrade lignin through sequential actions.
12.3.1 Lignin-Degrading Peroxidase
12.3.1.1 Manganese-Dependent Peroxidase
Manganese peroxidase (EC1. 11. 1.13, MnP) is an extracellular peroxidase, which was first found in P. chrysosporium more than 30 years ago, and is one of the most important lignin peroxidases [42, 95]. It was found that almost all white rot fungi contain MnP , such as Panus tigrinus, Trametes versicolor, Lenzites betulinus, Irpex lacteus, Lenzites gibbosa, Phlebia radiata, Fomitiporia mediterranea, Agaricus bisporus, Bjerkandera sp., Nematoloma frowardii, as well as species belong to Corticiaceae, Stereaceae, Hericiaceae, Ganodermataceae, Hymenochaetaceae, Polyporaceae, Strophariaceae, and Tricholomataceae [17]. Recently, more and more MnP was found in bacteria, yeast, and mold, such as Bacillus pumilus, Paenibacillus sp., Azospirillum brasilense, and Streptomyces psammoticus [25, 69, 90]. Generally, these microorganisms have multiple MnP isoenzymes, and there are at least two or more isomers.
The oxidation reaction of Mn2+ to Mn3+ catalyzed by MnP is H2O2-dependent. It takes Mn2+ as the preferred reducing substrate and turns it into highly active Mn3+. The MnP-catalyzed lignin degradation cycle starts when H2O2 is bound to MnP. However, excessive H2O2 will affect the activity of MnP [76]. The generated Mn3+ would chelate with organic acids produced during the fermentation process, such as oxalic acid, to form stable low-molecular-weight substances. Then, the chelated Mn3+ acting as an oxidant penetrates or diffuses into the cell wall of plant and nonspecifically attacks and oxidizes the phenol structure of lignin, thus leading to the generation of unstable phenoxy radicals [17]. The phenoxy radicals are further broken down spontaneously with the reaction of demethylation, alkyl–aryl bond cleavage, Cα–Cβ cleavage, and Cα oxidation [47].
In general, secondary metabolism caused by the nutrient limitation of nitrogen and/or carbon in the growing process of microbe will cause the secretion of peroxidase and the degradation of lignin. Sufficient Mn2+ is necessary for the secretion of MnP by microorganisms. The addition of Mn2+ can increase the activity of MnP, promote the growth of fungi, shorten reaching time of the maximum MnP activity, and further enhance the degradation of lignocellulose. Meanwhile, in some microorganisms, Mn2+ can stimulate the transcription levels of MnP and played a posttranscriptional role in MnP production [45, 77].
12.3.1.2 Lignin Peroxidase
Lignin peroxidase (EC 1.11.1.14, LiP) is an extracellular enzyme which can oxidize the non-phenolic aromatic compounds with redox potentials higher than 1.4 V. The first LiP was found in Phanerochaete chrysosporium, and then many white-rot fungi, such as Trametes versicolor, Bjerkandera sp., and Phlebia tremellosa, were reported that can produce LiP. In recent years, it has also been detected in some bacteria, such as Acinetobacter calcoaceticus, Streptomyces viridosporus, etc. [52]. Generally, microorganisms secrete several lignin peroxidase (LiP) isoenzymes, of which the relative composition and isoelectric point (pI) are greatly affected by microbial culture conditions [116]. The crystal structure of LiPs has been recognized. It belongs to glycosylase, which possesses iron (III) protoporphyrin IX (ferriprotoporphyrin IX) as the prosthetic group [60]. It can oxidize iron cytochrome C, suggesting that the enzyme can directly act on polymerized lignin. Therefore, it plays an important role in the oxidative degradation of lignin.
Similar to MnP, the catalytic reaction of LiP requires the participation of H2O2. The catalytic process of LiP can be regarded as a series of free radical chain reactions initiated by H2O2 produced by white-rot fungi. Under the catalysis of LiP, an electron is extracted from the aromatic substrate and oxidized to form an aromatic cation group which can be used as both a reaction group and a cation. Then, the substrate would be partially or completely oxidized into various derived low-molecular-weight compounds through series branch reactions. LiP is also currently the only known oxidase that can cleave Cα–Cβ and aromatic rings in an acellular state. However, different from MnP, LiP has low substrate specificity and can react with different lignin-like compounds. Meanwhile, LiP has high redox properties (about 1.2 V at pH 3), which can not only oxidize the typical phenol and aromatic amines but also can oxidize a series of other aromatic ethers and polycyclic aromatic compounds, as well as methoxy aromatic ring substrates without free phenolic group, through ionization reaction [113].
12.3.1.3 Versatile Peroxidase
Versatile peroxidase (EC 1.11.1.16, VP), considering as structural heterozygote between LiP and MnP, can directly oxidize the phenolic and non-phenolic aromatic compounds. The reported microorganisms that possess VP activities are major in genus of Pleurotus, Bjerkandera, Phanerochaete, Trametes, etc. [11, 19, 122, 139].
VPs have some catalytic properties of both LiP and MnP, resulting in an even wider range of substrates. It can catalyze the substrates with high and medium redox potentials, even azo dyes and other non-phenolic compounds [10, 40]. In addition, a Mn2+ binding site similar to MnP was in the structure of VP, which enables VP to participate in the oxidation of low and high redox potential substrates [38]. The degradation pathway of phenolic β-O-4 catalyzed by VP was identified as Cα–aryl cleavage rather than Cα–Cβ cleavage . In the VP catalyzed reaction of phenolic lignin dimers, neutral radicals were generated firstly by the oxidation of 4-OH position, and then the polymerization or depolymerization reactions were reacted depending on the functional group at the 5-position of the guaiacol group (G5). The side chain of lignin would cleave owing to the substitution of G5 with methoxy (S-O-4) [139]. Therefore, VP-producing microorganisms play an important role in the biodegradation of lignin and lignocellulose . Wen Kong et al. [64] found that although a lignin-degrading Physisporinus vitreus strain can secrete both laccase and VP, only VP can oxidize non-phenolic lignin compounds as well as the β-O-4 and 5–5′ dimers.
12.3.1.4 Dye-Decolorizing Peroxidase
Dye-decolorizing peroxidase (EC 1.11.1.19, DyP) was first found in Bjerkandera adusta (formerly named as Geotrichum candidum) [62]. They have low substrate specificity and can oxidize all typical peroxidase substrates, such as ABTS, DMP, adlerol, various anthraquinone dyes (e.g., Congo red, mordant black 9, reactive blue 5, etc.), and so on [20]. Presently, more and more DyP-producing fungi have been reported, such as Pleurotus ostreatus [32], Termitomyces albuminosus [54], and Irpex lacteus [28]. Especially, the popular reports of DyPs in bacteria, such as Rhodococcus jostii [104], Bacillus subtilis [84, 111], Kocuria rosea [94], and Thermobifida fusca [101], promote the deep researches and wide applications of DyP in lignin degradation.
DyPs also belong to the heme peroxidase family but with different structural folds from other types of heme peroxidases. They possess special reactivity to the oxidation of polycyclic dyes and phenolic compounds. The active site aspartic acid residue of DyP acts as a proton donor [118]. The catalytic characteristics on lignocellulose and lignin of DyPs are obviously similar to LiP and MnP. However, DyPs also have hydrolase or oxygenase activities [117]. Meanwhile, they can degrade different dyes at low pH (pH 3–4), especially anthraquinone dyes [20]. These characteristics make the research of VPs have significant meanings. Especially considering the high redox of VPS in fungi, the application of bacterial VPS in biotechnology will be more extensive in the future [49].
12.3.2 Lignin-Degrading Auxiliary
12.3.2.1 Glyoxal Oxidase
Glyoxal oxidase (EC 1.2.3.5, GLOX), belonging to radical copper oxidase family, is one of three known extracellular enzymes secreted by microorganisms under conditions of lignin decomposition [24]. The study of GLOX is mainly based on Phanerochaete chrysosporium. The catalytic mechanism of GLOX is that it can oxidize many simple dicarbonyl and hydroxycarbonyl compounds (especially glyoxal and methylglyoxal) to generate carboxylic acids coupling with the reduction of O2 to H2O2 [133]. Therefore, GLOX is inactive in vitro catalysis for lignin degradation unless being coupled with peroxidase as auxiliary enzymes, since H2O2 is necessary for the degradation of lignin catalyzed by peroxidase such as LiPs and MnPs [52, 59].
12.3.2.2 Aryl Alcohol Oxidase
Aryl alcohol oxidase (EC 1.1.3.7, AAO), also known as glucose–methanol–choline oxidase, was originally found in Polystictus versicolor. At present, more and more fungi have reported possessed AAO activity, such as Pleurotus species, Fusarium species, Geotrichum candidum, Amauroderma boleticeum, Phanerochaete chrysosporium, and Bjerkandera adusta, and even some bacteria, such as Sphingobacterium sp. ATM [68, 120].
AAO has two domains with non-covalently bound FAD cofactors [35]. The auxiliary effect of AAO on the degradation of lignin is mainly considered to be that it can catalyze the oxidative dehydrogenation of phenol and non-phenol aromatic alcohol, the formation of corresponding aldehydes from polyunsaturated primary alcohols or aromatic secondary alcohols, and the generation of H2O2 as by-products [46]. In addition, AAO can promote the depolymerization of lignin by cooperating with laccase, since it can reduce the free radical intermediates (such as guaiacol, erucic acid, etc.) generated from laccase oxidation [82].
12.3.2.3 Heme-Thiolate Haloperoxidases
Heme-thiolate haloperoxidases (HTHPs) are extracellular enzymes which use cysteine residue as the proximal axial ligand of heme. For a long time, chloroperoxidase (CPO) from the ascomycete Caldariomyces fumago was the only known halogenating enzyme. Later, another type of fungal HTHP was found in Agrocybe aegerita, named as Agrocybe aegerita peroxidase (APO, also called aromatic peroxygenases), which showed strong brominating and weak chlorinating as well as weak iodating activities. Considering it can convert veratryl alcohol into veratryl aldehyde at pH 7, this enzyme was called “alkaline lignin peroxidase” before Ullrich et al. recognized it as HTHP [48, 123].
HTHPs represent a unique oxidoreductase sub-subclass of heme proteins with peroxygenase and peroxidase activity, but it cannot oxidize the non-phenolic β-O-4 structure of lignin. They can transfer oxygen via hydrogen peroxide to aromatic and aliphatic substrates similar to cytochrome P450, rather than directly to O2 [98]. The auxiliary effect of fungal HTHP on lignin degradation was only found until recently. Ruiz-Dueñas et al. [107] found that the theme peroxides of Pleurotus ostreatus not only included cytochrome c peroxidase and lignin-degrading peroxidases (MnP and VP) but also included HTHP. This was the first description of HTHP in Pleurotus. Moreover, compared with the typical peroxidase, HTHP has surprisingly high reactivity against benzyl hydrogen and phenolic substrates [52, 128]. Now by accumulating the genome sequence of ascomycetes and basidiomycetes, the sequence library of HTHPs was gradually expanded [65].
12.3.2.4 Flavin Adenine Dinucleotide (FAD)-Dependent Glucose Dehydrogenase (GDH)
The flavin adenine dinucleotide (FAD)-dependent glucose dehydrogenases (EC 1.1.99.10, FADGDHs) belongs to the glucose–methanol–choline (GMC) oxidoreductase family. FAD is used as the major electron acceptor to catalyze the oxidation of first hydroxyl group of glucose and other sugar molecules [138]. The reported FADGDHs were mainly derived from fungal extracellular enzymes, membrane binding protein of Gram-negative bacteria, and cytosolic enzymes in some insects [36, 99]. The first reported FADGDHs from basidiomycetes were obtained by recombined GDH (PcGDH) from Pycnoporus cinnabarinus CIRM BRFM 137 in Aspergillus niger [105]. Compared with other glucose–methanol–choline (GMCs) oxidoreductases, the substrate binding domain of PcGDH is conserved. The enzyme does not use oxygen as the external electron acceptor and has the obvious ability of reducing the oxidized quinone or free radical intermediate, which makes the enzyme play an important role in decreasing the toxicity of quinones and protecting fungal cells from phenoxy. This provides a new way to detoxify the toxic compounds formed in the process of lignin degradation by PcGDH [105].
12.3.2.5 Pyranose 2-Oxidase
Pyranose 2-oxidase (EC 1.1.3.10, pyranose: 2-oxo oxidoreductase, P2OX) belongs to the glucose–methanol–choline oxidoreductase (GMC) family. It is a sugar oxidizing enzyme that can produce hydrogen peroxide during the glucose starvation period of Phanerochaete chrysosporium [30]. P2OX has been found in a variety of microorganisms, including Phanerochaete chrysosporium, Trametes versicolor, Oudemansiella mucida, Phlebiopsis gigantean, Pleurotus ostreatus, Polyporus obtusus, Trametes multicolor, Peniophora sp., Tricholoma matsutake, etc. [2, 23, 63, 83]. A new type bacterial P2OX (PaP2OX) was even identified from the lignin-degrading bacteria Pantoea ananatis. Different from other reported bacterial P2Ox, this enzyme showed homotetrameric spatial conformation that is similar to fungal P2Ox [143]. P2O was preferentially located in hyphal periplasmic space and prefer to oxidize aldopyranoses derived from cellulose and hemicellulose into ketoaldoses at C2 (also at C3) [66].
The catalytic process of P2OX can be divided into two steps, i.e., the formation of FADH intermediate by transferring hydride ion to N5 atom of isoalloxazine and the regeneration from dioxygen to FAD. It is worth noting that the benzoquinones and lignin-derived substances during lignin degradation can also be used for the regeneration of reduced FAD [2]. A large number of studies have shown that the synergistic effect of P2O and laccase can improve the degradation efficiency of lignin. It is attributed to the quinone-reducing activity of P2O which can reduce the polymerization of phenoxy radicals or quinoid intermediates in vivo. Ai et al. [2] found that during the catalysis of laccase, P2O can effectively inhibit the polymerization of lignin and even promote the lignin degradation by transferring electrons to various quinones and ABTS (2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) cation radicals, as well as quinoids generated by laccase catalysis.
12.3.3 Laccase (EC 1.10.3.2)
Laccase , also called blue polycopper oxidase, is a kind of copper-containing polyphenol oxidase. Laccase plays an important role in the biodegradation of lignin. In plants, laccase is an intracellular enzyme involved in lignin synthesis, while in microorganisms, laccase mainly participates in the lignin degradation as an extracellular enzyme. Laccase-secreting fungi mainly include Basidiomycota, Ascomycota, and Deuteromycota. Compared with fungi, laccase is the most popular ligninolytic enzyme in prokaryotes. Laccase gene has been found in Proteus, Actinomycetes, and Firmicutes [121].
Different from lignin peroxidase taking H2O2 as prerequisite condition, laccase can use oxygen molecules as the final electron acceptor to catalyze the oxidation, degradation, or polymerization of phenolic and non-phenolic substrates. The utilization of O2 instead of H2O2 as substrate results in a mild catalytic condition for lignin biodegradation. Meanwhile, in vitro, laccase synergistically degrades lignin with some quinone-reducing enzymes, such as pyranose oxidase, glucose oxidase, resveratrol oxidase , and cellobiose dehydrogenase [2, 33, 70, 78]. Due to its unique catalytic properties, laccase has been widely used in the high-value utilization of lignin [115].
There are four Cu2+ located at three binding domains in the active center of typical laccase, which have different electronic paramagnetic resonance (EPR) parameters. These copper ions synergistically mediate the catalytic oxidation of phenolic and non-phenolic substrates. However, in recent years, it has been found that there is a kind of atypical laccase in the complex laccase family. Anita et al. found a laccase that possessed two copper binding domains rather than three, which was called as small laccase or small laccase-like multicopper oxidase (LMCO) [3]. Meanwhile, it was found that in addition to copper ions, some other metal ions were combined on the binding domains of some kind of laccase. For example, the active center of Phlebia radiate laccase contains two Cu and one PQQ cofactor acting as T3Cu [93]. The active center of Pleurotus ostreatus laccase contains Cu, Fe, and Zn [85]. The active center of Phellinus ribis contains Cu, Mg, and Zn [5]. These differences in active centers caused the significant lignin degradability of laccase.
The oxidation lignin and its model compounds catalyzed by laccase are a one-electron reaction generating free radicals. Firstly, the substrate was catalyzed by laccase to generate unstable phenoxy radicals and continues to be converted to quinone in the consequent reactions [86]. In general, laccase can catalyze bond cleavage in low-molecular-weight phenol lignin model compounds, including ortho-diphenol and para-diphenol, aminophenol, methoxy-substituted phenol, thiol, polyphenol, polyamine, etc. In general, laccase can only oxidize the phenolic structure of lignin, accounting for 10% of the lignin structure. The rest of non-phenolic lignin structures with high redox potential could not be degraded by laccase unless by adding low-molecular-weight mediators, such as ABTS and HBT [88]. Recently, some atypical laccases have been found to directly oxidize non-phenolic lignin model compounds or even polycyclic aromatic hydrocarbons without relying on mediators. For example, a kind of atypical laccase, lacking the specific 600 nm band without the blue color (also called “yellow” or “white” laccase), was widely distributed in many microorganisms, such as Schizophyllum commune, Pleurotus ostreatus, Leucoagaricus gongylophorus, Daedalea flavida, Aureobasidium pullulans, and Lentinus squarrosulus [1, 13, 50, 87, 100, 112, 126]. These atypical laccases have important research and application significance for broadening the use of laccase and reducing the cost of laccase mediator catalytic system.
12.3.4 Other Enzymes
Besides the abovementioned ligninolytic enzymes , some enzymes reported recently would be promising options for lignin degradation. β-etherases (EC) are non-radical ligninolytic enzymes, which can selectively cleave β-O-4 aryl ether bonds in lignin. The conversion rate of lignin model compound GGE (1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenoxy) propane-1,3-diol) catalyzed by β-etherase from Sphingobium sp. SYK-6 was close to 100% [106]. However, except for Chaetomium and Sphingobium genus, the natural existence of β-etherase was rarely reported [97]. Another lignin-degrading enzyme called superoxide dismutases has been identified recently from Sphingomonas bacterium, which can degrade lignin and generate low-molecular-weight aromatic derivatives, such as guaiacol and demethylated guaiacol [39].
12.4 Major Metabolic Pathway of Lignin Degradation
It is very important to study the metabolism network of lignin and construct lignin-degrading system, which is beneficial for the bioconversion of lignin into high-value intermediate metabolites. The known catabolic pathways related to lignin degradation in white-rot fungus include benzoic acid metabolism, dioxin metabolism, phenol metabolism, polycyclic aromatic hydrocarbon (PAH) metabolism, aminobenzoic acid metabolism, naphthalene metabolism, toluene metabolism, fatty acid metabolism, ether ester metabolism, xylene metabolism, carbon metabolism, etc. [75]. Due to the biochemical versatility and high environmental adaptability, increasing attention has been paid on the catabolic pathways related to lignin degradation in bacteria. Different from fungi, the metabolism of lignin and its derived aromatic compounds by bacteria is completed in vivo, taking β-ketoadipate pathway as the main pathway [131].
As bioinformatics and genomic sequencing technologies are used to screen functional genes encoding lignin-degrading enzymes, more and more genes and their encoded metabolic pathways have been reported. For example, 29 lignin-degrading enzyme genes were recognized from Aspergillus fumigatus, including multicopper oxidase, lignin-modifying peroxidase, glucose–methanol–choline oxidoreductase, vanillyl alcohol oxidase, galactose oxidase, 1,4-benzoquinone reductase, monooxygenase, ferulic acid esterase, etc., which degraded lignin by participating in six catabolic pathways, namely, galactose metabolism, phenylalanine metabolism, pyruvate metabolism, benzoate metabolism, toluene metabolism, and aminobenzoic acid metabolism [71].
At least 15 gene sequences related to lignin degradation, including oxidase, copper oxidase, laccase, dioxygenase, decarboxylase, and so on, were found in Comamonas serinivorans C35. These enzymes undergo at least four lignin-degrading pathways, i.e., benzoate pathway, phenol pathway, p-hydroxyacetophenone pathway, and β-ketoadipate pathway, to depolymerize lignin into a variety of low-molecular-weight aromatic compounds, such as 3-methylbenzaldehyde, guaiacol, vanillic acid, vanillin, syringic acid, syringaldehyde, p-hydroxybenzoic acid, ferulic acid, etc. [144]. The recognition of these metabolic pathways provides a theoretical basis for the subsequent biological transformation of lignin through the regulation of metabolic pathways. This section would introduce the major metabolic pathways related to lignin degradation in microorganisms.
12.4.1 β-Aryl Ether Catabolic Pathways
β-Aryl ether is the most abundant structure in lignin. In bacteria, the study of aryl ether catabolic pathways is mainly based on Sphingomonas paucimobilis , whose metabolic pathways for lignin dimmers are shown in Fig. 12.1. First, DNA-dependent lignin dehydrogenase (LigD) catalyzed the α-hydroxy groups to generate the corresponding ketones. Then, a reductive ester (ether) bond cleavage reaction took place under the action of glutathione-dependent β-esterase (LigEFG, belonging to glutathione-S-transferase superfamily) [9]. The derived ketone compounds would be consequently oxidized to vanillic acid through the oxidation of γ-hydroxyl group to carboxylic acid. In the metabolic pathway of S. paucimobilis SYK-6, vanillate was demethylated to protocatechuate under the catalysis of LigM (tetrahydrofolate-dependent demethylase), producing 5-methyl tetrahydrofolate as a by-product (as shown in Fig. 12.2). It can also be oxidized by C–C bond cleavage reaction in a manner similar to β-oxidation with the generation of lignin-derived products, dominating of methyl ketone (acetovanillone). The latter can be further used as the mediator of fungal laccase to promote the lignin degradation [146]. Besides S. paucimobilis SYK-6, β-aryl ether catabolic pathway was also found in Pseudomonas acidovorans, P. putida, and Rhodococcus jostii rHA1 [9].
β-aryl ether metabolic pathways in Sphingomonas paucimobilis for lignin and lignin-derived compounds. LiP lignin peroxidase, LigD DNA-dependent lignin dehydrogenase, LigEFG β-etherase (GST), LigM tetrahydrofolate-dependent demethylase [9]
The biphenyl metabolic pathways of lignin and lignin-derived compounds [145]. LigZ gene encoding OH-DDVA dioxygenase, LigX gene encoding non-heme iron-dependent demethylase, LigZ gene encoding extradiol dioxygenase, LigY gene encoding C–C hydrolase, LigW/W2 gene encoding decarboxylases, LigM DDVA, 5,5′-dehydrodivanillate, OH-DDVA 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl, 5CVA 5-carboxyvanillate, PCA protocatechuate
In white-rot fungi, P. chrysosporium catalyzed β-aryl ether model compounds by Cα–Cβ oxidative cleavage catalyzed by lignin peroxidase catalyses, with the production of vanillin as a product, as well as other oxidation products like benzyl ketone. Then vanillin was oxidized by vanillate dehydrogenase to generate vanillate and by-product 2-hydroxyacetaldehyde. The 2-hydroxyacetaldehyde might be further oxidized to oxalic acid, which would be used to complex with Mn2+ to promote the lignin degradation catalyzed by manganese peroxidase.
12.4.2 Biphenyl Catabolic Pathways
Biphenyl mainly exists between two guaiac units, accounting for about 10–15% in the molecular structure of lignin, only inferior to that of aryl ether. Biphenyl catabolic pathway is popular in lignin-degrading microorganisms, which mainly oxidized biphenyl and chlorinated biphenyl to 2,3-dihydroxybiphenyl by oxidative cleavage (Fig. 12.2) [145]. Firstly, non-heme iron-dependent demethylase (LigX) catalyzed the demethylation of 5,5′-dehydrodivanillate (DDVA) to 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl (OH-DDVA). Then the extradiol dioxygenase (LigZ) was catalyzed the meta-cleavage of OH-DDVA, and the ring-opened product was hydrolyzed by C–C hydrolase (LigY) to generate 5-carboxyvanillate (5-CVA). Two decarboxylases (LigW) which can convert 5-CVA into the central intermediate vanillate were found in Sphingomonas sp. SYK-6 and catalyzed vanillate into protocatechuate (PCA). The metabolic pathway of biphenyls was also common in fungi. Yang and Zhang [134] found that both the laccase and MnP in Trametes sp. SQ01 have the ability to convert the conjugated diene of 2-hydroxy-6-oxygen −6-phenyl-2,4-hexadienoic acid (HOPDA) to monoene, so that the hydroxyl group on C disappears, resulting in the hydrolysis of HOPDA. The degradation of biphenyl by white-rot fungus mainly depends on oxidative cleavage. In vitro, using 1-hydroxybenzotriazole (HBT) as mediator, the Trametes versicolor laccase can significantly degrade the phenolic/non-phenolic biphenyl model compounds simulating 5–5′ type condensed lignin substructures [12].
12.4.3 Ferulic Acid Catabolic Pathway
Ferulic acid is a phenylpropane compound and is an important intermediate in the biosynthesis of lignin, which connects the benzene ring of lignin through ester bond. Ferulic acid structure accounts for about 1.5% of the total plant cell wall. It was reported that amounts of microorganisms possessed two types of ferulic acid side chain cleavage. In Sphingomonas SYK-6, Pseudomonas fluorescens AN103, Pseudomonas sp. HR199, P. putida WCS358, and Amycolatopsis sp. HR167, two enzymes were found participating in the cleavage of ferulic acid side chains, i.e., feruloyl-coenzyme A (CoA) synthetase and feruloyl-CoA hydratase/lyase [79]. Firstly, the COA was transferred to carboxyl group of ferulate under the catalysis of feruloyl-coenzyme A (COA) synthetase. The resulting feruloyl-CoA was then hydrated to form 4-hydroxy-3-methoxy-phenoxy-þ-hydroxypropionyl-CoA (HMPHP-CoA) under the catalysis of feruloyl-CoA hydratase/lyase (FerB). Finally, vanillin and acetyl-CoA were generated by cleaved HMPHP-CoA (Fig. 12.3). There is another ferulic acid metabolism pathway catalyzed by non-oxidative decarboxylase in some microorganisms . It removes one carbon from the side chain of ferulic acid, thereby forming 4-hydroxy-3-methoxystyrene. For example, Cupriavidus sp. B-8 can directly remove the carboxyl group in ferulic acid to generate corresponding aromatic compounds owing to containing of ferulic acid decarboxylase. The 4-vinyl aromatic compound generated by decarboxylation is reduced to 4-ethylphenol under the co-effect of vinylphenol reductase and NADPH. Then 4-ethylphenol was further converted to vanillic acid and consequently degraded to protocatechuic acid that enters into the ring-open reaction [142].
The ferulic acid catabolic pathway in Sphingomonas SYK-6 [79]. FerA feruloyl-CoA synthetase gene, FerB feruloyl-CoA hydratase/lyase gene (ferB), PCA protocatechuate, LigM tetrahydrofolate (H4folate)-dependent demethylase
12.4.4 Tetrahydrofolate-Dependent O-Demethylation Catabolic Pathway
Vanillate and syringate are important intermediate metabolites in the biodegradation of lignin-derived aromatic compounds. There are two ways for microorganisms to metabolize vallinate and finally produce protocatechuate (PCA) that can be further oxidized by protocatechuic acid 3,4-dioxygenase through the β-ketoadipate pathway via intradiol oxidative cleavage (Fig. 12.4) [109].
Tetrahydrofolate-dependent O-demethylation pathway of vanillate, syringate, and 3-O-MGA linked with H4folate-mediated C1 metabolism in S. paucimobilis SYK-6 [109]. DesA gene encoding O-demethylase, LigM gene encoding tetrahydrofolate (H4folate)-dependent demethylase, 3-MGA 3-O-methylgallate, PCA protocatechuate
One of the tetrahydrofolate-dependent O-demethylation catabolic pathways is vallinate metabolism depending on O-demethylase, which mainly occurs in aerobic bacteria (such as Pseudomonas and Acinetobacter). For example, the vanillate-degradation reaction catalyzed by vanillate-O-demethylase in A. dehalogenans firstly transferred the methyl of vanillate to the corrinoid protein by methyl transferase I; the latter then transferred methyl to tetrahydrofolate (H4folate) by methyltransferase II. The other tetrahydrofolate-dependent O-demethylation catabolic pathway is a metabolic system relying on tetrahydrofolate (H4folate)-dependent demethylase (LigM), which is mainly reported in anaerobic bacteria, including Acetobacterium dehalogenans, Acetobacterium woodii, and Moorella thermoacetica. In this pathway, syringate was firstly transformed into 3-O-methylgallate (3MGA) under the action of syringate O-demethylase (encoded by DesA). Then gallate was formed by catalyzing 3MGA with tetrahydrofolate (H4folate)-dependent demethylase (LigM).
12.4.5 3-Methyl Gallate Catabolic Pathway
There was three proposed 3-methyl gallate (3-MGA) catabolic pathway (Fig. 12.5): (1) converting 3-MGA to 4-oxalomesaconate (OMA) via gallate in the reaction catalyzed by vanillate/3MGA O-demethylase (LigM) and gallate dioxygenase (DesB) or PCA 4,5-dioxygenase (LigAB), which appears to be the major pathway of 3-MGA; (2) converting 3-MGA to OMA via 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate (CHMOD) by 3-MGA 3,4-dioxygenase (DesZ) and a hydrolase; and (3) converting 3-MGA to 2-pyrone-4,6-dicarboxylate (PDC) by PCA 4,5-dioxygenase (LigAB) and 3-MGA 3,4-dioxygenase (DesZ) [79, 80].
12.4.6 Protocatechuate (PCA) Catabolic Pathway
Many metabolic pathways in the lignin-degrading microbes can produce vanillin or its oxidation product vanillate. These compounds further demethylated to produce protocatechuate (PCA) which would be degraded by non-heme-o-diphenol dioxygenase through oxidative ring opening. There are three known non-heme-o-diphenol dioxygenases, i.e., PCA-3,4-dioxygenase (3,4-PCD), PCA-4,5-dioxygenase (4,5-PCD), and PCA-2,3-dioxygenase (2,3-PCD). 3,4-PCD catalyzes the meta-cleavage between two phenolic hydroxyl groups, while the other two enzymes catalyze ortho-cleavage of adjacent groups of phenolic hydroxyl groups (Fig. 12.6).
In bacteria (mainly Gram-negative bacteria), such as SYK-6 and Bacillus Ligniniphilus L1, protocatechuate (PCA) was converted to 4-carboxy-2-hydroxy-muconate semialdehyde (CHMS) through ortho-cleavage catalyzed by protocatechuate-4,5-dioxygenase (LigAB) [6, 129]. Then CHMS is further converted to OMA (4-oxalomesaconate) by CHMS dehydrogenase (LigC). Under the catalysis of PDC hydrolase (LigI), OMA hydratase (LigJ), and CHA aldolase (LigK), CHMS was further converted to pyruvic acid and oxaloacetic acid, which would finally enter into the tricarboxylic acid cycle . In other bacteria (mainly Gram-positive bacteria), such as Pseudomonas putida and calcium acetate Acinetobacter, protocatechuate is oxidized by 3,4-PCD to generate cis-hexanedioic acid and then further degraded by β-ketoadipate metabolic pathway or ortho-cleavage pathway.
12.4.7 Diarylpropane Catabolic Pathways
The degradation of diarylpropanes by fungi has been well studied, which was attributed to the Cα–Cβ cleavage catalyzed by lignin peroxidase. In Phanerochaete chrysosporium, 1-(3,4-dimethoxyphenyl)-2-phenylethylene glycol (dimethoxyhydrobenzoin) (DMHB) was catalyzed by lignin peroxidase to obtain either benzaldehyde and α-hydroxy (dimethoxybenzyl) radical or resverataldehyde and α-hydroxybenzyl radicals. The diarylpropane degradation pathway in bacteria was different from that in fungi. For example, in Pseudomonas paucimobilis TMY1009, the interphenyl double bond in the dimeric lignin model compounds was degraded by dioxygenase with the generation of lignostilbene. Then the latter was further degraded by lignostilbene-α, β-dioxygenase to generate two vanillins which then enters the vanillin metabolic pathway (Fig. 12.7) [9, 58].
12.4.8 Phenylcoumarane and Pinoresinol Lignin Component Catabolic Pathway
Degradation pathways of phenylcoumarane and pinoresinol in Phanerochaete Chrysosporium and Fusarium solani have been revealed. The first step in the metabolic pathway of lignin and lignin-derived components was both referred to the α-hydroxylation [22]. The α-hydroxylated product can generate benzyl ketone by ring opening. Therefore, it was proposed that in bacteria the degradation of phenylcoumarin may be catalyzed by lignostilbene-α, β-dioxygenase. For example, P. xanthomonas degraded the alkylated phenylcoumarin firstly by side chain oxidation, then by heterocyclic oxidation to form a furan, and finally by oxidative cleavage of the Cα–Cβ bond [89]. The filamentous fungal Fusarium solani M-13-1 would break down the phenolic coumarin by directly breaking the Cα–Cβ bond to form 5-acetylvanillin [91]. The degradation of benzyl ketone can produce diarylpropane skeleton and then degrade through diarylpropane catabolic pathway. As for the degradation process of pinoresinol, in F. solani m-13-1, the cleavage of C–O bond was catalyzed according to the oxidation of benzyl group, thus resulting in the generation of monocyclic ketone as intermediate. Further oxidation of the aryl–alkyl group produced a carboxyl group and a corresponding lactone [57].
Sphingomonas SYK-6 can also degrade phenylcoumarin model compounds and rosinol model compounds, but the related genes were not found [9, 79].
12.5 Biocatalytic Conversion of Lignin
Systems biology provides powerful tools to reveal the basic metabolic pathways of lignin. These cognitions are applied to the construction of engineered strains using synthetic biology technology. The lignin-degrading engineered strains were used to produce various value-added products from lignin or lignin-derived aromatic compounds.
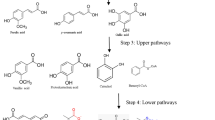
When catalyzed by lignin-degrading enzyme system, lignin is degraded by microorganisms through “upper pathways” to generate heterogeneous mixture of aromatic monomers which can be used as carbon and energy sources by many microorganisms. This process acts as a “biological funnel” to reduce the heterogeneity of carbon for catabolism [55]. It would be a great opportunity if we focus on the production of lignin-derived fuels and chemicals by selectively converting heterogeneous mixture of lignin-derived molecules instead of single intermediate. On the other hand, the lignin-derived aromatic compounds may inhibit the microbial growth, thereby hindering the value-added conversion of lignin. Metabolic engineering and synthetic biology can efficiently reduce the product inhibition. At present, the lignin-derived products through microbial degradation pathway mainly include the secondary metabolic products such as vanillin (vanillin) or cis,cis-muconic acid accumulation through the deletion of genes (blue) and the metabolic pathway of triacylglycerol lipid or polyhydroxybutyrate (green) accumulated by first metabolism (green) (as shown in Fig. 12.8). This section will introduce the typical metabolic engineering cases of lignin.
Production of bioproducts from microbial degradation pathways [8]. The metabolic pathways responsible for accumulation of small molecule products vanillin or cis,cis-muconic acid (in dashed line) and the accumulation of triacylglycerol lipids or polyhydroxybutyrate (in solid line)
12.5.1 Metabolic Engineering for the Production of Lignin-Derived Aromatic Compounds
Lignin, as an aromatic polymer, has always been considered as a renewable source of aromatic chemicals in the future. In principle, the deep research and disclosure of microbial metabolic pathways and expression genes of lignin enable researchers to design biological metabolic pathways for the production of lignin-derived chemicals from renewable materials. In recent years, more and more metabolic pathways converting lignin and its derivatives into aromatic chemicals have been constructed and some engineered strains have been obtained.
12.5.1.1 Vanillin
Vanillin is a high-value chemical that has been widely used in food/condiment industry. More than 99% of vanillin produced in industry is synthesized from petrochemical products or lignin derivatives. At present, there are commercial methods to produce vanillin from lignosulfonate through chemical way. Although lignin can generate aromatic compounds through pyrolysis or chemical catalysis, such methods usually generate complex product mixtures with a total yield of 5–15%. Nowadays, especially in the food industry, increasing interest has been put on the natural and healthy ingredients. Therefore, the alternative source of natural vanillin from biotechnology process has attracted more and more attentions. The most attractive methods to produce vanillin include biosynthesis using natural aromatic compounds like ferulic acid and de novo biosynthesis from primary metabolites like glucose. It has reported that there were several microorganisms that can convert ferulic acid into vanillin, including Pseudomonas, Rhodococcus, Pycnoporus cinnabarinus, etc. The intermediate vanillin is produced by undesired catabolism caused by vanillin dehydrogenase. In order to obtain high yield and concentration of vanillin by preventing vanillin from being oxidized to vanillin acid, Fleige et al. [37] constructed the Amycolatopsis sp. ATCC 39116 vanillin dehydrogenase deletion mutant (ΔVDH). This strain cannot grow on vanillin anymore and cannot secrete vanillin dehydrogenase. When ferulic acid was used as substrate, vanillin concentration was enhanced by 2.3 times, while the vanillin content was greatly reduced. Rhodococcus jostii rHA1, a lignin decomposing bacterium, could grow to high cell density on the smallest medium containing lignocellulose and could tolerate the production of toxic aldehyde metabolites. The vdh gene (ro02986) and vanA gene (ro4165), encoding vanillin dehydrogenase and vanillin demethylase, respectively, were found in R. jostii RHA1. Sainsbury et al. [108] constructed R. jostii rHA1 mutant strain (ΔVDH gene deletion strain RHA045) which deleted vanillin dehydrogenase gene using predictive gene deletion method. The mutant strain could not grow when vanillin is used as sole organic substrate but could grow on vanillic acid. For the ΔVDH mutant, when it grows in the medium containing 2.5% wheat straw lignocellulose and 0.05% glucose, the quantity and intensity of p-hydroxybenzaldehyde, vanillin, and protocatechuic acid (4,5-dihydroxybenzoic acid) in the metabolites were all increased significantly. After being cultivated for 144 h, vanillin was accumulated up to 96 mg/L, accompanied by a small amount of ferulic acid and 4-hydroxybenzaldehyde.
12.5.1.2 Monolignols
Lignols, including coumarin alcohol, caffeinol, coniferol, and mustard alcohol, are important metabolites involved in the lignin biosynthesis of plants, and their derivatives have many physiological and pharmaceutical functions. Chemical reduction route has been developed for the production of monolignols from the corresponding phenylpropionic acid [102]. However, these methods have the disadvantages of expensive raw materials, harsh reaction conditions, low total yield, etc.
There have been some studies on the biosynthesis of coumarin by selecting effective enzymes. Then, the pathway could be further expanded to produce caffeinol and terpineol by introducing hydroxylase and methyltransferase. Jansen et al. [51], for the first time, constructed a complete artificial phenylpropane pathway of p-coumarin in E. coli by extending the metabolic pathway of L-tyrosine with the combination of genes from various plants and microorganisms. This pathway was co-expressed with four enzymes including L-tyrosine ammonia lyase (RsTAL) from Rhodobacter sphaeroides, p-coumaric acid CoA ligase (Pc4CL) from Petroselinum crispum, cinnamoyl-CoA reductase (ZmCCR) from Zea mays, and cinnamyl alcohol dehydrogenase (ZmCAD) in E. coli. Firstly, L-tyrosine is deaminated by RsTAL to generate p-coumaric acid which was further activated by CoA through p-coumaric acid CoA ligase (4CL) and consequently was reduced to p-coumaric acid by ZmCCR and ZmCAD. By co-expression of RsTAL, Pc4CL, ZmCCR, and ZmCAD, p-coumaryl alcohol could be directly generated from LB by the constructed strain.
However, the productivity of the artificial pathway was low which might be attributed to the low TAL activity, since the production of p-coumaryl alcohol was increased by five times when cultivating E. coli with 1 mM coumaric acid as substrate. Phosphorothioate-based ligase-independent gene cloning (PLICing) method has become an attractive method characterized as single-gene, enzyme-free, and sequence-independent cloning. During the assembly of DNA fragments, the Operon-PLICing method changes the interval between the Shine–Dalgarno sequence and the START codon to balance the expression of all genes in the metabolic pathway at the translation level, thereby maximizing the concentration of the target product and providing an opportunity [7]. Based on PLICing method, Jennifer et al. [53] developed Operon-PLICing method for the rapid assembly of p-coumaryl alcohol synthetic pathways. They heterologously expressed the encoding genes of the four enzymes required for the complete metabolic pathway in E. coli. With Operon-PLICing, 81 different clones were constructed within a few days, each of which carried a different p-coumaryl alcohol operon. The p-coumarol concentration of the selected five optimal mutants was ranging from 48 to 52 mg L−1.
High catalytic activity of enzymes is the prerequisite for effective production of p-coumaryl alcohol [53]. In order to increase the production of p-coumarol, Chen et al. [15] screened TAL, 4CL1, CCR, and AD with high catalytic activity from different origins to construct a new synthetic pathway. By the combination of RgTAL from Pseudomonas glutamicum, the At4CL1 from A. thaliana, the LlCCR from Lactobacillus leucocephala, and the ScADH6 from Saccharomyces cerevisiae, the constructed strain can effectively transform the phenylpropionic acids into corresponding alcohols. The titer of p-coumaryl alcohol generated by the constructed strain was up to 656.0 ± 9.9 mg L−1 (Fig. 12.9).
Biosynthetic pathways of monolignols derived from lignin [15]. TAL tyrosine ammonia lyase, 4CL1 p-coumarate-CoA ligase, CCR cinnamoyl-CoA reductase, ADH6 alcohol dehydrogenase, HpaBC 4-hydroxypropanoic acid 3-hydroxylase , CCoAOMT caffeoyl-CoA O-methyltransferase, COMT caffeate 3-O-methyltransferase
12.5.2 Metabolic Engineering for Biomaterial Production from Lignin
12.5.2.1 Muconate/Muconic Acid
With the increase of oil price and the decrease of resources, people are increasingly interested in the production of bioplastics from renewable resources through environmentally friendly fermentation. Cis,cis-muconic acid (MA) is one of the most valuable diunsaturated dicarboxylic acids of industrial value. It can be further converted into basic material and monomer of commercial polymer materials, such as adipic acid or terephthalic acid, which are widely used in cosmetics, pharmaceutical, and food industries.
In recent years, many different functional engineering strains have been designed, such as Pseudomonas putida KT2440, Amycolatopsis species ATCC 39116, and E. coli, to produce MA either via biosynthesis from glucose and glycerol or via biotransformation from aromatic synthesis of biotransformed MA from glucose, glycerin, and even aromatic compounds. The production of MA from aromatic compounds is of great significance because it requires only a few biochemical reaction to achieve 100% molar yield. So far, it has been proved that different aromatic compounds (including benzoate, toluene, catechol , phenol, p-coumarate guaiacol) can be used in the production of MA. More importantly, the strains that can use aromatic compounds to produce MA provide the possibility for the production of MA from lignin-derived hydrolysate in the future.
12.5.2.1.1 Construction of Engineered C. glutamicum for MA Production
Most biotechnology processes that use Corynebacterium glutamate cell factories rely on sugar causing the competition with human nutrition. The β-ketoadipate pathway is the main pathway for assimilation of lignin-derived aromatic compounds, which indicated that engineered C. glutamicum strains can produce MA from lignin-based aromatic compounds if the gene encoding muconate cycloisomerase was deleted. Berker et al. deleted the gene encoding muconate cycloisomerase from the genome of wild-type C. glutamicum ATCC 13032 to block the catabolism of small aromatics at the MA level resulting in the accumulation of MA [56].
Contrary to the wild type, the obtained mutant C. glutamicum MA-1 can no longer grow on the aromatic compounds benzoic acid, catechol, and phenol as the sole carbon sources. In contrast, by adding a small amount of glucose, the consumption of aromatic compounds and the growth of strain were observed. Within 24 h, 5 mM phenol, 10 mM catechol, and 20 mM benzoic acid were completely transformed by C. glutamicum MA-1. The mutated strain C. glutamicum MA-2 was further obtained through constitutive overexpression, which could fasten the conversion of catechol into MA. During batch feeding, the mutant strain MA-2 accumulated 85 g L−1 mA from catechol within 60 h, and the maximum yield was 2.4 g L−1 h−1. Moreover, engineering strain MA-2 successfully converted lignin-derived hydrolysate into MA, reaching 12.5 mm (1.8 g L−1) within 27 h.
12.5.2.1.2 Construction of Genetic Engineering P. putida for MA Production
In P. putida , muconate is produced by catechol-1,2-dioxygenase, which is a catechol branch of the phthalate pathway. Protocatechuate is metabolized through another branch of the β-ketoadipate pathway, which does not use MA as an intermediate. Given that many lignin derivatives will be metabolized through one of these aromatic intermediates, Vardon et al. [124] replaced the gene encoding protocatechin-3,4-dioxygenase with the gene encoding protocatechin decarboxylase from Enterobacter cloacae . This enabled the conversion of protocatechuic acid and upstream metabolites to catechol while eliminating the further catabolism of protocatechuic acid to β-ketoadipate. This resulted in the transition of aromatic compounds from catechol and protocatechuate branches of the β-ketoadipate pathway to muconate. Acetate was used as carbon source and energy source to evaluate the muconate productivity of engineered P. putida KT2440-CJ103 using lignin-derived monomers, which can successfully convert catechol, phenol, and benzoate into muconate via catechol branch and convert protocatechuate, turpentine oil, ferulate, vanillin, caffeine, p-coumarate, and 4-hydroxybenzoate via protocatechuate branch.
Taking p-coumaric acid as carbon source, the cis-muconate yield of P. putida KT2440-CJ103 was 13.5 g/L when cultivated in a fed-batch bioreactor for 78.5 h. Alkaline pretreated liquor, mainly containing p-coumarate, ferulate, glycolate, and acetate, was used for the cultivation of P. putida KT2440-CJ103, which indicated that glycolate and acetate were quickly consumed and used as source of carbon and energy required for cell growth and a yield of 0.7 g L−1 cis-muconate was obtained after 24 h.
12.5.2.1.3 Construction of Genetic Engineering E. coli for MA Production
Microorganisms with the ability to metabolize aromatic compounds, such as Sphingomonas SYK-6, can take syringate and vanillin (main lignin-derived aromatic compounds) as carbon sources and metabolize them into small molecular metabolites, such as protocatechuate or catechol. The vanillin catabolic pathway is responsible for providing SYK-6 with sufficient nicotinamide adenine dinucleotide (NADH) and nicotinamide adenine dinucleotide phosphate (NADPH), which is critical for microbial growth. Sphingomonas SYK-6 has been proved to be extremely highly tolerant to high pH, which is a desirable characteristic for the high-value utilization of lignin. Moreover, it was found that the decomposition of lignin is a mechanism required for SYK-6 to survive [125]. The unique lignin-catabolizing characteristics of SYK-6 made it attractive in the high-value utilization of lignin. However, it was difficult for SYK-6 to be used as a host for lignin valorization since there were too many genes needed to be knocked out and overexpressed. Wu et al. [127] used vanillin as raw material which was derived from hydrogen peroxide catalyzed Kraft lignin to produce cis,cis-muconic acid by a newly developed pathway. Based on substrate consumption, Pseudomonas putida mt-239 containing the gene encoding catechol 1,2-dioxygenase (CatApmt2) obtained the highest yield of cis,cis-muconic acid (314 mg L−1), followed by Acinetobacter calcoaceticus 40 containing the gene encoding catechol 1,2-dioxygenase (CatAac) (238 mg L−1).
12.5.2.2 Pyruvate
Pyruvate is the key intermediate for the sugar metabolism of all biological cells and the mutual conversion of various substances in vivo. Owing to containing active ketone and carboxyl groups, pyruvate is widely used in chemical, pharmaceutical, food, agriculture, environmental protection, and other fields as a basic chemical raw material. It has been developed as the fundamental strategies for the metabolic engineering of the bio-based production of amino acids, alcohols, terpenoids, lactate, etc. In aerobic organisms, the aromatic products (such as benzoate and p-coumarate benzoate and p-coumarate) first degraded aerobically into intermediates such as protocatechuate and catechol. Then the intermediates would be cleaved by the O2-dependent dioxygenase. However, according to the difference position of adjacent hydroxyl groups (ortho-position or meta-position), the oxygenase would catalyze the aromatic ring cleavage of catechol and protocatechuate through different degradation pathways to generate different combinations of succinate, acetyl-CoA, and pyruvate. Then the products would be metabolized through parallel pathways and finally entry into the TCA cycle. The ortho cleavage pathway of catechol and protocatechuate produces succinate and acetyl-CoA, while, the meta-cleavage pathway of catechol generates pyruvate and acetyl-CoA. The products of 2,3-meta-cleavage pathway and 4,5-meta-cleavage pathway of protocatechuate were different; the former generate pyruvate and acetyl-CoA, while the latter produce two pyruvate molecules [55].
The metabolic pathway of the target product derived from lignin degradation could be optimized. Generally, the meta-cleavage pathway of catechol and PCA is superior to the ortho-cleavage pathway. Therefore, the pyruvate yield from lignin aromatic molecules produced by a reconstructed strain by replacing the P. putida KT2440 endogenous catechol ortho-degradation pathway with the exogenous meta-cleavage pathway from P. putida MT-2 was increased by approximately 10%. Moreover, the pyruvate production would be increased by nearly five times if replacing the endogenous protocatechuate ortho-cleavage pathway with the meta-cleavage pathway from Sphingobium sp. SYK-6. Meanwhile, the pyruvate can further be converted into L-lactic acid aerobically, which will enhance the total pyruvate production consequently [55].
12.5.2.3 Polyhydroxyalkanoates (PHA)
Plastic products are the necessities of people’s daily life and industrial and agricultural production. The widely used petroleum-based plastics cause serious environmental pollution and consume a lot of unrenewable resources. Polyhydroxyalkanoate (PHA) is a kind of environmentally friendly plastics, which can be completely biodegraded. The physical and chemical properties and processing performances of PHA are similar to traditional plastics. Therefore, PHA is an ideal substitute for petroleum-based plastics and has broad application prospects. Actually, PHA has been widely used in coatings, films, biomedical materials, biocompatible drug delivery, and organic/inorganic composite bioplastics [131].
Some microbes can produce PHA, and then PHA can be reserved as carbon source under specific nutritional constraints. Therefore, the microbial synthesis of PHAs is the major method for the industrial preparation of PHA . It has been found that PHA can be transformed from lignocellulose by microorganisms such as Ralstonia eutropha, Pseudomonas putida, and Bacillus megaterium [16]. Many bacteria have developed complex metabolism for lignin conversion with advanced enzyme systems for PHA production. Therefore, screening high-yield strains and choosing cheap, renewable, and wide-ranging wastes as substrates for PHA synthesis are of great significance for reducing production cost of PHA, recycling, and valorization of wastes.
The β-ketoadipate pathway also plays an important role in lignin conversion, because aromatic compounds derived from lignin can be used to synthesize high-molecular-weight compounds, including lipids and polyhydroxybenzoates (PHA). As the most thoroughly studied Pseudomonas, P. putida KT2440 had various metabolic functions. It had acyl-CoA synthetase that encodes fadD gene, β-oxidation pathway that provides 3-OH-aryl-CoA, and polymerization–depolymerization system that integrates two polymerases (PhaC1 and PhaC2) and a depolymerase (PhaZ). However, P. putida KT2440 specifically catalyzed the complete assimilation of phenylacetyl-CoA or benzoyl-CoA generated from these polyesters through two β-oxidation pathways in the cell. It was found that blocking either fadD or PhaC1 can inhibit the synthesis and accumulation of plastic polymers in cells. By disrupting PhaC2, the storage quantity of polymers could be reduced by two-thirds. The blocking of PhaZ could hinder the metabolic pathway of polymers, thus reducing its production. The productivity of plastic polymer in the mutant that absence of glyoxylate cycle or β-oxidation pathway was relatively higher than that in the wild strain [41]. The aromatic model compounds and heterogeneous lignin hydrolytes derived from alkaline pretreated lignin can be converted into medium-chain-length polyhydroxyalkanoates (mcl-PHAs). When they were used as substrate for P. putida KT2440, 36% of PHA was accumulated in the cell of P. putida after 48 h [72]. Olivera et al. [92] also proposed that the simple mutation or deletion of genes involved in the β-oxidation pathway in P. putida could accumulate amount of PHAs that account for more than 55% of the cell dry weight of mutants. Genetically bacteria Pseudomonas putida was used to produce new type of bioplastic containing aromatic or aliphatic and aromatic monomer mixture. The mutation (−) or deletion (Δ) of some genes in the β-oxidation pathway (fadA−, fadB−, ΔfadA, or Δfad BA mutants) can lead to the accumulation of abnormal homopolymers or copolymers in cells. The morphology of microorganisms changed significantly, since these macromolecular substances occupied 90% of the cytoplasm. Introducing blockade into the β-oxidation pathway or other related catabolic pathways allows mutant strains to synthesize polymers different from wild-type strains, to accumulate some intermediate products that are rapidly decomposed in wild type, and to accumulate the terminal decomposition products (such as phenylacetic acid, phenylbutyric acid, trans-cinnamic acid, or its derivatives) in the fermentation broth.
References
Ademakinwa, A. N., & Agboola, F. K. (2016). Biochemical characterization and kinetic studies on a purified yellow laccase from newly isolated Aureobasidium pullulans NAC8 obtained from soil containing decayed plant matter. Journal of Genetic Engineering & Biotechnology, 14(1), 143–151.
Ai, M. Q., Wang, F. F., Zhang, Y. Z., et al. (2014). Purification of pyranose oxidase from the white rot fungus Irpex lacteus and its cooperation with laccase in lignin degradation. Process Biochemistry, 49(12), 2191–2198.
Anita, S., Aggarwal Neeraj, K., Sharma, A., et al. (2015). Actinomycetes: A source of lignocellulolytic enzymes. Enzyme Research, 2015, 1–15.
Arias, M. E., Blanquez, A., Hernandez, M., et al. (2016). Role of a thermostable laccase produced by Streptomyces ipomoeae in the degradation of wheat straw lignin in solid state fermentation. Journal of Analytical & Applied Pyrolysis, 122, 202–208.
Augustin, C. M., Parvu, M., Damian, G., et al. (2012). A “yellow” laccase with “blue” spectroscopic features, from Sclerotinia sclerotiorum. Process Biochemistry, 47(6), 968–975.
Barry, K. P., & Taylor, E. A. (2013). Characterizing the promiscuity of LigAB, a lignin catabolite degrading extradiol dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry, 52(38), 6724–6736.
Blanusa, M., Schenk, A., Sadeghi, H., et al. (2010). Phosphorothioate-based ligase-independent gene cloning (PLICing): An enzyme-free and sequence-independent cloning method. Analytical Biochemistry, 406(2), 141–146.
Bugg, T. D., & Rahmanpour, R. (2015). Enzymatic conversion of lignin into renewable chemicals. Current Opinion in Chemical Biology, 29, 10–17.
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., et al. (2011). Pathways for degradation of lignin in bacteria and fungi. Natural Product Reports, 28(12), 1883–1890.
Camarero, S., Sarkar, S., Ruiz-Duenas, F. J., et al. (1999). Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. Journal of Biological Chemistry, 274(15), 10324–10330.
Carabajal, M., Kellner, H., Levin, L., et al. (2013). The secretome of Trametes versicolor grown on tomato juice medium and purification of the secreted oxidoreductases including a versatile peroxidase. Journal of Biotechnology, 168(1), 15–23.
Castro, A. I. R. P., Evtuguin, D. V., & Xavier, A. M. B. (2003). Degradation of biphenyl lignin model compounds by laccase of Trametes versicolor in the presence of 1-hydroxybenzotriazole and heteropolyanion [SiW11VO40]5−. Journal of Molecular Catalysis B: Enzymatic, 22(1–2), 13–20.
Chaurasia, P. K., Yadav, R. S. S., & Yadava, S. (2014). Purification and characterization of yellow laccase from Trametes hirsuta MTCC-1171 and its application in synthesis of aromatic aldehydes. Process Biochemistry, 49(10), 1647–1655.
Chen, Y., Liyuan, C., Chongjian, T., et al. (2012). Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process. Bioresour Technology, 123, 682–685.
Chen, Z., Sun, X., Li, Y., et al. (2017). Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metabolic Engineering, 39, 102–109.
Chen, Z., & Wan, C. (2017). Biological valorization strategies for converting lignin into fuels and chemicals. Renewable & Sustainable Energy Reviews, 73, 610–621.
Chi, Y., Wu, S., & Yu, C. (2019). Regulation of Mn2+on three manganese peroxidase genes Lg-mnp1, 2, and 3 at transcriptional level. Journal of Jilin Agricultural University, 41(5), 540–552.
Chong, G. G., Huang, X. J., Di, J. H., et al. (2018). Biodegradation of alkali lignin by a newly isolated Rhodococcus pyridinivorans CCZU-B16. Bioprocess & Biosystems Engineering, 41(4), 501–510.
Coconi-Linares, N., Magaña-Ortíz, D., Guzmán-Ortiz, D. A., et al. (2014). High-yield production of manganese peroxidase, lignin peroxidase, and versatile peroxidase in Phanerochaete chrysosporium. Applied Microbiology & Biotechnology, 98(22), 9519–9519.
Colpa, D. I., Fraaije, M. W., & Bloois, E. (2014). DyP-type peroxidases: A promising and versatile class of enzymes. Journal of Industrial Microbiology & Biotechnology, 41(1), 1–7.
Crowford, D. L. (1983). Lignin degradation by Streptomyces viridosporus: Isolation and characterization of a new polymeric lignin degradation intermediate. Applied & Environmental Microbiology, 45(3), 898–904.
Cui, F., & Dolphin, D. (1992). Iron porphyrin catalyzed oxidation of lignin model compounds: The oxidation of veratryl alcohol and veratryl acetate. Canadian Journal of Chemistry, 70(8), 2314–2318.
Daniel, G., Volc, J., & Kubatova, E. (1994). Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Applied and Environmental Microbiology, 60(7), 2524–2532.
Daou, M., & Faulds, C. B. (2017). Glyoxal oxidases: Their nature and properties. World Journal of Microbiology and Biotechnology, 33(5), 87–97.
de Oliveira, P. L., Duarte, M. C. T., Ponezi, A. N., et al. (2009). Purification and partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz J Microbiol, 40, 818–826.
Duan, J., Huo, X., Du, W. J., et al. (2016). Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp WJDL-Y2. Letters in Applied Microbiology, 62(1), 55–62.
Duan, J., Liang, J. D., Du, W. J., et al. (2014). Biodegradation of Kraft lignin by a bacterial strain Sphingobacterium sp. HY-H. Advanced Materials Research, 955–959, 548–553.
Duan, Z., Rui, S., Liu, B., et al. (2018). Comprehensive investigation of a dye-decolorizing peroxidase and a manganese peroxidase from Irpex lacteus F17, a lignin-degrading basidiomycete. AMB Express, 8(1), 119–134.
Elsalam, H. E. A., & Bahobail, A. S. (2016). Lignin biodegradation by thermophilic bacterial isolates from Saudi Arabia. Biological Chemistry Science, 7, 1413–1424.
Eriksson, K. E., Pettersson, B., Volc, J., et al. (1986). Formation and partial characterization of glucose-2-oxidase, a H2O2 producing enzyme in Phanerochaete chrysosporium. Applied Microbiology & Biotechnology, 23(3–4), 257–262.
Fan, Y., Zhang, Z., Wang, F., et al. (2019). Lignin degradation in corn stover catalyzed by lignin peroxidase from Aspergillus oryzae broth: Effects of conditions on the kinetics. Renewable Energy, 130, 32–40.
Faraco, V., Alessandra, P., Giovanni, S., et al. (2007). Identification of a new member of the dye-decolorizing peroxidase family from Pleurotus ostreatus. World Journal of Microbiology & Biotechnology, 23(6), 889–893.
Feng, H., Jing, F., Lu, X., et al. (2001). Synergistic effects of cellobiose dehydrogenase and manganese-dependent peroxidases during lignin degradation. Chinese Science Bulletin, 46(23), 1956–1962.
Feng, H., Sun, Y., Zhi, Y., et al. (2015). Lignocellulose degradation by the isolate of Streptomyces griseorubens JSD-1. Functional & Integrative Genomics, 15(2), 163–173.
Fernández, I. S., Ruíz-Dueñas, F. J., Santillana, et al. (2009). Novel structural features in the GMC family of oxidoreductases revealed by the crystal structure of fungal aryl-alcohol oxidase. Acta Crystallographica, 65(11), 1196–1205.
Ferri, S., Kojima, K., & Sode, K. (2011). Review of glucose oxidases and glucose dehydrogenases: A bird's eye view of glucose sensing enzymes. Journal of Diabetes Science and Technology, 5(5), 1068–1076.
Fleige, C., Hansen, G., Kroll, J., et al. (2013). Investigation of the Amycolatopsis sp. Strain ATCC 39116 vanillin dehydrogenase and its impact on the biotechnical production of vanillin. Applied and Environmental Microbiology, 79(1), 81–90.
Francisco, J. R.-D., & ángel, T. M. (2009). Microbial degradation of lignin: How a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microbial Biotechnology, 2(2), 164–177.
Gall, D. L., Ralph, J., Donohue, T. J., et al. (2017). Biochemical transformation of lignin for deriving valued commodities from lignocellulose. Current Opinion in Biotechnology, 45, 120–126.
Garcia-Ruiz, E., Mate, D., Gonzalez-Perez, D., et al. (2014). Directed evolution of ligninolytic oxidoreductases: From functional expression to stabilization and beyond. In S. Riva & W. D. Fessner (Eds.), Cascade biocatalysis: Integrating stereoselective and environmentally friendly reactions (pp. 1–22). Weinheim: Wiley.
Garcia, B., Olivera, E. R., Minambres, B., et al. (1999). Novel biodegradable aromatic plastics from a bacterial source: genetic and biochemical studies on a route of the phenylacetyl-coa catabolon. Journal of Biological Chemistry, 274(41), 29228–29241.
Glenn, J. K., & Gold, M. H. (1985). Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Archives of Biochemistry and Biophysics, 242, 329–341.
Godden, B., Ball, A. S., Helvensteian, P., et al. (1992). Towards elucidation of the lignin degradation pathway in actinomycetes. Journal of General Microbiology, 138, 2441–2448.
Grzegorz, J., Anna, P., Justyna, S., et al. (2017). Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. Fems Microbiology Reviews, 41(6), 1–22.
Hakala, T. K., Kristiina, H., Pekka, M., et al. (2006). Differential regulation of manganese peroxidases and characterization of two variable MnP encoding genes in the white-rot fungus Physisporinus rivulosus. Applied Microbiology & Biotechnology, 73(4), 839–849.
Hernández-Ortega, A., Ferreira, P., & Martínez, A. T. (2012). Fungal aryl-alcohol oxidase: A peroxide-producing flavoenzyme involved in lignin degradation. Applied Microbiology & Biotechnology, 93(4), 1395–1410.
Higuchi, T. (2004). Microbial degradation of lignin: Role of lignin peroxidase, manganese peroxidase, and laccase. Proceedings of the Japan Academy Ser B Physical and Biological Sciences, 80(5), 204–214.
Hofrichter, M., & René, U. (2006). Heme-thiolate haloperoxidases: Versatile biocatalysts with biotechnological and environmental significance. Applied Microbiology and Biotechnology, 71(3), 276–288.
Hofrichter, M., Ullrich, R., Pecyna, M. J., et al. (2010). New and classic families of secreted fungal heme peroxidases. Applied Microbiology & Biotechnology, 87(3), 871–897.
Ike, P. T. L., Moreira, A. C., Almeida, d., et al. (2015). Functional characterization of a yellow laccase from Leucoagaricus gongylophorus. SpringerPlus, 4(1), 654.
Jansen, F., Gillessen, B., Mueller, F., et al. (2014). Metabolic engineering for p-coumaryl alcohol production in Escherichia coli by introducing an artificial phenylpropanoid pathway. Applied Biochemistry and Biotechnology, 61, 646–654.
Janusz, G., Pawlik, A., Sulej, J., et al. (2017). Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiology Reviews, 41(6), 941–962.
Jennifer, A., Philana, V. S. W., Sokolowsky, S., et al. (2016). Combinatorial optimization of synthetic operons for the microbial production of p-coumaryl alcohol with Escherichia coli. New Biotechnology, 33, S26–S27.
Johjima, T., Ohkuma, M., & Kudo, T. (2003). Isolation and cDNA cloning of novel hydrogen peroxide-dependent phenol oxidase from the basidiomycete Termitomyces albuminosus. Applied Microbiology and Biotechnology, 61(3), 220–225.
Johnson, C. W., & Beckham, G. T. (2015). Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metabolic Engineering, 28, 240–247.
Judith, B., Kuhl, M., Kohlstedt, M., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microbial Cell Factories, 17(1), 115.
Kamaya, Y., Nakatsubo, F., Higuchi, T., et al. (1981). Degradation of d,l-syringaresinol, a β-β′ linked lignin model compound, by Fusarium solani M-13-1. Archives of Microbiology, 129(4), 305–309.
Kamoda, S., & Saburi, Y. (1993). Cloning, expression, and sequence analysis of a lignostilbene- α,β -dioxygenase gene from Pseudomonas paucimobilis TMY1009. Journal of the Agricultural Chemical Society of Japan, 57(6), 926–930.
Kersten, P. J. (1990). Glyoxal oxidase of Phanerochaete chrysosporium: Its characterization and activation by lignin peroxidase. Proceedings of the National Academy of Sciences of the United States of America, 87(8), 2936–2940.
Khare, S., & Prakash, O. (2017). Current developments in biotechnology and bioengineering: Production, isolation and purification of industrial products. Journal of Cleaner Production, 158, 380–381.
Khatami, S., Deng, Y., Tien, M., et al. (2019). Formation of water-soluble organic matter through fungal degradation of lignin. Organic Geochemistry, 135, 64–70.
Kim, S. J., & Shoda, M. (1999). Purification and characterization of a novel peroxidase from Geotrichum candidum Dec1 involved in decolorization of dyes. Applied and Environmental Microbiology, 65(3), 1029–1035.
Koker, T. H. D., Mozuch, M. D., Cullen, D., et al. (2004). Isolation and purification of pyranose 2-oxidase from Phanerochaete chrysosporium and characterization of gene structure and regulation. Applied & Environmental Microbiology, 70(10), 5794–5800.
Kong, W., Fu, X., Wang, L., et al. (2017). A novel and efficient fungal delignification strategy based on versatile peroxidase for lignocellulose bioconversion. Biotechnology for Biofuels, 10(1), 218.
Kour, D., Rana, K. L., Yadav, N., et al. (2019). Agriculturally and industrially important fungi: Current developments and potential biotechnological applications. In A. N. Yadav, S. Singh, S. Mishra, & A. Gupta (Eds.), Recent advancement in white biotechnology through fungi: Volume 2: Perspective for value-added products and environments (pp. 1–64). Cham: Springer.
Kujawa, M., Ebner, H., Leitner, C., et al. (2006). Structural basis for substrate binding and regioselective oxidation of monosaccharides at C3 by pyranose 2-oxidase. Journal of Biological Chemistry, 281(46), 35104–35115.
Kukolya, J., Dobolyi, C., & Hornok, L. (1997). Isolation and identification of thermophilic cellulolytic actinomycetes. Acta Phytopathologica et Entomologica Hungarica, 32(1–2), 97–107.
Kumar, V. V., & Rapheal, V. S. (2011). Induction and purification by three-phase partitioning of aryl alcohol oxidase (AAO) from Pleurotus ostreatus. Applied Biochemistry and Biotechnology, 163(3), 423–432.
Kupryashina, M. A., Selivanov, N. Y., & Nikitina, V. E. (2012). Isolation and purification of Mn-peroxidase from Azospirillum brasilense SP245. Applied Biochemistry and Microbiology, 48(1), 17–20.
Leonowicz, A., Rogalski, J., Jaszek, M., et al. (1999). Cooperation of fungal laccase and glucose l-oxidase in transformation of Björkman lignin and some phenolic compounds. Holzforschung, 53(4), 376–380.
Li, X. (2019). Pathway study of lignin degradation by Aspergillus fumigatus from buffalo rumen, Vol. Master, Huazhong Agricultural University.
Linger, J. G., Vardon, D. R., Guarnieri, M. T., et al. (2014). Lignin valorization through integrated biological funneling and chemical catalysis. Proceedings of the National Academy of Sciences of the United States of America, 111(33), 12013–12018.
Liu, G., & Qu, Y. (2018). Engineering of filamentous fungi for efficient conversion of lignocellulose: Tools, recent advances and prospects. Biotechnology Advances, 37(4), 519–529.
Liu, Z. H., Le, R. K., Kosa, M., et al. (2019). Identifying and creating pathways to improve biological lignin valorization. Renewable and Sustainable Energy Reviews, 105, 349–362.
Ma, R., Zhao, F., & Zhao, H. (2017). Reveals the pathway of lignin degradation by studied the transcriptome of Pleurotus ostreatus fermented with corn straw. Feed Research, 9, 42–48.
Manavalan, T., Manavalan, A., & Heese, K. (2015). Characterization of lignocellulolytic enzymes from white-rot fungi. Current Microbiology, 70, 485–498.
Martínez, A. T. (2002). Molecular biology and structure-function of lignin-degrading heme peroxidases. Enzyme & Microbial Technology, 30(4), 425–444.
Marzullo, L., Cannio, R., Giardina, P., et al. (1995). Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. Journal of Biological Chemistry, 270(8), 3823–3827.
Masai, E., Katayama, Y., & Fukuda, M. (2007). Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Journal of the Agricultural Chemical Society of Japan, 71(1), 1–15.
Masai, E., Sasaki, M., Minakawa, Y., et al. (2004). A novel tetrahydrofolate-dependent o-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. Journal of Bacteriology, 186(9), 2757–2765.
Mathews, S. L., Pawlak, J., & Grunden, A. M. (2015). Bacterial biodegradation and bioconversion of industrial lignocellulosic streams. Applied Microbiology and Biotechnology, 99, 2939–2954.
Mathieu, Y., Piumi, F., Valli, R., et al. (2016). Activities of secreted aryl alcohol quinone oxidoreductases from Pycnoporus cinnabarinus provide insights into fungal degradation of plant biomass. Applied and Environmental Microbiology, 82, 2411–2423.
Michael, B., Sabine, B., Dorothee, H. P., et al. (2004). Crystal structure of pyranose 2-oxidase from the white-rot fungus Peniophora sp. Biochemistry, 43(37), 11683–11690.
Min, K., Gong, G., Woo, H. M., et al. (2015). A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Scientific Reports, 5, 8245–8252.
Min, K. L., Kim, Y. H., Kim, Y. W., et al. (2001). Characterization of a novel laccase produced by the wood-rotting fungus Phellinus ribis. Archives of Biochemistry and Biophysics, 392(2), 279–286.
Moreno, A. D., Ibarra, D., Eugenio, M. E., et al. (2020). Laccases as versatile enzymes: From industrial uses to novel applications. Journal of Chemical Technology & Biotechnology, 95(3), 481–494.
Mukhopadhyay, M., & Banerjee, R. (2015). Purification and biochemical characterization of a newly produced yellow laccase from Lentinus squarrosulus MR13. Biotech, 5(3), 227–236.
Munk, L., Sitarz, A. K., Kalyani, D. C., et al. (2015). Can laccases catalyze bond cleavage in lignin? Biotechnology Advances, 33(1), 13–24.
Nakatsubo, F., Kirk, T. K., Shimada, M., et al. (1981). Metabolism of a phenylcoumaran substructure lignin model compound in ligninolytic cultures of Phanerochaete chrysosporium. Archives of Microbiology, 128(4), 416–420.
Niladevi, K. N., & Mangrove, P. P. (2005). Actinomycetes as the source of ligninolytic enzymes. Actinomycetologica, 19, 40–47.
Ohta, M., Higuchi, T., & Iwahara, S. (1979). Microbial degradation of dehydrodiconiferyl alcohol, a lignin substructure model. Archives of Microbiology, 121(1), 23–28.
Olivera, E., Carnicero, D., Jodra, R., et al. (2001). Genetically engineered Pseudomonas: A factory of new bioplastics with broad applications. Environmental Microbiology, 3(10), 612–618.
Palmieri, G., Giardina, P., Bianco, C., et al. (2000). Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Applied & Environmental Microbiology, 66(3), 920–924.
Parshetti, G. K., Supriya, P., Dayanand, C. K., et al. (2012). Industrial dye decolorizing lignin peroxidase from Kocuria rosea MTCC 1532. Annals of Microbiology, 62(1), 217–223.
Paszczyński, A., Huynh, V. B., & Crawford, R. (1986). Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Archives of Biochemistry and Biophysics, 244(2), 750–765.
Peralta, R.M., da Silva, B.P., Gomes Côrrea, et al.., 2017. Charpt 5 Enzymes from basidiomycetes-peculiar and efficient tools for biotechnology. In G. Brahmachari (Ed.), Biotechnology of microbial enzymes. Springer, 1–64. Academic, 119–149.
Pere, P., Domínguez, D. M. P., & Anett, S. (2015). From gene to biorefinery: Microbial β-etherases as promising biocatalysts for lignin valorization. Frontiers in Microbiology, 6, 916.
Piontek, K., Ullrich, R., Liers, C., et al. (2010). Crystallization of a 45kDa peroxygenase/peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron. Acta Crystallographica, 66(6), 693–698.
Piumi, F., Levasseur, A., Navarro, D., et al. (2014). A novel glucose dehydrogenase from the white-rot fungus Pycnoporus cinnabarinus: production in Aspergillus niger and physicochemical characterization of the recombinant enzyme. Applied Microbiology and Biotechnology, 98, 10105–10118.
Pozdniakova, N. N., Turkovskaia, O. V., Iudina, E. N., et al. (2006). Yellow laccase from the fungus Pleurotus ostreatus D1: purification and characterization. Applied Biochemistry & Microbiology, 42(1), 56–61.
Rahmanpour, R., Rea, D., Jamshidi, S., et al. (2016). Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Archives of Biochemistry & Biophysics, 594, 54–60.
Ralph, J., Helm, F. R., Quideau, S., et al. (1992). Lignin-feruloyl ester cross-links in grasses. Part 1. Incorporation of feruloyl esters into coniferyl alcohol dehydrogenation polymers. ChemInform, 24(21), 2961–2969.
Rinaldi, R., Jastrzebski, R., Clough, M. T., et al. (2016). Paving the way for lignin valorisation: Recent advances in bioengineering, biorefining and catalysis. Angewandte Chemie International Edition, 55(29), 8164–8215.
Roberts, J. N., Singh, R., Grigg, J. C., et al. (2011). Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry, 50(23), 5108–5119.
Rola, B., Pawlik, A., Frąc, M., et al. (2015). The phenotypic and genomic diversity of Aspergillus strains producing glucose dehydrogenase. Acta Biochimica Polonica, 62(4), 747–755.
Rosini, E., Allegretti, C., Melis, R., et al. (2016). Cascade enzymatic cleavage of the beta-O-4 linkage in a lignin model compound. Catalysis Science & Technology, 6, 2195–2205.
Ruiz-Dueñas, F. J., Fernández, E., Martínez, M. J., et al. (2011). Pleurotus ostreatus heme peroxidases: An in silico analysis from the genome sequence to the enzyme molecular structure. Comptes Rendus Biologies, 334(11), 795–805.
Sainsbury, P. D., Hardiman, E. M., Ahmad, M., et al. (2013). Breaking down lignin to high-value chemicals: The conversion of lignocellulose to vanillin in a gene deletion mutant of Rhodococcus jostii RHA1. Acs Chemical Biology, 8(10), 2151–2156.
Sainsbury, P. D., Mineyeva, Y., Mycroft, Z., et al. (2015). Chemical intervention in bacterial lignin degradation pathways: Development of selective inhibitors for intradiol and extradiol catechol dioxygenases. Bioorganic Chemistry, 60, 102–109.
Sánchez, C. (2009). Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnology Advances, 27(2), 185–194.
Santos, A., Mendes, S., Brissos, V., et al. (2013). New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Applied Microbiology & Biotechnology, 98(5), 2053–2065.
Sharma, M., Chaurasia, P. K., Yadav, A., et al. (2016). Purification and characterization of a thermally stable yellow laccase from Daedalea flavida MTCC-145 with higher catalytic performance towards selective synthesis of substituted benzaldehydes. Russian Journal of Bioorganic Chemistry, 42(1), 59–68.
Sigoillot, J. C., Berrin, J. G., Bey, M., et al. (2012). Fungal strategies for lignin degradation. Advances in Botanical Research, 61, 263–308.
Stephanie, L. M., Joel, P., & Amy, M. G. (2015). Bacterial biodegradation and bioconversion of industrial lignocellulosic streams. Applied Microbiology & Biotechnology, 99(7), 2939–2954.
Stevens, J. C., & Shi, J. (2019). Biocatalysis in ionic liquids for lignin valorization: Opportunities and recent developments. Biotechnology Advances, 37(8), 107418.
Suderman, R. J., Dittmer, N. T., Kanost, M. R., et al. (2006). Model reactions for insect cuticle sclerotization: Cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochemistry and Molecular Biology, 36(4), 353–365.
Sugano, Y., Matsushima, Y., Tsuchiya, K., et al. (2009). Degradation pathway of an anthraquinone dye catalyzed by a unique peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation, 20(3), 433–440.
Sugano, Y., Muramatsu, R., Ichiyanagi, A., et al. (2007). DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family. Journal of Biological Chemistry, 282(50), 36652–36658.
Suman, S. K., Dhawaria, M., Tripathi, D., et al. (2016). Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. International Biodeterioration & Biodegradation, 112, 12–17.
Tamboli, D. P., Telke, A. A., Dawkar, V. V., et al. (2011). Purification and characterization of bacterial aryl alcohol oxidase from Sphingobacterium sp. ATM and its uses in textile dye decolorization. Biotechnology & Bioprocess Engineering, 16(4), 661–668.
Tian, J. H., Pourcher, A. M., Bouchez, T., et al. (2014). Occurrence of lignin degradation genotypes and phenotypes among prokaryotes. Applied Microbiology & Biotechnology, 98(23), 9527–9544.
Tsukihara, T., Yoichi, H., Takahito, W., et al. (2006). Molecular breeding of white rot fungus Pleurotus ostreatus by homologous expression of its versatile peroxidase MnP2. Applied Microbiology and Biotechnology, 71(1), 114–120.
Ullrich, R., Nüske, J., Scheibner, K., et al. (2004). Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Applied & Environmental Microbiology, 70(8), 4575.
Vardon, D. R., Franden, M. A., Johnson, C. W., et al. (2015). Adipic acid production from lignin. Energy & Environmental Science, 8(2), 617–628.
Varman, A. M., He, L., Follenfant, R., et al. (2016). Decoding how a soil bacterium extracts building blocks and metabolic energy from ligninolysis provides road map for lignin valorization. Proceedings of the National Academy of Sciences, 113(40), E5802.
Wang, S. N., Chen, Q. J., Zhu, M. J., et al. (2018). An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization. Biochimie, 148, 46–54.
Wu, W., Dutta, T., Varman, A. M., et al. (2017). Lignin valorization: Two hybrid biochemical routes for the conversion of polymeric lignin into value-added chemicals. Scientific Reports, 7(1), 8420.
Xiaoshi, W., René, U., Martin, H., et al. (2015). Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. Proceedings of the National Academy of Sciences of the United States of America, 112(12), 3686–3691.
Xie, C. (2016). The study of lignin degradation by Bacillus ligniniphilus L1, Master Degree, Jiangsu University.
Xu, J., & Yang, Q. (2010). Isolation and characterization of rice straw degrading Streptomyces griseorubens C-5. Biodegradation, 21(1), 107–116.
Xu, R., Zhang, K., Liu, P., et al. (2018a). Lignin depolymerization and utilization by bacteria. Bioresource Technology, 269, 557–566.
Xu, Z., Qin, L., Cai, M., Hua, W., & Jin, M. (2018b). Biodegradation of kraft lignin by newly isolated Klebsiella pneumoniae, Pseudomonas putida, and Ochrobactrum tritici strains. Environmental Science and Pollution Research, 25, 14171–14181.
Yamada, Y., Wang, J., Kawagishi, H., et al. (2014). Improvement of ligninolytic properties by recombinant expression of glyoxal oxidase gene in hyper lignin-degrading fungus Phanerochaete sordida YK-624. Microbiology & Fermentation Technology, 78(12), 2128–2133.
Yang, X., & Zhang, X. (2016). Transformation of biphenyl intermediate metabolite by manganese peroxidase from a white rot fungus SQ01. Acta Microbiologica Sinica, 56(6), 1044–1055.
Yang, Y. (2012). The screening of Aspergillus-Streptomyces-Pleurotus and their coimmoblized biodegradation of alkali lignin, Vol. Doctor, Dalian University of Technology.
Yang, Y. S., Zhou, J. T., Lu, H., et al. (2011). Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation, 22(5), 1017–1027.
Yang, Y. S., Zhou, J. T., Lu, H., et al. (2012). Isolation and characterization of Streptomyces spp. strains F-6 and F-7 capable of decomposing alkali lignin. Environmental Technology, 33(22–24), 2603–2609.
Yoshida, H., Sakai, G., Mori, K., et al. (2015). Structural analysis of fungus-derived FAD glucose dehydrogenase. Scientific Reports, 5(1), 13498.
Zeng, J., Mills, M. J., Simmons, B., et al. (2017). Understanding factors controlling depolymerization and polymerization in catalytic degradation of β-ether linked model lignin compounds by versatile peroxidase. Green Chemistry, 19(9), 2145–2154.
Zeng, J., Singh, D., Laskar, D. D., et al. (2011). Deconstruction of wheat straw lignin by Streptomyces viridosporus as insight into biological degradation mechanism. In AIChE Annual Meeting.
Zeng, J., Singh, D., Laskar, D. D., & Chen, S. (2013). Degradation of native wheat straw lignin by Streptomyces viridosporus T7A. International Journal of Environmental Science & Technology, 10(1), 165–174.
Zhang, H. (2012). Study on biodegradation of lignin and lignin model compounds by Cupriavidus sp. B-8, Master Degree, Central South University.
Zhang, K., Huang, M., Ma, J., et al. (2018). Identification and characterization of a novel bacterial pyranose 2-oxidase from the lignocellulolytic bacterium Pantoea ananatis Sd-1. Biotechnology Letters, 40(5), 871–880.
Zhang, P. (2017). Characterization and metabolic mechanism of lignin biodegradation by Comamonas serinivorans C35. Master Degree, Jiangsu University.
Zhang, X., Peng, X., & Eiji, M. (2014). Recent advances in Sphingobium sp. SYK-6 for lignin aromatic compounds degradation – A review. Acta Microbiologica Sinica, 54(8), 854–867.
Zhang, Y. (2013). The mechanism of lignin and related mode benzene compounds degradation by Pandoraea sp. B-6 and Cupriavidus basilensis B-8, Doctor Degree, Central South University.
Zhu, D., Zhang, P., Xie, C., et al. (2017). Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnology for Biofuels, 10(1), 44.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Qiu, W. (2021). The Route of Lignin Biodegradation for Its Valorization. In: Liu, ZH., Ragauskas, A. (eds) Emerging Technologies for Biorefineries, Biofuels, and Value-Added Commodities. Springer, Cham. https://doi.org/10.1007/978-3-030-65584-6_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-65584-6_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-65583-9
Online ISBN: 978-3-030-65584-6
eBook Packages: EnergyEnergy (R0)