Abstract
A fungus strain F-3 was selected from fungal strains isolated from forest soil in Dalian of China. It was identified as one Aspergillus sp. stain F-3 with its morphologic, cultural characteristics and high homology to the genus of rDNA sequence. The budges or thickened node-like structures are peculiar structures of hyphae of the strain. The fungus degraded 65% of alkali lignin (2,000 mg l−1) after day 8 of incubation at 30°C at pH 7. The removal of colority was up to 100% at 8 days. The biodegradation of lignin by Aspergillus sp. F-3 favored initial pH 7.0. Excess acid or alkali conditions were not propitious to lignin decomposing. Addition of ammonium l-tartrate or glucose delayed or repressed biodegradation activities. During lignin degradation, manganese peroxidase (28.2 U l−1) and laccase (3.5 U l−1)activities were detected after day 7 of incubation. GC-MS analysis of biodegraded products showed strain F-3 could convert alkali lignin into small molecules or other utilizable products. Strain F-3 may co-culture with white rot fungus and decompose alkali lignin effectively.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lignin, next to cellulose, is the second most abundant compound in plant biomass. And it is also known for resistance to microbial degradation because of its high molecular weight and presence of various biologically stable linkages. Microbial lignin degradation is drawing attention as an alternative to pulping or enzymatic hydrolysis of lignocellulosic materials. Delignification of wood by the alkaline pulping process produces millions of tons of kraft lignin annually of which little has been recovered and used. Studies on the biodegradation of lignin have significant implications on our understanding of the global carbon cycle and development of an environmentally friendly technique for selective removal of lignin in paper-making. Furthermore, the effective bioconversion of lignin is of importance to fully utilize it for new materials or energy. There was a breakthrough in the field of lignin biodegradation in 1983 when fungal ligninases and their hydrogen peroxide requirement were described by Tien and Kirk (1983). Since that time, biodegradation of lignin by white rot fungi and their lignolytic enzymes have been widely studied (Hatakka 2001; Pointing 2001; Baborová et al. 2006; Baldrian et al. 2005; Asgher et al. 2008). Most lignin research on bioconversion and biodegradation has been on wood-rot fungi, particularly white-rot fungi, and not on other lignolytic organisms. However, because of slow growth rates; easily counteracted by other lower microorganisms; damageable enzymes and production costs for them, etc., the direct use of single white rot fungi and their enzymes in industrial processes is still very difficult now. Molds derive energy not through photosynthesis but from the organic matter in which they live. Typically, molds secrete hydrolytic enzymes, mainly from the hyphal tips. These enzymes degrade complex biopolymers such as starch, cellulose and lignin into simpler substances which can be absorbed by the hyphae. In this way, molds play a major role in causing decomposition of organic material, enabling the recycling of nutrients throughout ecosystems. Members of the molds, as majority of microscopic forms, inhabit lignin-rich environments, and their relative lignin-degrading potential is worth of being studied. It was shown that some of them, such as Aspergillus fumigatus, A. japonicus, A. niger, and A. terreus, Penicillium simplicissimum, were capable of degrading both aromatic and carbohydrate components of water-soluble lignocarbohydrate complexes (LCC) or Kraft lignin (Milstein et al. 1984; Zeng et al. 2006). Although the function of mold species was noticed, few studies have so far described mold species occurring biodegradation of alkali lignin.
In this context, a fungus strain F-3 was isolated and characterized by its morphology and by rDNA sequencing. Its capacity to biodegrade alkali lignin was tested in liquid cultures and co-cultures with one white rot fungus—Pleurotus ostreatus strain G5. The biodegraded residues of alkali lignin were analyzed with GC-MS.
Materials and methods
Chemicals and Media
All chemical reagents were of analytical or higher grade. Standard lignin: Sigma; alkali lignin was purchased from Wuhan East China chemicals co. Ltd (Prepared from alkaline pulping wastewater, brown powder, mineral content ≤1.5%, pH 8–10, water content ≤5%, filtered through 80 mesh).
Medium: Isolation and storage medium: potato dextrose agar (PDA) medium. Screening medium (or selection medium): Crawford’s liquid medium containing 2.0 g l−1 alkali lignin (Crawford et al. 1982). Biodegradation medium (or called as transparent medium) g per litre: Glucose 10, KH2PO4 2, MgSO4 0.5, (NH4)2SO4 0.5, VB1 0.002, alkali lignin 2.0.
Screening and characteristics of fungi
Soil samples were taken from the forest zone of Dalian, China, and used for isolation on plates of PDA medium with conventional dilution method (Barnett and Hunter 1972). The soil conditions are as follows: soil type—brown soil; pH 6–7; organic matter ≥75; N-total 2.5–4.3. Their ability to produce lignin modifying activities was then determined using screening medium. The isolates were inoculated to the screening medium and incubated at 30°C on a rotary shaker at 150 rpm. The decolourization of cultures was measured as biodegradation of alkali lignin. The most active isolate was further characterized. For macro-morphological observations, isolates were cultivated on plate and tubes of PDA medium at 28°C in the dark for 7 days. For microscopic observations, mounts were made in lactic acid from colonies or a drop of alcohol was added to remove air bubbles and excess conidia. Scanning Electron Microscopy (SEM) was performed using a JEM-1200EX electron microscope.
rDNA sequence analysis of strain F-3
Total DNA samples were used to amplify the ITS DNA (18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence gene) using TaKaRa Fungi Identification PCR Kit with TaKaRa Thermal Cycler Dice TP600 (TaKaRa BIO INC).The amplification products were gel purified using TaKaRa Agarose Gel DNA Purification Kit Ver. 2.0 (Code No. DV805A). DNA sequencing were performed using Seq Forward Primer (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) with an ABI PRISMtm 3730XL DNA Sequencer, and the ABI Prism Big Dye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystem).
The partial sequence was subjected to BLAST analysis for homology analysis using the online option available at www.ncbi.htm.nih.gov/BLAST (Altschul et al. 1997). The relative sequences from Blast results were aligned with ClustalX1.8 (Thompson et al. 1997) and constructed phylogenic trees with MEGA4.2 (Tamura et al. 2007).
Biodegradation of alkali lignin by strain F-3
Biodegradation of alkali lignin: Dispense 100 ml of the transparent or screening medium with 2,000 mg l−1 of alkali lignin into each 250 ml Erlenmeyer flask. The flasks were then plugged by cotton caps and the caps were again covered by non-absorbing paper with rubber band. The flasks were labeled accordingly and were sterilized by autoclaving at 110°C for 20 min. After autoclaving, the flasks were allowed to cool down at room temperature. 0.5 cm2 mycelia of the selected strain from 7-day-old agar slants of PDA, served to inoculate Erlenmeyer flasks. The time course of lignin degradation was followed in shaking flasks at 30°C on a rotary shaker at 150 rpm. Disappearance of lignin was monitored by aseptically removing 5-ml samples for measurement of ultraviolet (UV) absorption spectra at 280 nm after centrifugation at 12,000 rpm for 10 min (Lundquist et al. 1977).
The colority of cultures was determined according to the CPPA standard method (1974).
All assays were performed at least in duplicate, with the corresponding controls. Both uninoculated media (called “blanks”) and inoculated autoclaved vials (called “controls”) were used.
Ligninolytic enzymes activity
Lignin peroxidase (LiP, EC1.11.1.14) activity was determined as described by Tien and Kirk (1984). The reaction mixture (in a total volume of 2 ml) in 0.125 mM sodium tartrate-HCl buffer (pH 4.5) contained 1.5 ml of extracellular culture fluid and 2.5 mM veratryl alcohol, 0.5 mM H2O2. The reaction was monitored by measuring the change in A310 for 5 min. Manganese peroxidase (MnP, EC1.11.1.1) activity was determined as described by Glenn and Gold (1985). MnP reaction mixture (in a total volume of 2 ml) in 50 mM succinate-lactate buffer (pH 4.5) contained 0.1 mM Manganese(II) sulfate, 0.1 mM H2O2, 0.25 mM phenol red. The reaction was monitored by measuring the change in A610 for 5 min. And laccase (Lac, EC1.10.3.2) activity was determined as described by Niku-Paavola et al. (1990). The laccase reaction mixture (in a total volume of 2 ml) in 100 mM acetate buffer (pH 4.0) contained 1.5 ml of extracellular culture fluid and 0.5 mM ABTS as the substrate. The reaction was monitored by measuring the change in A420 for 5 min. One unit of enzymes activity was defined as the amount catalyzing 1 μmol of substrates oxidized per minute of reaction.
GC-MS analysis of degraded products
Sample preparation for the GC-MS analysis: 60 ml supernatant of centrifuged biodegraded liquids was adjusted pH to 1.0–2.0 with hydrochloric acid, and centrifuged at 12,000 rpm (17,000 g) for 10 min. The acidified supernatant was extracted three times with 20 ml ethyl acetate. The ethyl acetate phase extracts were dehydrated with sodium sulfate anhydrous and mixed then evaporated under vacuum in a rotary apparatus at 40°C. The residue was dissolved in 1.2 ml pyridine. 100 μl pyridine solvent was derivatized with 20 μl BSTFA (BSTFA + TMCS 99:1 Supelco, USA) in microreaction vials of 2 ml volume with mininert valves (Supelco Inc., Bellefonte, PA, USA) and kept at 50°C for 20 min. For the analysis an Agilent GC-MS system (6890N/5975B) with a quadrupole mass spectrometer and a capillary gas-chromatograph were used. The fused silica capillary column (30 m × 0.25 mm i.d.) was coated with SE 30 (0.25 μm). Helium was used as carrier gas (0.7 bar) at a flow rate of 1.0 ml/min. The splitter/injector was kept at 300°C with a split ratio of 5:1. The column temperature was programmed as follows: 100°C (4 min) − 4°C/min − 280°C (3 min). The ion source temperature was 250°C, the emission current 0.6–0.7 mA. The chromatograms were recorded by ion monitoring in the m/e range 50–500.
Results and discussion
Isolation and identification of strain F-3
Bacteria can hardly grow well in most of forest soil because of a low pH value, and so the fungi are the main decay producers. They will cause decay of lignin-containing dead leaves.
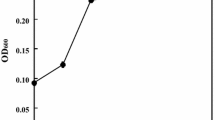
One strain F-3 was selected from the isolates of fungi from forest soil with selection medium. After day 8 of incubation, the fungus degraded 55% of alkali lignin, subsequently the discoloration rate slowed down gradually. The maximum biodegradation of alkali lignin was up to 65% (Fig. 1) at 12 days of incubation. Growth rate of F-3 on PDA medium is moderately rapid and texture of colonies is downy gradually to powdery; and colonies are 8–15 mm in diameter for 7 days (Fig. 2). Surface colony color ranges from white, pale pink, gray, to blue–green, while the reverse is uncolored. From the colony, it appears to be similar to Penicillium or A. fumigatus to some extent. Microscopic observations of appearance: mycelium composed of hyaline, branched, septate, smooth-walled hyphae with width of 3–12 μm. Conidiophores are smooth—walled and brittle, hyaline to pale brown, and with length ranging from 170–500 μm, width 5–12 μm; conidial heads are loosely radiate, and with width 30–60 μm; Conidiogenous cells biseriate; vesicles spathulate to subclavate, 15–20 μm in diameter; metulaes 10–12 × 4.5–5.5 μm; phialides 8.5–10 × 4.5–5.5 μm; metulaes and phialides covering from 60% to almost the entire vesicle; conidia are globose, blue green, more or less rough-walled, and 3.5–4 μm in diameter; budges or thickened node-like structures 7–20 μm in diameter (Figs. 3, 4). The ITS region is a convenient universal marker for fungal species identification. The result of the partial ITS region DNA sequence of strain F-3 alignment based on BLAST analysis revealed that the isolate was identified as Aspergillus with high homology. There are many budges in hyphae of the strain F-3. This is a peculiar structure of hyphae compared with known species of Aspergillus. So we named it Aspergillus sp. strain F-3. The sequence was registered in the NCBI GenBank Data Library under the accession number GQ149340. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Felsenstein 1985). The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980) and are in the units of the number of base substitutions per site. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007).The evolutionary relations of the strain among the genus of Aspergillus were shown in the phylogenetic tree (Fig. 5).
Phylogenic tree based on ITS region gene sequences showing relationships among strain F-3 and the most close type strain species of Aspergillus. Numbers at nodes indicate percentages of bootstrap support based on a neighbor-joining analysis of 1,000 resampled datasets. Bar, 0.001 substitutions per nucleotide position
There are many budges or round and ovate thickened node-like structures in hyphae of strain F-3. It was recognized that the budges in the tip of hyphae were relative to the growth restarting (Horio and Oakley 2005). In general, node-like structures were less abundant in mycelia under natural conditions. These cell-like bulges are suggestive of intercalary spores. However, these are not reported to occur in the organism before, and no crosswalls (indicative of intercalary spore formation) are found. Perhaps the nodes represent storage material in the hyphae and occur only when certain metabolic products are excessive, depending on the food source (Murphy et al. 1974). Luengo et al. (1986) found bulges were formed in the hyphae of a high penicillin-producing strain of Penicillium chrysogenum AS-P-78. When penicillin was accumulated in the broth, the size and number of bulges increase during the fermentation in parallel with penicillin accumulation. Several species of the genus Aspergillus form sexual spores within minute spherical shells (cleistothecia) with an envelope of thick-walled globose cells (hülle cells). It was found that lignin-decomposing enzyme—laccase localizes in hülle cells and cleistothecial primordia of Aspergillus nidulans (Hermann et al. 1983). So the presence of bulges or node-like structures in hyphae of strain F-3, may be relative to some products, such as lignin-decomposing enzymes.
Biodegradation of lignin and relative enzyme activities
As a result of isolation and screening, one of lignin degrading Aspergillus sp. strain F-3 was selected on the basis of its color reduction efficiency in selection broth containing alkali lignin (2,000 mg l−1). The conditions for lignin degradation by fungi are varied for different species. The optimal pH was 4.5 and 6.0 for Sporotrichum pulverulentum (Kirk et al. 1978) and P. ostreatus B1 in bubble-column reactor (BCR) (Zhao et al. 2008) respectively. For Aspergillus sp. strain F-3, its biodegradation of lignin favored initial pH 7.0 (Fig. 6). Generally, the pH value of industrial wastewater which contains lignin such as cotton pulp black liquor is neutral to alkali. Strain F-3 could function under higher pH value than other fungi, which showed its prospective importance of using in industry.
The biodegradation of alkali lignin by fungi is always influenced by usable carbon sources. Studies have shown that degradation of alkali lignin by Aspergillus sp. strain F-3 was under C-limited condition, and depressed by glucose. The decolorization of alkali lignin could be delayed for 4 days by glucose at low concentration of 1 g l−1 whereas the removal rate was up to that of control at last. The biodegradation of lignin could be repressed with higher content of glucose (Fig. 7).Similar results are reported in other researches. A. japonicus OM-4 degraded the lignin of the high molecular size soluble LCC (lignocarbohydrate complexes) and many phenolic compounds efficiently in the absence of additional carbon sources (Milstein et al. 1981). Aspergillus wentii could degrade aqueous fraction (AL-A) of alkali isolated straw lignin (AL) under C-limited condition (50% decrease in OD281) (Janshekar et al. 1982). The activity of both endoglucanase and exoglucanase of A. terreus could be reduced by addition of carbon sources (Emtiazi et al. 2001). But there are different effects in other fungi. Both starch and phenolic compound(s) present in potato extract are capable of inducing laccase synthesis in Pleurotus florida (Nirmalendu et al. 1999).
Addition of ammonium l-tartrate at low concentration to selection medium could show relatively high degradation activity of lignin whereas its value was less than that of the control (Fig. 8). Alkali lignin was efficiently decomposed by strain F-3 in the biodegradation medium with addition of 1.0 g l−1 yeast extract. In contrast, the ligninolytic activities of P. ostreatus strain G5 was repressed (Fig. 9). It shows that at the early stage at 2–8 days, there was a significant biodegradation of lignin in culture, removal rate of lignin increased quickly up to its maximum 65% after day 8 of incubation. Ligninolytic enzymes of white-rot fungi are generally produced under N-limited media (Janshekar et al. 1982; Zhao et al. 2008). The fungi A. wentii, Chaetomium cellulolyticum, Humicola fuscoatra and Sporotrichum thermophile decreased the lignin absorbance only to a smaller extent (16–20%) under sufficient nitrogen supply. P. ostreatus was the poorest strain in this respect (Janshekar et al. 1982). The correlation of biodegradation of lignin by Aspergillus sp. strain F-3 to N-sources is different from that of white-rot fungi to some extent. Compared with white-rot fungi—P. ostreatus strain G5, Aspergillus sp. F-3 is not sensitive to nitrogen source in the beginning of alkali lignin degradation. As the control with white rot fungus—P. ostreatus strain G5, the removal rates of lignin and colority by F-3 were higher than that of the former, about 65–70%.
Biodegradation of lignin by mixed cultivations of S. pulverulentum, C. cellulolyticum, P. ostreatus, A. wentii showed generally poorer results than those of pure cultures. This shows that the lignin degrading ability of different species may not be of a cumulative nature and hence can not be intensified by using a heterogenous culture of these species (Janshekar et al. 1981). As Aspergillus and white rot fungi are natural habitats in lignocellulosic materials, an attempt of co-cultures of P. ostreatus G5 − Aspergillus sp. F-3 showed strain F-3 could co-grow with G5 and decompose alkali lignin. In Fig. 10, slightly higher degrading rates appeared in co-cultures systems of white rot fungus-Aspergillus after day 7 of incubation. The removal of colority was higher than that of lignin. But on day 12, co-cultures systems did not have an advantage on removal of lignin. The immobilization of A. niger and Phanerochaete chrysosporium on polyurethane foam is considered. The different morphology of the obtained bioparticles seriously modify the productivity of citric acid and extracellular peroxidases (lignin peroxidase, LiP, and manganese peroxidase, MnP) by A. niger and P. chrysosporium, respectively. The best results are obtained in both cases when fungi developed inside the cube foam (Sanromán et al. 1996). Submerged pure cultures which are either suspended or immobilized on synthetic support material have been used for most of studies. Although immobilized cultures have been shown to be very effective in dye decolorization (Palma et al. 1999; Zhang et al. 1999), the aging process of the mycelium may limit the length of the process (Palma et al. 1999). The ability of T. versicolor to produce enzymes during growth was exploited in the investigation of long-term decolorization activity of T. versicolor in sequencing batch reactors (Borchert and Libra 2001). By shearing suspended pellets of T. versicolor in each cycle, high and stable degrees of decolorization of reactive textile dyes were repeatedly found without decrease in activity over time in pure culture. However, decolorization efficiency decreased because bacterial contamination occurred easily. Judy et al. (2003) investigated four strategies for their potential in optimizing the use of white rot fungi in nonsterile culture for treating wastewater containing textile dyes. He considered that the two strategies of tolerance of low pH and production of extracellular enzymes of white rot fungi did not show potential. The third strategy, the limitation of nitrogen, had potential as limited nitrogen source could suppress bacterial growth in some extent. And the fourth strategy, which was considered as the most potential in further development, was the addition of cost-effective fungi inocula in processing system. From our results, strain F-3 could co-exist with white rot fungus and degrade lignin efficiently. In any case, this is of importance to put them in industrial use because community of microorganisms is surely better than one species or strain in decomposition of organic pollutants based on micro-ecology principle.
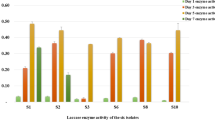
For some other microorganisms, they are capable of degrading lignin and may contain ligninolytic enzymes. It was shown (Emtiazi et al. 1999) that A. terreus had peroxidase and laccase activities when grown on wheat straw and removed COD(chemical oxygen demand) of textile dyes effluent. The enzyme activities of Aspergillus sp. strain F-3 were measured by us. There were activities of manganese peroxidase (MnP), and laccase (Table 1) in biodegradation culturing solution. LiP activity was not detected in the strain. The ligninolytic enzymes of strain F-3 are only partial supports for its strong biodegradation ability of alkali lignin as the enzymes activities were not detected in high level. There may be other enzymes connected with the degrading process (Emtiazi et al. 2001). The mechanism of strain F-3 for decomposing lignin still needs further study.
GC-GS analysis of products decomposed
GC-MS is used in the present study because it has been proved to be a suitable technique for analyzing low molecular weight compounds released from lignin due to degradation (Kirk et al. 1978; Janshekar et al. 1982). Small molecular products could be realized through GC-GS analysis of products degraded by Aspergillus sp. strain F-3. The results showed that alkali lignin could be effectively degraded by F-3 and be converted into broken phenol (C3–C8) and other compounds (Figs. 11, 12). Jouni’s study showed the α-hydroxyl groups in the propane chain of both phenolic and nonphenolic tetramers were first oxidized symmetrically in two successive steps to give monoketones and diketones (Jouni et al. 1987), then these ketone metabolites were decomposed through Cα–Cβ cleavage. The metabolic route may resemble Jouni’s results according to products degraded by F-3. But some products, such as 1,4-anthracenediol, were intricate now. They may be metabolites of F-3 or resultant products from alkali lignin. Butyl cyclohexyl phthalate may be transformed from abietic acid, which is the most abundant of several closely-related organic acids that constitute most of rosin, the solid portion of the oleoresin of coniferous trees (Table 2). The pathway and mechanism of alkali lignin decomposed by strain F-3 still need further studies. However F-3 is possessed of potential value in bioconversion and biodegradation of lignin and relative compounds.
Conclusions
The newly isolated Aspergillus sp. strain F-3 has peculiar node-like structures in its hyphae. This morphological character is different from present known species or strains of Aspergillus. A hypha extends only at its tip and the tapered region involved in this process is called the extension zone during its growth. The budges in the tip of hyphae were relative to the growth restarting. These cell-like bulges are different from suggestive intercalary spores, as no crosswalls were found. So the presence of bulges or node-like structures in hyphae of strain F-3, may be relative to the accumulation of some products, which may participate in alkali lignin-decomposing process.
Aspergillus sp. strain F-3 is possessed of ligninolytic enzymes activities and is capable of degrading alkali lignin alone. GC-GS analysis of products decomposed by strain F-3 showed alkali lignin can be degraded to small molecules or converted into utilizable products.
The strain can effectively degrade alkali lignin under co-culture system of Pleurotus—Aspergillus. It implied their potential values in industrial use. Molds grow rapidly and with high antibacterial ability in natural environment. Many different mold species vary enormously in their tolerance to temperature and humidity extremes. Certain molds can survive in harsh conditions such as the snow-covered soils of Antarctica, refrigeration, highly acidic solvents, and even petroleum products such as jet fuel. The system of co-exist of Pleurotus—Aspergillus may keep degradation activity under non-sterilized conditions with its relative strong antimicrobial contamination.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Asgher M, Kausar S, Bhatti HN, Shah SAH, Ali M (2008) Optimization of medium for decolourization of Solar golden yellow R direct textile dye by Schizophyllum commune IBL-06. Int Biodeterior Biodegrad 61:189–193
Baborová P, Moder M, Baldrian P, Cajthamlová K, Cajthaml T (2006) Purification of a new manganese peroxidase of the white-rot fungus Irpex lacteus and degradation of polycyclic aromatic hydrocarbons by the enzyme. Res Microbiol 157:248–253
Baldrian P, Valaskova V, Meerhautova V, Gabriel J (2005) Degradation of lignocellulose by Pleurotus ostreatus in the presence of copper, manganese lead and zinc. Res Microbiol 156:670–676
Barnett HL, Hunter B (1972) Illustrated genera of imperfect fungi, 3rd edn. Burgess Publishing Company, Minneapolis
Borchert M, Libra JA (2001) Decolorization of reactive dyes by the white rot fungus Trametes versicolor in sequencing batch reactors. Biotechnol Bioeng 75:313–321
CPPA (1974) Technical section standard method H5P. Canadian Pulp and Paper Association, Montreal
Crawford DL, Sutherland JB, Pommeto AL III, Miller JM (1982) Production of an aromatic aldehyde oxidase by Streptomyces viridosporus. Arch Microbiol 131:351–355
Emtiazi G, Nahvi I, Salehbaig M (1999) Production of cellulose (exoglucanse) by fungi in different media. Res Bull Isfahan Univ 1:15–28
Emtiazi G, Naghavi N, Bordbar A (2001) Biodegradation of lignocellulosic waste by Aspergillus terreus. Biodegradation 12:259–263
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Glenn JK, Gold MH (1985) Purification and characterisation of an extracellular Mn(II)-dependent peroxidase from the lignin degrading basidiomycete Phanerochaete chrysosporium. Arch Biochem Biophys 242:329–341
Hatakka A (2001) Biodegradation of lignin. In: Steinbüchel A, Hofrichter M (eds) Biopolymers. Lignin, humic substances, and coal, vol 1. Wiley-VCH, Weinheim, pp 129–180
Hermann TE, Kurtz MB, Champe SP (1983) Laccase localized in hülle cells and cleistothecial primordia of Aspergillus nidulans. J Bacteriol 154(2):955–964
Horio T, Oakley BR (2005) The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol Biol Cell 16:918–926
Janshekar H, Brown C, Fiechter A (1981) Determination of biodegraded lignins by ultraviolet spectrophotometry. Anal Chim Acta 130:81–91
Janshekar H, Haltmeier T, Brown C (1982) Fungal degradation of pine and straw alkali lignins. Eur J Appl Microbiol Biotechnol 14:174–181
Jouni J, Jukka P, Mirjasalkinoja S (1987) Initial steps in the pathway for bacterial degradation of two tetrameric lignin model compounds. Appl Environ Microbiol 53(11):2642–2649
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirk TK, Schultz E, Connors WJ, Lorenz LF, Zeikus JG (1978) Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporiurn. Arch Microbiol 117:277–285
Libra JA, Borchert M, Banit S (2003) Competition strategies for the decolorization of a textile-reactive dye with the white-rot fungi Trametes versicolor under non-sterile conditions. Biotechnol Bioeng 82(6):736–744
Luengo JM, Dominguez A, Cantroal JM, Martin JF (1986) Formation of bulges associated with penicillin production in high-producing stains of Penicillium chrysogenum. Curr Microbiol 13:203–207
Lundquist K, Kirk TK, Connors WJ (1977) Fungal degradation of kraft lignin and lignin sulfonates prepared from synthetic 14C lignin. Arch Microbiol 112:291–296
Milstein O, Vered Y, Gressel J, Flowers HM (1981) Biodegradation of wheat straw lignocarbohydrate complexes (LCC) II. Fungal growth on aqueous hydrolysate liquors and particulate residues of wheat straw. Eur J Appl Microbiol Biotechnol 13:117–127
Milstein OA, Haars A, Sharma A, Vered Y, Shragina L, Trojanowski J, Flowers HM, Gressel J, Hcittermann A (1984) Lignin degrading ability of selected Aspergillus spp. Appl Biochem Biotechnol 9:393–394
Murphy JA, Campbell LL, Pappelis AJ (1974) Morphological observations of Diplodia maydis on synthetic and natural substrates as revealed by scanning electron microscopy. Appl Microbiol 27(1):232–250
Niku-Paavola ML, Raaska L, Itävaara M (1990) Detection of white rot fungi by a non-toxic stain. Mycol Res 94:27–31
Nirmalendu D, Tapas Kumar C, Mina M (1999) Role of potato extract in extracellular laccase production of Pleurotus florida. J Basic Microbiol 39(5–6):299–303
Palma C, Moreira MT, Mielgo I, Feijoo G, Lema JM (1999) Use of a fungal bioreactor as a pretreatment or post-treatment step for continuous decolorization of dyes. Water Sci Technol 40:131–136
Pointing SB (2001) Feasibility of bioremediation by white rot fungi. Appl Microbial Biotechnol 57:20–33
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanromán A, Feijoo G, Lema JM (1996)Immobilization of Aspergillus niger and Phanerochaete chrysosporium on polyurethane foam. Prog Biotechnol 11:132–135 (Immobilized cells—basics and applications, Proceedings of an international symposium organized under auspices of The Working Party on Applied Biocatalysis of the European Federation of Biotechnology Noordwijkerhout)
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tien M, Kirk TK (1983) Lignin-degrading enzymes from himenomycete Phanerochaete chrysosporium Burds. Science 221:661–663
Tien M, Kirk TK (1984) Lignin degrading enzyme from Phanerochaete chrysosporium: purification, characterization and catalytic properties of a unique H2O2 requiring oxygenase. Proc Natl Acad Sci USA 81:2280–2284
Zeng GM, Yu HY, Huang HL, Huang DL, Chen YN, Huang GH, Li JB (2006) Laccase activities of a soil fungus Penicilliums implicissimum in relation to lignin degradation. World J Microbiol Biotechnol 22(4):317–324
Zhang FM, Knapp JS, Kelvin NT (1999) Development of bioreactor systems for decolorization of Orange II using white rot fungus. Enzym Microb Technol 24:48–53
Zhao LH, Zhou JT, Lv H, Zheng CL, Yang YS, Sun HJ, Zhang XH (2008) Decolorization of cotton pulp black liquor by Pleurotus ostreatus in a bubble-column reactor. Bull Environ Contam Toxicol 80:44–48
Acknowledgments
This work was supported by National Key Scientific and Technology Project for Water Pollution Treatment (2008 ZX07208-004-2); Projects (IRT0813) supported by Program for Changjiang Scholars and Innovative Research Team in University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, Y.S., Zhou, J.T., Lu, H. et al. Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 22, 1017–1027 (2011). https://doi.org/10.1007/s10532-011-9460-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-011-9460-6