Abstract
Genebanks seek to understand the partitioning of genetic diversity among species, populations, and individuals in their collections since this informs decisions for adopting the most effective sampling strategy. Recent reports have suggested that diploid wild species have much less heterogeneity within populations than cultivated forms. We here review past empirical phenotypic trait variation data and examine previous and new DNA marker datasets. We also examine simulation datasets and calculations designed to mimic the effects of artificial biases against wild species heterozygosity due to ascertainment, ploidy, and allele frequencies. Trait data suggests large practical variation exists within populations. Similarly, DNA markers on multiple individuals within diploid wild potato species populations show that substantial heterogeneity in a species is partitioned within populations. Simulations illustrate that biases due to ascertainment, ploidy, and allele frequencies account for much of the apparent homogeneity of wild diploid potato species.
Resumen
Bancos de germoplasma buscan entender la organización de la diversidad genéticadentro de especies, poblaciones e individuos en sus colecciones, ya que esto podriainformar que decisiones adoptar para un muestreo más efectivo. Reportes recienteshan sugerido que las especies silvestres diploides tienen menos heterogeneidaddentro de poblaciones que las formas cultivadas. Aquí revisamos previos datosempiricos de variación en caracteres fenotípicos y examinamos grupos de datosprevios y nuevos de marcadores de ADN. También examinamos grupos de datosde simulación y cálculos diseñados para imitar los efectos de sesgos artificiales conimpacto en heterozigosidad de las especies silvestres debido determinación, ploidíay frecuencia de alelos. Datos de caracteres sugieren que existe variación prácticagrande dentro de las poblaciones. Similarmente, los marcadores de ADN enmúltiples individuos dentro de poblaciones de especies diploides de papasilvestre muestran que niveles sustanciales de heterogeneidad en especies seencuentran distribuidos dentro de poblaciones. Las simulaciones ilustran que lossesgos debidos a determinación, ploidía y frecuencia de alelos explican en granmedida la aparente homogeneidad de las especies silvestres diploides de papa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Importance of Assessing Diversity
Potato is a clonal crop, but has about 100 wild species relatives. Maintaining these wild relatives in the genebank as botanical seed populations has several practical advantages over in vitro or tuber clonal propagation. It is also important to accurately characterize the diversity among individuals within these populations, since this informs decisions for adopting the most effective approaches to sampling when collecting, maintaining and evaluating germplasm.
Emphasis on Diversity within Species
Great effort has been expended by numerous taxonomists over many decades in an effort to partition potato diversity into species. This includes studies of many kinds based on physical traits as well as DNA markers. The most recent taxonomy reduced the number of species by about half (Spooner et al. 2014). This sacrificed some level of information for the goal of having a scheme in which potato species names are more stable, and all populations in the genebank having a definite species name, rather than being uncertain or hybrids (Bamberg et al. 2018). Thus, species names should already establish the first hierarchy of diversity, and the genebank manager should be able to confine concerns about diversity sampling to variation among populations within species and individuals within populations within species. The question of how to efficiently sample populations has often been addressed by selecting “core” collections (reviewed in Bamberg and del Rio 2014 and Bamberg et al. 2016). One can also assess populations within species in the genebank by how their total DNA marker diversity has accumulated over real time as populations were added, or how many markers we would expect to miss in a single average population (Bamberg and del Rio 2016).
Context for Measuring the Partitioning of Diversity within Populations—Index of Variation
Hosaka and Hanneman (1991) proposed an index of variation (IV) as the percent of variable markers of all those detected. But fixed markers may be irrelevant. They might be common to all potatoes or even all plants. We proposed IV2 (Bamberg et al. 2000) in which variation within a population is measured in the context of only markers known to be variable within the species. Thus, if all populations are identical and have within them the total variation found within the species, they would have an IV2 of 100%. At the other extreme, if a population is completely homogeneous (e.g., strict inbreeding species with a single genotype per population), individuals within populations do not vary at all for traits or markers that vary within the species, and so all have an IV2 of 0%.
Review of Heterogeneity – DNA Markers
Hardigan et al. (2015) reported that wild diploid species have very low heterozygosity, but that was on the basis of tuberosum cultivar-derived SNP loci for which there is ascertainment bias. For example, the primitive outcrossing diploid S. jamesii had the least apparent heterozygous loci at <1%. Similarly, Aversano et al. (2015) sequenced an individual of the diploid outcrossing species S. commersonii, claiming the observed average 1.5% SNP heterozygosity was very low compared to cultivars, but also not accounting for the ascertainment bias of comparing random SNPs to selected tuberosum-derived SNPs of tetraploid cultivated potato. In contrast, RAPD data from our early work (Bamberg and del Rio 2004, Bamberg et al. 2009), and a follow-up study with new data using the same tuberosum-derived SNP markers (Bamberg et al. 2015b) showed S. jamesii populations had essentially the maximum detectable heterozygosity when adjusted for ascertainment bias. They had IV2 of 50-70%, at least as much as the 56% (Hirsch et al. 2013) observed in the average tuberosum cultivar (where 100% of these markers were known to be variable by design). Similarly, Haynes et al. (2017) used SSRs (presumably without ascertainment bias) to show that 10 natural diploid S. chacoense populations had at least as much heterozygosity as tetraploid cultivars (after adjustment was made for the ploidy difference). Bisognin and Douches (2002) used SSRs and isozymes to detect high levels of heterogeneity within populations.
Review of SNP Frequency Reports in Potato
The review of Sood et al. (2017, p. 65) concludes potato is extremely heterozygous, citing various estimates of 1% to 5% SNPs per base pair. For example, Hardigan et al. (2017) did a GBS study of numerous wild species, concluding that SNP diversity of potato was over 1%, much higher than reported in other crops, particularly inbreds. Leisner et al. (2018) sequenced an inbred clone of wild diploid (normally outcrossing) species S. chacoense, similarly showing that some chromosomal regions that had ostensibly resisted becoming homozygous (presumably still being in their natural non-inbred form) had about 1.5% SNP per base pair. The homogenized regions’ SNP frequency was only one-tenth as much, roughly as expected by chance following several generations of selfing, and approaching known inbreds like tomato and soybean at about one SNP per 1000 bp (see Simko et al. 2006). On the other hand, Li et al. (2018) also did a GBS study of numerous potato species concluding that potato actually has about 20-fold less SNP frequency than these other reports, attributing the discrepancy to improper SNP filtering (Huang et al. 2018, Hardigan et al. 2018).
Review of Heterogeneity – Traits
There is evidence of high phenotype variation within populations of wild diploid potato species. This is the variability relevant to potato breeding, so it would indeed be curiously unhelpful if a reasonable analysis of DNA markers concluded that individuals within diploid potato germplasm populations are very homozygous and uniform.
The following are examples in diploid wild species with 5% or less within-population SNP heterozygosity according to Hardigan et al. (2015) for species not known to be selfers.
S. berthaultii... highly significant variation for multiple Colorado Potato Beetle resistance parameters (Bamberg et al. 1996).
S. boliviense… variation for folate (Robinson et al. 2019).
S. chacoense... highly significant variation for multiple CPB resistance parameters (Bamberg et al. 1996), leptine glycoalkaloid (Sinden et al. 1986), in vitro rooting characteristics (Christensen et al. 2017), seed proteins (Hosaka and Hanneman 1991).
S. microdontum… highly variable tuber calcium level (Bamberg et al. 1998), large late blight differences (Douches et al. 2001), tuber greening under illumination (Bamberg et al. 2015a), presence of Crazy Sepal mutant (Bamberg 2006).
S. bulbocastanum… resistance to nematodes (Brown et al. 1989), late blight (Lokossou et al. 2010), Zebra chip (Cooper and Bamberg 2014), seed proteins (Hosaka and Hanneman 1991).
S. bulbocastanum, cardiophyllum, pinnatisectum, circaeifolium... resistance to late blight, black leg, CPB (Chen et al. 2003), seed proteins (Hosaka and Hanneman 1991).
S. jamesii… tuber antioxidants (Hale et al. 2008).
S. commersonii… resistance to early blight (Jansky et al. 2008).
Unpublished late blight evaluations sponsored by the genebank conducted at multiple locations also identified wide ranges of resistance within single populations of S. microdontum, S. berthaultii and S. okadae.
Materials and Methods
Empirical Assessment of Heterozygosity within Populations
We sought to re-assess heterozygosity within populations in the context of the whole diversity within model species. Thus, we calculated the IV2 value, the proportion of loci polymorphic within populations of all loci polymorphic in the species (see Table 1).
S. jamesii heterogeneity was assessed with RAPD markers on individuals within 7 genebank populations, and AFLP markers on 28 individuals of PI 275169 from the genebank, 34 individuals of a large natural population in Ida Canyon near Sierra Vista in southern Arizona, and 78 individuals from an extremely large natural population in Navajo Canyon of Mesa Verde National Mesa Verde National Park near Cortez in southwestern Colorado. The species context was usually comprehensive, being derived from an AFLP marker set on 128 genebank populations sampled as bulks of at least 25 individuals as described in Bamberg et al. (2016).
S. microdontum is a species with an array of remarkable qualities for breeding (Bamberg and del Rio 2014) and genome analysis suggests it has had a surprisingly large introgression to cultivars (Hardigan et al. 2017). This species’ heterozygosity was assessed by AFLP on 23 plants from genebank population PI 473170 in the context of the core set of 50 populations identified previously by Bamberg and del Rio (2014).
Factors that Bias Estimation of within-Population Diversity
Various factors can lead to underestimation of diversity. These include bias due to ascertainment (Bamberg et al. 2015b), comparing populations with different allele frequencies (Bamberg and del Rio 2004), and different ploidy (Meirmans et al. 2018). We examined each of these by calculating and modeling the bias when artificial differences appear between two samples that are actually the same except for the bias factor.
Ascertainment Bias
For ascertainment bias simulation, we isolated the variable of proportion of heterozygous loci. We used the Randbetween function of Excel® to compose 10 genotypes of 5,000 loci as though they were drawn from populations with heterozygosity at 2%, 5% and 12%. DARwin software (Perrier and Jacquemoud-Collet 2006) was used to visualize the result as radial plots based on genetic similarities calculated with simple matching coefficient and UPGMA clustering. This analysis included a single random 50% heterozygosity individual representing populations with maximum detectable heterozygosity serving as the root.
Allele Frequencies Bias
With all other factors equal, multiple samples of single individuals from one population will appear to be more similar than single individuals drawn from different populations. We used approximate parameters of AFLP results reported for diploid S. brevicaule in Bryan et al. (2017). Simulation data was created using the Index and Randbetween functions of Excel. For example, a cell with the formula… =INDEX(A1:A8, RANDBETWEEN(1,8))… returns the value in a randomly chosen cell from the array (A1:A8). To isolate the effect of different allele frequencies, 20 individual genotypes were drawn at random for which each of 200 polymorphic loci had the same random allele frequency (that is, as though they were sibs from the same population). Then 20 individuals were assigned genotypes for 200 polymorphic loci where each locus could be a different allele frequency (that is, as though there were individuals from different populations). We repeated this using 2,000 polymorphic loci. These simulations were based on random allele frequencies from 1% to 99%, but empirical evidence (Bryan et al. 2017, Bamberg and del Rio 2004) suggests that real allele frequencies tend to be at the extremes, so we added a simulation of random genotypes for which 200 polymorphic loci could have allele frequencies x only as (20% > x > 80%). In these simulations, the proportion and identity of polymorphic loci was identical for all populations. This means that all simulated populations are identical in terms of the loci possessing the allele, with the only difference being the frequency of the allele at those loci. Thus, if multiple individuals from the same population had been bulked, it would appear to be identical to the bulk of individuals from all individuals of the different populations (since all the same alleles would be included in both bulks). We also calculated the apparent GS of two single plants drawn from the same population (polymorphic loci have the same random allele frequency for both plants) versus those drawn from different populations (polymorphic loci have different random allele frequencies for each plant) by averaging GS for 50,000 random loci.
Bias Due to Ploidy
Assessing ploidy bias does not require simulation. One need only calculate the average percent heterozygous loci individuals for all possible allele frequencies for both diploids and tetraploids, then average the 4x/2x ratio. We made this calculation using Excel.
Results and Discussion
Empirical Assessments of Heterozygosity within Populations
Table 1. presents new and previous empirical data on wild species. High IV2 was observed. Thus, of the polymorphic loci detected in a species, many are polymorphic within populations of that species. Combined with potentially low marker frequencies, the average risk of miss-sampling a polymorphic locus with a single plant is high. So, any practical evaluation aimed at mining traits variable within these populations would not be expected to be very efficient if based on single plant samples per population.
Simulations of the Effects of Biases
Ascertainment Bias
A SNP locus is codominant, genotyping for the wild allele and, potentially, a second mutant allele. They are mapped mutants in functional genes of S. tuberosum (cultivars), so are not random and may not be neutral. Using SNPs from the Infinium 8303 Potato SNP array to measure diversity will introduce a cultivated potato “ascertainment bias” (Bamberg et al. 2015b). The concern of this bias is that populations less related to the germplasm from which SNPs were developed necessarily appear less heterogeneous, even potentially reversing the actual order of diversity of two populations (Lachance and Tishkoff 2013). A simple illustration with animal physical descriptors shows why ascertainment bias needs to be adjusted by calculating the IV2 statistic: Descriptors used to measure diversity in a certain bird species would likely be applicable to most birds. But mammals would appear to be very homogenous as an average of bird descriptors because they have zero variation for many descriptors such as feather length, egg weight, beak color, etc. To use bird descriptors fairly to assess the relative diversity in mammals, the denominator in the ratio of variable versus fixed descriptors has to be descriptors for which variation is known to occur in both birds and mammals (e.g., lifespan, body temperature, diet). Within potato, we would expect the most primitive species like jamesii (Sarkinen et al. 2013) to have their apparent heterozygosity depressed the most by ascertainment bias.
If potato averages only a few percent of polymorphic loci across the whole genome, a plant tested with a random sample of all possible loci will obviously appear unrealistically distinct and homozygous in comparison to a plant tested with a set of custom loci all known to be polymorphic within its own genepool. Thus, comparable estimates of diversity within populations of potato species relatively unrelated to tuberosum must level the playing field by considering only loci for which variation has been shown to be possible as variation among populations.
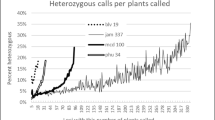
Figure 1 illustrates the simulation for which polymorphic loci were artificially restricted to 2%, 5%, and 12%, representing about the observed heterozygosity reported in Hardigan et al. (2015, Figure 4) for very primitive wild diploids, typical wild diploids, and primitive cultivated landrace diploids, respectively. Ten genotypes of each were randomly selected only on the basis of observed heterozygosity level that could be depressed by ascertainment bias. As expected, individuals cluster by their degree of ascertainment bias. Specifically, when percent heterozygous loci randomly decreases, individuals appear increasingly homogeneous and distinct from individuals with greater percent heterozygosity. Thus, ascertainment bias would make the very primitive species S. jamesii appear very homogeneous despite empirical evidence of both high DNA and trait variability (Table 1).
One can easily calculate and graph the pattern of apparent similarity between and among populations as ascertainment bias varies from none to 100%. An example is provided in Supplemental Figure S.1b.
Bias Due to Small Samples of Low and Different Allele Frequencies
Bryan et al. (2017) used AFLP markers on an array of wild potato species to compare the variation among numerous individuals in a species—about 20 plants from a single population and single plants from each of about 20 different populations. It was assumed that since within-population individuals tightly clustered with each other, one could conclude that any single random plant from that cluster was a good representative of the genetic identity of the population. But single individuals within a population have a special biased relationship with each other, being drawn from a single distribution of loci which each have a single allele frequency. This means that all of the samples would present exactly the same genotype if assessed as a bulk of multiple plants using a dominant DNA marker like AFLP. Figure 2 shows the effect of common versus different allele frequencies. For 200 polymorphic loci at all random allele frequencies, individuals drawn at random from the same population (each locus has the same random allele frequency, the white circles) will always appear much more similar to each other than individuals from different populations (where each locus in each individual can be from a different allele frequency-- black squares). Remember that this effect is completely due to bias, since all these simulated individuals are polymorphic at exactly the same loci so would appear to be identical in bulk because all the alleles would be captured regardless of their individual frequency—each set has exactly the same alleles that are present in the other. Supplemental Figure S2b illustrates that increasing the number of polymorphic loci observed to 2,000 does not remove the bias, but increases it. Supplemental Figure S2c illustrates that bias also increases if one considers only extreme allele frequencies at 200 loci, which is more realistic according to empirical assessments of allele frequencies at real polymorphic loci in wild potato species.
PCA plots of relationship of individual genotypes chosen at random from 200 polymorphic loci at all random frequencies, isolating allele frequency bias. Individuals represented by circles have random allele frequencies in common (as if from a single population) while individuals represented as squares are drawn at random from random allele frequencies (as if from different populations)
We can easily estimate absolute similarities of the two different groups of plants with calculations on simulated genotypes. When there are 50,000 polymorphic loci and genotypes are drawn at random, two random plants deviate very little from the expected 50% Simple Matching Coefficient similarity as individuals from different populations and 66% similarity as individuals from the same population. If allele frequencies are restricted to the extremes (that is, allele frequencies tend to be close to fixed or rare rather than balanced), the bias making individuals from the same population look artificially similar becomes even more extreme. For example, when allele frequencies (x) are at the extremes (20% > x > 80%), individuals drawn randomly from the same population appear to be very close to 83% similar, and when allele frequencies are very extreme (10% > x > 90%), individuals drawn randomly from the same population will appear to be very close to 95% similar (random individuals from different populations remain at an average 50% similarity).
Ploidy Bias
Most DNA markers have two alleles by design. If allele frequencies are a and b, the chance of sampling random heterozygous diploid individuals will be the complement of the homozygote frequency = 1-(a2 + b2). For tetraploids, it is 1-(a4 + b4). So if a diploid and tetraploid population had the same allele frequencies for all polymorphic loci, the chance of detecting heterozygosity in a single plant is obviously much greater for tetraploids. If known polymorphic loci have random allele frequencies, the average advantage of a tetraploid in detecting heterozygotes is very close to 1.8 times as much as its corresponding diploid form—higher if allele frequencies are extreme. Figure 3 shows the percent heterozygous loci observed in a variety of classes of potato germplasm (Hardigan et al. 2017, Fig 2D) and how very primitive diploid “outgroups”, other wild diploid species, and cultivated diploid landraces are not remarkably less heterozygous when the adjustment is made for ploidy bias by multiplying by a factor of 1.8. Again, the magnitude of this bias will depend on the actual proportions of allele frequencies, with the advantage of detecting heterozygosity in tetraploids increasing as the balance of allele frequency departs from 50%:50%. Calculations based on the observed allele frequencies of the S. jamesii population from Navajo Canyon in Table 1 indicate that one would indeed expect about 1.8 times as much heterozygosity to be detected in a single tetraploid individual as a single comparable diploid. In the study of Haynes et al. (2017), diploid S. chacoense heterozygosity estimates needed to be increased by a factor of 1.3 to remove ploidy bias and allow a fair comparison to the heterozygosity of tetraploid cultivars.
Conclusions
When biases are corrected for, marker evidence is in harmony with trait evidence that primitive diploid potato species can be quite heterogeneous within their populations. It follows that accurate collection, evaluation and preservation of diversity depends on sampling an adequate number of individuals within populations.
Abbreviations
- USPG:
-
US Potato Genebank
References
Aversano, R., F. Contaldi, M.R. Ercolano, V. Grosso, M. Iorizzo, F. Tatino, L. Xumerle, A.D. Molin, C. Avanzato, A. Ferrarini, M. Delledonne, W. Sanseverino, R.A. Cigliano, S. Capella-Gutierrez, T. Gabaldón, L. Frusciante, J.M. Bradeen, and D. Carputo. 2015. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. The Plant Cell 27: 954–968.
Bamberg, J.B. 2006. Crazy sepal: A new floral Sepallata-like mutant in the wild potato Solanum microdontum bitter. American Journal of Potato Research 83: 433–435.
Bamberg, J.B., and A.H. del Rio. 2004. Genetic heterogeneity estimated by RAPD polymorphism of four tuber-bearing potato species differing by breeding system. American Journal of Potato Research 81: 377–383.
Bamberg, J.B., and A.H. del Rio. 2014. Selection and validation of an AFLP marker Core collection for the wild potato Solanum microdontum. American Journal of Potato Research 91: 368–375.
Bamberg, J.B., and A.H. del Rio. 2016. Accumulation of genetic diversity in the US potato Genebank. American Journal of Potato Research 93: 430–435.
Bamberg, J.B., C.A. Longtine, and E.B. Radcliffe. 1996. Fine screening Solanum accessions for resistance to Colorado potato beetle. American Journal of Potato Research 73: 211–223.
Bamberg, J., J. Palta, L. Peterson, M. Martin, and A. Krueger. 1998. Fine screening potato (Solanum) species germplasm for tuber calcium. American Journal of Potato Research 75: 181–186.
Bamberg, J., C. Singsit, A.H. del Rio, and E.B. Radcliffe. 2000. RAPD analysis of genetic diversity in Solanum populations to predict the need for fine screening. American Journal of Potato Research 77: 275–278.
Bamberg, J.B., A.H. del Rio, and Rocio Moreyra. 2009. Genetic consequences of clonal versus seed sampling in model populations of two wild potato species indigenous to the USA. American Journal of Potato Research 86: 367–372.
Bamberg, J., R. Navarre Moehninsi, and J. Suriano. 2015a. Variation for tuber greening in the diploid wild potato Solanum microdontum. American Journal of Potato Research 92: 435–443.
Bamberg, J.B., A. del Rio, J. Coombs, and D. Douches. 2015b. Assessing SNPs versus RAPDs for predicting heterogeneity in wild potato species. American Journal of Potato Research 92: 276–283.
Bamberg, J.B., A.H. del Rio, D. Kinder, L. Louderback, B. Pavlik, and C. Fernandez. 2016. Core collections of potato (Solanum) species native to the USA. American Journal of Potato Research 93: 564–571.
Bamberg, J.B., A. del Rio, S. Jansky, and D. Ellis. 2018. Ensuring the genetic diversity of potatoes. In: Achieving sustainable cultivation of potatoes no. 26, Vol.1 (Ed. prof. Gefu Wang-Pruski). Burleigh-Dodds science publishers. Chapter 3: 57–80.
Bisognin, D., and D. Douches. 2002. Genetic diversity in diploid and tetraploid late blight resistant potato germplasm. HortScience 37: 178–183.
Brown, C.R., H. Mojtahedi, and G.S. Santo. 1989. Comparison of reproductive efficiency of Meloidogyne chitwoodi on Solanum bulbocastanum in soil and in vitro tests. Plant Disease 73: 957–959.
Bryan, G.J., K. McLean, R. Waugh, and D.M. Spooner. 2017. Levels of intra-specific AFLP diversity in tuber-bearing potato species with different breeding systems and Ploidy levels. Frontiers in Genetics 8: 119. https://doi.org/10.3389/fgene.2017.00119.
Chen, Q., L.M. Kawchuck, D.R. Lynch, M.S. Goettel, and D.K. Fujimoto. 2003. Identification of late blight, Colorado potato beetle, and blackleg resistance in three Mexican and two south American wild 2x(1EBN) Solanum species. American Journal of Potato Research 80: 9–19.
Christensen, C., L. Zotarelli, K.G. Haynes, and J. Colee. 2017. Rooting characteristics of Solanum chacoense and Solanum tuberosum in vitro. American Journal of Potato Research 94: 588–598.
Cooper, W.R., and J.B. Bamberg. 2014. Variation in Bactericera cockerelli (Hemiptera: Triozidae) oviposition, survival, and development on Solanum bulbocastanum germplasm. American Journal of Potato Research 91: 532–537.
Douches, D.S., J.B. Bamberg, W. Kirk, K. Jastrzebski, B.A. Niemira, J. Coombs, D.A. Biognin, and K.J. Fletcher. 2001. Evaluation of wild Solanum species for resistance to the US-8 genotype of Phytophthora infestans utilizing a fine-screening technique. American Journal of Potato Research 78: 159–165.
Hale, A.L., L. Reddivari, M.N. Nzaramba, J.B. Bamberg, and J.C. Miller Jr. 2008. Interspecific variability for antioxidant activity and phenolic content among Solanum species. American Journal of Potato Research 85: 332–341.
Hardigan, M., J. Bamberg, C. Robin Buell, and D. Douches. 2015. Taxonomy and genetic differentiation among wild and cultivated germplasm of Solanum sect. Petota The Plant Genome 8 (1): 16.
Hardigan, M.A., F.P.E. Laimbeer, L. Newton, E. Crisovan, J.P. Hamilton, B. Vaillancourt, K. Wiegert-Rininger, J.C. Wood, D.S. Douches, E.M. Farre, et al. 2017. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proceedings of the National Academy of Sciences USA 114: E9999–E10008.
Hardigan, M.A., F.P.E. Laimbeer, J.P. Hamilton, B. Vaillancourt, D.S. Douches, E.M. Farre, R.E. Veilleux, and C.R. Buell. 2018. Avoiding “one-size-fits-all” approaches to variant discovery. Proceedings of the National Academy of Sciences USA Letter. 115:E6394–E6395.
Haynes, K.G., H.E.M. Zaki, C.T. Christensen, E. Ogden, L.J. Rowland, M. Kramer, and L. Zotarell. 2017. High levels of Heterozygosity found for 15 SSR loci in Solanum chacoense. American Journal of Potato Research 94: 638–646.
Hirsch, C.N., C.D. Hirsch, K. Felcher, J. Coombs, D. Zarka, A. Van Deynze, W. De Jong, R.E. Veilleux, S. Jansky, P. Bethke, D.S. Douches, and C.R. Buell. 2013. Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 (Bethesda) 3: 1003–1013.
Hosaka, K., and R.E. Hanneman Jr. 1991. Seed protein variation within accessions of wild and cultivated potato species and inbred Solanum chacoense. Potato Research 34: 419–428.
Huang, B., D.M. Spooner, and Q. Liang. (2018). Genome diversity of potato. Proceedings of the National Academy of Sciences 115:July 10 letter.
Jansky, S., R. Simon, and D.M. Spooner. 2008. A test of taxonomic predictivity: Resistance to early blight in wild relatives of cultivated potato. Phytopathology 98: 680–687.
Lachance, J., and S.A. Tishkoff. 2013. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. Bioessays 35: 780–786.
Leisner, C.P., J.P. Hamilton, E. Crisovan, N.C. Manrique-Carpintero, A.P. Marand, L. Newton, G.M. Pham, J. Jiang, D.S. Douches, S.H. Jansky, and C.R. Buell. 2018. Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. The Plant Journal 94: 562–570.
Li, Y., C. Colleoni, J. Zhang, Q. Liang, Y. Hu, H. Ruess, R. Simon, Y. Liu, H. Liu, G. Yu, E. Schmitt, C. Ponitzki, G. Liu, H. Huang, F. Zhan, L. Chen, Y. Huang, D. Spooner, and B. Huang. 2018. Genomic analyses yield markers for identifying agronomically important genes in potato. Molecular Plant 11: 473–484.
Lokossou, A., H. Rietman, M. Wang, P. Krenek, H. van der Schoot, B. Henken, R. Hoekstra, V. Vleeshouwers, E. van der Vossen, R. Visser, E. Jacobsen, and B. Vosman. 2010. Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Molecular Plant-Microbe Interactions 23: 1206–1216.
Meirmans, P.G., S. Liu, and P.H. van Tienderen. 2018. The analysis of polyploid genetic data. Journal of Heredity 109: 283–296.
Perrier, X., Jacquemoud-Collet, J.P. 2006. DARwin software. http://darwin.cirad.fr/
Robinson, B.R., C.G. Salinas, P.R. Parra, J.B. Bamberg, R.I. Diaz de la Garza, and A. Goyer. 2019. Expression levels of the Y-Glutamyl hydrolase I gene predict vitamin B9 content in potato tubers. Agronomy 9: 734. https://doi.org/10.3390/agronomy9110734.
Sarkinen, T., L. Bohs, R. Olmstead, and S. Knapp. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evolutionary Biology 13: 214.
Simko, I., K.G. Haynes, and R.W. Jones. 2006. Assessment of linkage disequilibrium in potato genome with single nucleotide polymorphism markers. Genetics 173: 2237–2245.
Sinden, S., L. Sanford, and K. Deahl. 1986. Segregation of Leptine glycoalkaloids in Solanum chacoense bitter. Journal of Agricultural and Food Chemistry 34: 372–377.
Sood, S., V. Bhardwaj, S.K. Pandy, and S.K. Chakrabarti. 2017. Potato genetic resources. In: S. K. Chakrabarti et al. (eds.). The Potato Genome, Compendium of Plant Genomes. https://doi.org/10.1007/978-3-319-66135-3_.
Spooner, D.M., M. Ghislain, R. Simon, S.H. Jansky, and T. Gavrilenko. 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Botanical Review 80: 283–383.
Acknowledgements and Perspectives
We thank the UW Peninsular Agricultural Research Station program and staff for their assistance. Doyle’s “The Greek Interpreter” (1893) recounts Holmes’ contention that the creativity of “art in the blood” was responsible for his remarkable powers of deduction. It is intriguing to note that Avery, who first published the discovery of DNA as the basis of heredity (1944) similarly started his academic career as a student of music. Thus, when visionary scientists created the US Potato Genebank (1948) its value as a collection of potato heredity encoded in the language of DNA was a very new idea. Now after 75 years, hundreds of thousands of research papers, and the elevation of this esoteric chemical to a household term, we continue to look for creative ways to study and interpret the basic characteristics of DNA that relate to mining exotic germplasm to improve the potato crop.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 114 kb)
Rights and permissions
About this article
Cite this article
Bamberg, J., del Rio, A. Assessing under-Estimation of Genetic Diversity within Wild Potato (Solanum) Species Populations. Am. J. Potato Res. 97, 547–553 (2020). https://doi.org/10.1007/s12230-020-09802-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12230-020-09802-3