Abstract
Current research energies are fixated on the synthesis of environmentally friendly and non-hazardous products, which include finding and recognizing biosurfactants that can substitute synthetic surfactants. Microbial biosurfactants are surface-active compounds synthesized intracellularly or extracellularly. To use biosurfactants in various industries, it is essential to understand scientific engagements that demonstrate its potentials as real advancement in the 21st century. Other than applying a substantial effect on the world economic market, engineered hyper-producing microbial strains in combination with optimized cultivation parameters have made it probable for many industrial companies to receive the profits of 'green' biosurfactant innovation. There needs to be an emphasis on the worldwide state of biosurfactant synthesis, expression of biosurfactant genes in expressive host systems, the recent developments, and prospects in this line of research. Thus, molecular dynamics with respect to genetic engineering of biosynthetic genes are proposed as new biotechnological tools for development, improved synthesis, and applications of biosurfactants. For example, mutant and hyper-producing recombinants have been designed efficaciously to advance the nature, quantity, and quality of biosurfactants. The fastidious and deliberate investigation will prompt a comprehension of the molecular dynamics and phenomena in new microorganisms. Throughout the decade, valuable data on the molecular genetics of biosurfactant have been produced, and this solid foundation would encourage application-oriented yields of the biosurfactant production industry and expand its utilization in diverse fields. Therefore, the conversations among different interdisciplinary experts from various scientific interests such as microbiology, biochemistry, molecular biology, and genetics are indispensable and significant to accomplish these objectives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Science, mysteries, phenomena, discoveries, innovations, and genetic engineering are required to invigorate novel methods in synthesizing microbial products. Surfactants of microbial origin are plausible substitution to chemically synthesized ones due to mindfulness on the need to protect the environment [1]. Most commercial synthetic surfactants produced are toxic, carcinogenic, and non-environmentally friendly [2]. In recent years, research efforts in scientific and industrial communities have centered on environmentally friendly bio-products, namely biosurfactants (BioSs). BioSs are surface-active compounds (heterogeneous groups of secondary metabolites) which could reduce interfacial tension (IFT) and surface tension (ST), thus making it have potential benefits over chemical surfactants for their utilization [3, 4]. BioSs are advantageous over synthetic surfactants due to their effectiveness, specificity, high biodegradability, low toxicity, biocompatibility, and pollution control applications. These attributes allow their biotechnological applications in cosmetic and pharmaceutical products, food additives, agriculture, and the petroleum industry [1, 5, 6]. The other promising properties include emulsification, excellent detergency, foaming, enhanced oil recovery, and metal sequestration. They also play essential roles in microbial adhesion to the interfaces, changing surface-active phenomena, spreading, wetting, flocculation/dispersion, flotation, and solubility of hydrophobic organic compounds [5, 7].
For example, there are factors, such as low product yields, high costs, severe foaming, and complex procedures originating from set-ups during cultivation that limit industrial and biotechnological applications of BioSs [8]. The connections between these biomolecules' production conditions, their structures, and capabilities are central components of their improvement in industry, environment, and biotechnology [9]. Thus, future research's central part is the improved synthesis of BioSs with high-value properties by genetic engineering or modifications. Accordingly, low product yields that limit biotechnological and environmental applications permitted the increasing consideration in the genetic engineering relative to improving the synthesis of BioS production as of late [8]. The BioS synthesis by certain microorganisms may be ascribed to the presence of specific genes or enzymes activated in the presence of hydrocarbons and other carbon substrates [10]. It is pertinent to note that bacteria are capable of synthesizing BioSs while degrading hydrocarbon compounds. Nevertheless, relatively little is known about the genetics, molecular features, and functional characterization of the degradative and BioS synthetic systems [11,12,13].
It is worthy to note that the industrial and biotechnological applications of genetically enhanced and hyper-producing recombinant strains have not been appropriately established, even with hyper-BioS producers being reported [14]. The development of hyper-producing microbial strains is carried out through the utilization of new-age biotechnology to screen potent proteins and enzymes present in microorganisms, which may help improve their outcome as bio-products [10, 15]. This is realized through the use of different advances inclusive of introduction or the manipulation of genetically engineered microorganisms, single/naked genes, and operons, in silico computation for the discovery of novel metabolic pathways, bio-prospecting using high throughput screening, genome mining, and metagenomic screening [15, 16]. Thus, the significant focus therein in BioS research is the utilization of conventional genetic improvement approaches. Genetic engineering involves the modifications of microbial genetic materials to obtain new or improved product capabilities of biotechnological and environmental importance. The advancement in BioS surface activity and cost-effectiveness of BioS synthesis is also the general objective for developing hyper-producing microorganisms [17]. There are different hyper-producing microbial strains that have developed for improved BioS activity such as B. subtilis (pHT43-comXphrC), B. subtilis fmbR, B. subtilis fmbR-1, B. subtilis THY-7, Bacillus subtilis THY-7/Pg3-srfA, B. subtilis TS593, B. subtilis TS662, B. subtilis, B. subtilis 168S1, 168S2 [18,19,20,21]. In recent years, many studies on enhancing the production of BioS have been of interest with a focus on the use of inexpensive substrates, optimization of medial components and growth conditions, fermentation strategies, statistical model optimization of process and environmental parameters, as well as optimization of downstream processes [13, 22, 23]. Reductions in BioS production total costs will hinge on developing cheaper strategies, including the use of low-cost substrates and the use of genetically engineered microbial strains for enhanced production yields. The possible use and improved application of these hyper-producers, in addition to novel cost-effective bioprocesses, throw the challenges and offer prospects for BioS competitiveness in the world market. This review paper provides a general synopsis on the biosynthesis and genetically engineering strategies for improving BioS synthesis with emphasis on the usage of the novel, recombinant, and hyper-producer microbial strains in bioremediation applications.
Biosurfactant Classifications
BioSs are produced by an extensive diversity of microorganisms and possess structures of different chemical and surface properties [6]. Microorganisms can produce different types of BioSs, which include glycolipids (mannosylerythritol, rhamnolipids, sophorolipids, xylolipid, cellobiose lipids trehalose lipids), lipopeptides (subtilisin, viscosin, serrawettin, surfactin, polymyxin, iturin), polysaccharide–protein complexes, flavolipid, phospholipids, fatty acids, polymeric surfactants (liposan, alasan, emulsan), and lipids [6, 24]. The most frequently produced low molecular weight surface-active compounds are glycolipids and lipopeptides. The other group that has often been used substitutively with BioSs to represent surface-active biomolecules is bioemulsifiers [25]. The different types of BioSs and the structural compositions are discussed and provided below.
Glycolipids
Numerous glycolipids, encompassing simple fatty acids esterified to a carbohydrate moiety have been defined from different microorganisms [26]. Their structural composition differs from simple sugars with fatty acyl substituents to complex carbohydrates that can successively be connected to aromatic compounds, nucleosides, or terpenoids, in addition to having different connection points to “un”, “mono”, “poly”, unsaturated fatty acids through glycosidic or ester linkages. Glycolipid BioS structures include rhamnolipids, sophorolipids, mannosylerythritol, trehalose lipids, and xylolipids.
Rhamnolipids
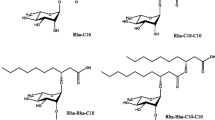
Rhamnolipids are glycolipid BioS, synthesized by different microorganisms and not limited to Pseudomonas aeruginosa, Renibacterium salmoninarum, Serratia rubidaea, Burkholderia thailandensis, Acinetobacter sp. YC-X 2 during secondary metabolism [27,28,29,30,31]. An oily glycolipid BioS synthesized formerly by Pseudomonas pyocyanea was also first discovered in 1946. Edwards and Hayashi [32] explained the chemical structure of the rhamnolipid as glycosides comprising of one (mono-rhamnolipids) (Fig. 1a) or two (di-rhamnolipids) (Fig. 1b) rhamnose sugars connected by an O-glycosidic bond to lipid moieties. The hydrophobic component of the rhamnolipid group consists of one or two bonds. Still, in uncommon cases, three β-hydroxy unsaturated fatty chains might be single, double, or triple bonded and possess different lengths of C8 to C16. The hydrophilic component of rhamnolipid comprises single- or twofold rhamnose sugars connected by an α-1, 2-glycosidic bond [33].
Sophorolipid
Sophorolipid is a glycolipid complex that is synthesized by a couple of non-pathogenic yeast species. Sophorolipid comprises of hydrophilic sugar head called sophorose and a hydrophobic unsaturated fat of 16 or 18-carbon chain length. Sophorose encloses glucose of disaccharide group connected by an irregular β-1, 2 bonds acetylated on the 6′- as well as 6″-positions [34]. The carboxylic end of sophorolipid could be either lactonized (Fig. 1c) or an acidic form of sophorolipid (Fig. 1d).
Mannosylerythritol and cellobiose lipids
Mannosylerythritol lipids are functional glycolipids also synthesized abundantly by yeast strains. They comprise fatty acids joined to 4-O-β-D-manno-pyranosyl erythritol or 1-O-β-D-manno-pyranosyl erythritol as the hydrophilic head group (Fig. 1e) [35]. Cellobiose lipid is another glycolipid BioSs with the significant product recognized as 16-O-(2″,3″,4″,6′-tetra-O-acetyl-β-cellobiosyl)-2-hydroxyhexadecanoic acid (Fig. 1f). Yeasts and mycelia organisms are shown to produce a few extracellular glycolipids, including cellobiose, and mannosylerythritol lipids [36].
Trehalolipids
Trehalolipids are made from unsaturated fatty acid group length (hydrophobic components) in a blend with carbohydrate group (hydrophilic component) (Fig. 1g). The hydrophobic parts of trehalolipids are vastly different, comprising hydroxylated branched fatty acids of varying chain lengths and aliphatic acids. The amounts of the hydrophobic chain in every molecule of trehalose lipids are usually mono-, di-, and tetra-esters, separately connected to long-chain unsaturated fats by an ester bond [26].
Xylolipids
Xylolipid is another class of BioS discovered recently with molecular composition of methyl-2-O-methyl–d-xylopyranoside, a hydrophilic component connected to hydrophobic parts of the octadecanoic acid (Fig. 1h) [37].
Lipopeptides
Lipopeptides are biomolecules comprising a lipid connected to a peptide group (small chains of amino acid monomers joined by peptide bonds). Lipopeptides are synthesized by various bacterial genera such as Bacillus, Streptomyces, Pseudomonas, and fungi such as Aspergillus [38]. Lipopeptides have received substantial consideration for their antimicrobial and surfactant properties. Bacillus subtilis produced acyclic lipopeptide surfactin, which is one of the most recognized BioSs [38]. The major lipopeptide groups are further discussed below.
Surfactin
Surfactin group is the most prominent Lipopeptide (Fig. 2a), which comprises a peptide loop of seven different amino acids (L-valine, two L-leucine, L-aspartic acid, glutamic acid, and two D-leucines) and a hydrophobic fatty acid chain, of 13 to 15 carbon length. Surfactin has shown potent antibacterial, antitumoral, antibiofilm, and antiviral activities as well as bioremediation process and environmental applications in recent studies [39, 40].
The chemical structures of (a) surfactin, (b) iturin, and (c) fengycin biosurfactant. The cyclic lipopeptide contains fatty acid chain linked with amino acids. The compound subordinate in each group originates from various amino acid constituents [41]
Iturin
Another lipopeptide group with a hydrophobic fatty acid joined by an amide bond to a peptide moiety (a constituent amino acid residual constituent) is iturin [41]. They possess a typical arrangement and show variability at four different positions (Fig. 2b) [42]. The various groups associated with iturin include bacillopeptin, mycosubtilin, iturin A, C, D, E, bacillomycin D, F, and L, respectively. Iturin, as a lipopeptide group, has also been useful in antimicrobial, pharmaceutical, and biotechnological applications [42].
Fengycin
Fengycin is another set of lipopeptide groups with a lipidic fraction and ten amino acids connected to an N-terminal end. Iturin and surfactin contrast from this group due to different amino acids, such as allo-threonine and ornithine [41]. Like iturin group, fengycin possesses robust antifungal action; it inhibits the development of a wide variety of plant pathogens and application in improved diesel biodegradation. The diverse array of the peptide component (variations with trademark Alanine-Valine di morphy positioned at six in the peptide ring) also authorizes the characterization of a new fengycin B into fengycin family (Fig. 2c).
Fatty Acids and Phospholipid
Fatty acid and phospholipid are unsaturated BioS of C12 to C14 lengths and complex unsaturated fatty acids comprising hydroxyl groups and alkyl branches. Different bacteria produce vast numbers of fatty acids and phospholipid surfactants as the fatty acids are suitable as BioSs due to their surface activity [38].
Flavolipid
A class of BioS with stable interfacial activity and emulsifying capacity is represented as flavolipids. This group’s polar end (Fig. 3) possesses two cadaverine molecules and citric acid, which is somewhat dissimilar to the polar groups in other reported BioSs. Flavolipid BioS is of interest due to their potentiality in environmental, biotechnology, industrial applications [43].
Polymeric Biosurfactants
Polymeric BioSs are generally high atomic weight biopolymers, with characteristics, for example, rigidity, increased thickness, and shear resistance. Emulsan and liposan, synthesized by Acinetobacter calcoaceticus and Candida lipolytica, respectively, are the best-studied polymeric BioS [38]. Different cases of particulate BioS are extracellular vesicles of microbial cells, which aid hydrocarbon emulsification [44]. Emulsan holds a backbone comprising a 2-amino-2-deoxy-hexuronic acid, amino sugars, glucose, fatty acids, and galactosamine (2-amino-2-deoxy-galactose) connected to the main chain through amide and ester bonds (Fig. 4) [45].
The structural composition of emulsan, a major microbial surface-active compound with high molecular weight [44]
Functional Characterization of Different Biosurfactant Biosynthetic Genes in Microorganisms
The molecular characterization and biosynthetic regulation of Bacillus subtilis lipopeptide BioS [46] and Pseudomonas aeruginosa glycolipid (rhamnolipid) BioS were the first to be reported [47]. Additionally, the molecular characteristics of other BioSs that have also been described include iturin and lichenysin from Bacillus species [48, 49], emulsan from Acinetobacter species [50], arthrofactin from Pseudomonas species [51], and mannosylerythritol lipids from Candida [48]. The biosynthetic characterization of other less-known BioSs such as viscosin, amphisin, serrawettin, hydrophobin, lokisin, and tensin is mostly non-existent [48].
Surfactin Synthetase Genes
The molecular characterization and biosynthetic regulation of surfactin ensue through a non-ribosomal peptide synthetase mechanism. The step includes a multienzyme peptide synthase complex, which comprises four enzymatic subunits SrfA, SrfB, SrfC, and SrfD. SrfA carries out the activation and addition of amino acids, namely Glu, Leu, and D-Leu. At the same time, srfB encodes synthetases that catalyze Val, Asp, and D-Leu's proliferation and activation. Subsequently, the thioesterase type 1 motif necessary for peptide termination and Leu is activated by SrfC (Fig. 5). Finally, the SrfD located terminally and encodes for thioesterase type II required for the lactonization process. The SrfA operon consists of a sfp gene encoding the phosphopantetheinyl transferase required for surfactin synthetase activation [52, 53] These enzymes are called surfactin synthetases needed for surfactin biosynthesis and are coded by srf operon [46].
Lichenysin Synthetase Operon
The non-ribosomal peptide synthetases (NRPSs) group, also known as multimodular peptide synthetases, is responsible for synthesizing lichenysin [54]. There is description of seven amino acid activation- thiolation, two epimerization, and one thioesterase domain in a lichenysin synthetase operon which is similar to surfactin and other peptide synthethases [48, 49]. There are recognition, activation, and incorporation of respective amino acids, namely L-Gln, L-Leu, D-Leu, L-Val, L-Asp, D-Leu, and L-Ile/L-Val by different functional modules such as LchAA, LchAB, and LchAC (Fig. 5). During lichenysin synthesis, the starting unit is LchAA, and modules in LchAB and LchAC initiate peptide chain elongation. Additionally, the terminal end of LchAC, specifically the putative thioesterase, is responsible for the cyclization and release of the peptide product. In contrast, the stimulation and initiation of the BioS synthesis are carried out by LchA-TE [55].
Non-Ribosomal Peptide Synthetase
The non-ribosomal peptides are assembled by NRPS enzymes comprising modules that are responsible for the sequential selection, activation, and condensation of precursor amino acids, fatty acids, alpha-keto acids, and alpha-hydroxy acids, as well as polyketide-derived units [56, 57]. These peptides' structural diversity is typically cyclic or branched compounds comprising small heterocyclic rings, proteinogenic amino acids, and other uncommon variations in the peptide backbone [58]. The functional characterization of the BioS biosynthesis gene clusters involved in directing the non-ribosomal synthesis of bioactive compounds in Bacillus amyloliquefaciens, Bacillus subtilis, and Bacillus tequilensis has been discussed in previous reports [59,60,61].
Iturin Synthetase Genes
The four open reading frames, ituD, ituA, ituB, and ituC constitute a significant component of iturin synthetase operon. The specific deficiency in iturin A production was confirmed due to the disruption of putative malonyl coenzyme A transacylase encoded by the ituD gene [62]. However, ituC and ituB genes encode the peptide synthetase that has one epimerization domain, thioesterase domain that helps in peptide cyclization, and two adenylation domains [48, 62]. On the other hand, the peptide synthetase encoded by the ituB gene possesses four amino acid adenylation domains. Finally, the ituA gene contains prominent features of three functional areas homologous to aminotransferase, β-ketoacyl synthetase, and amino acid adenylation [48].
Arthrofactin Synthetase Gene Cluster
The overall modular architecture of arthrofactin synthetase gene cluster follows the collinearity rule as revealed by cloning and recombinant technology. There are three genes of arthrofactin operon, namely, arfA, arfB, and arfC which encodes ArfA, ArfB, and ArfC with two, four, and five functional modules representing cyclic lipo-undecapeptide BioS. There is no epimerization domain in each of these modules but features condensation, adenylation, and thiolation domains that are characteristic for such multienzymes [51].
Emulsan Synthetase Genes
There are five different emulsan synthetase genes (wza, wzb, wzc, wzx, wzy) required for the biosynthesis of emulsan by Acinetobacter lwoffii RAG-1 [50]. The importance and establishment of wzc (protein tyrosine kinase) and wzb (protein tyrosine phosphatase), respectively, were later confirmed in the emulsan synthetase cluster. There was an emulsan-defective phenotype due to the deletion in either of the two genes [63].
Rhamnosyl-Synthetase Genes
The biosynthesis of rhamnolipid is involved by three main enzymatic reactions with β-oxidation confirmed to play a significant role in rhamnolipid production (Fig. 6) [47]. The substrate needed for both mono- and di-rhamnolipids is utilized and activated by rhamnose moiety, dependent on the RmlBCAD pathway. This process is further encoded by the catalytic activity of the enzyme AlgC and RmlBCAD operon. In the rhamnose sugar precursor synthesis, the typical D-glucose molecule is converted to D-glucose-1-phosphate catalyzed by phosphomannomutase enzyme AlgC, participating in the biosynthesis of glucose and rhamnose needed for the formation of core-LPS [64]. The process then follows the synthesis of dTDP-D-glucose by enzyme, RmIA. The RmIB further converts the dTDP-D-glucose to dTDP-4-oxo-6-deoxyl-D-glucose by RmIB and subsequent conversion to dTDP-6-deoxyl-L-deoxyl-4-hexulose by enzyme RmIC. The RmID enzymes convert dTDP-6-deoxy-L-lyso-4-hexulose to dTDP-L-rhamnose. The dTDP-D-glucose and dTDP-6-deoxyl-L-deoxyl-4-hexulose are rhamnosyl-transferases RhIB and RhIC substrates, needed for the mono- and di-rhamnolipid biosynthesis. Hypothetically, RhIG enzyme functions by relaying fatty acid synthesis intermediates into the rhamnolipid pathway [47, 53].
Alasan Synthetase Genes
The complex anionic polysaccharide containing three proteins, namely AlnA, AlnB, and AlnC, as well as covalently bound alanine (apoalasan), are components of Acinetobacter radioresistens KA53 alasan. The recombinant protein E. coli OmpA has an amino acid sequence homologous to that of the recombinant protein AlnA [65]. Likewise, the family of antioxidant enzymes known as peroxiredoxins has strong homology to AlnB amino acid sequence. Additional information about the mode of action of Alasan BioS is anticipated from the unknown genetic detail of AlnC [66].
Physiology, Pathways, and Kinetics of Biosurfactant Production
Biosurfactant Physiology and Metabolic Pathways
BioSs are synthesized through intracellular or extracellular adhesion to microbial cells when cultured in liquid medium containing immiscible substrate as a source of carbon and energy. Microbial cell function associated with BioS is not understood fully, as speculations have been made about their application in the emulsification of hydrophobic organic pollutants with low solubility [67]. The role played by BioS is enabled via reducing the surface tension between the interphase, thus increasing the availability of substrate for metabolism and uptake [25, 68,69,70]. The different pathways for the biosynthesis of BioSs are discussed below, ranging from glycolipids (rhamnolipids, sophorolipids, phospholipids, mannosylerythritol, trehalose lipids), lipopeptide (surfactin) including polymeric BioSs (emulsan). BioSs are amphiphilic compound, comprising both hydrophilic polar and hydrophobic non-polar joined ends. Microorganisms exploit the hydrophilic polar moieties for cell metabolism, whereas the utilization of hydrocarbon portion is entirely dependent on the hydrophobic moieties [67, 71]. The study on respective metabolic pathways gives an understanding of how BioSs are synthesized from different substrates. The synthesis of precursors for BioS production involves different metabolic pathways which are dependent on carbon substrates provided initially and utilized in the production culture medium. In the synthesis of glycolipids, the flow of the primary carbon source (carbohydrates) is regulated by the lipogenic pathways, while glycolytic pathway, on the other hand, enabled the formation of the hydrophilic moiety (Fig. 7a) [67]. A significant precursor of carbohydrates (glucose 6-phosphate) present in the hydrophilic component of glycolipid BioS is made from the degradation of carbohydrate substrates such as glucose or glycerol. Subsequently, acetyl-CoA is produced from pyruvate, which in turn gives malonyl-CoA in addition with oxaloacetate. Thus, this process is followed by conversion into an important precursor for the synthesis of lipids, namely fatty acids [71]. In a situation where petroleum hydrocarbons are utilized as the substrate source, the mode of action is principally engaged to both the gluconeogenesis and lipolytic pathways, thereby allowing its usage to produce sugars or fatty acids (Fig. 7b) [67].
The intermediate metabolism identified with the synthesis of glycolipid biosurfactant precursors with (a) different carbohydrate substrates with enzymes (i) phosphofructokinase; (ii) pyruvate kinase; and (iii) isocitrate dehydrogenase responsible for the flow of carbon (b) hydrocarbon substrates with enzymes (iv) isocitrate lyase; (v) malate synthase; (vi) phosphoenolpyruvate carboxykinase; and (vii) fructose-1-phosphatase responsible for the flow of carbon
So far, the biosynthesis of sophorolipids BioS includes the successive transfer of activated glucose molecules (UDP-glucose) to a hydroxyl acid in reactions catalyzed by two separate glycosyltransferases. The acetyltransferase further acetylates the glucose molecule, and the fatty acid constituents can be produced by modifying hydrocarbons or de novo from acetate in the growth medium [72]. In the case of mannosylerythritol biosynthesis, the genes required were formerly acknowledged on smut fungi named Ustilago maydis, which yields mannosylerythritol inclusive of cellobiose lipids [73]. Mannosylerythritol BioS is synthesized through different enzymatic reactions. The enzyme mannosyltransferase required in the synthesis of mannosylerythritol is encoded by emt1, while mat1 translates an acetyltransferase catalyzing the mannosylerythritol acetylation at both the C-4′ and C-6′ hydroxyl groups of mannoses. In addition, an acyltransferase is required for the acylation of mannosylerythritol, which is encoded by mac1 [73, 74].
On the other hand, trehalose (trehalolipids) biosynthesis encompasses glucose transfer from UDP-glucose to glucose-6-phosphate to synthesize trehalose-6-P-UDP. This is synthesized through the catalytic capability of trehalose-6-phosphate synthase. Subsequently, a free disaccharide catalyzed by trehalose-6-phosphate phosphatase is generated by de-phosphorylation [75]. Besides, the synthesis of phospholipid occurs in the cytosol corresponding to the membrane that is coupled with proteins that act in allocation (flippase and floppase) and synthesis (acyl transferases, phosphatase, and choline phosphotransferase). Ultimately, the phospholipids containing vesicles destined for the cytoplasmic cellular sprout out on its exterior. The exoplasmic cellular membrane also generates the release of phospholipids BioS on its inner leaflet [76].
Alternatively, the biosynthesis of surfactin, which is one of the prominent lipopeptide BioSs, ensues through a non-ribosomal peptide synthetase mechanism [46]. This process involves joining amino acids into the surfactin's peptide component catalyzed by surfactin synthase through a thiotemplate mechanism. This includes amino acid activation by ATP and assemblage of amino acids into a peptide chain. Using an acyltransferase enzyme, the lipopeptide BioS is then formed by linking the hydroxyl fatty acid to a peptide group [77]. There is also a second pathway responsible for regulating the biosynthesis of surfactin (expression of SrfA). For example, B. subtilis encodes for eight of these [PhrA, PhrC (CSF), PhrE, PhrF, PhrG, PhrH, PhrI, and PhrK] and 11 aspartyl-phosphate phosphatase proteins (RapA to RapK) as shown in Fig. 8 [78]. The activity of the co-transcribed Rap proteins is inhibited by Phr peptides. The RapC which is responsible for the de-phosphorylation of ComA is proportional to the concentration of PhrC. However, there is usually repression of surfactin synthesis when there is a high intracellular concentration of PhrC. In this regard, the BioS production is subsequently dependent on the SpoOK (permease) required for peptide transfer across the membrane [79]. The process involved in srfA gene expression is dependent on RapC low concentrations to improve the availability of phosphorylated ComA, thereby triggering transcription by binding the promoter region. Thus, srfA gene expression is further regulated by repressor proteins such as AbrB and GTP as well as other transcriptional regulators such as DegU and sensor CodY [78].
The genetic regulation for the biosynthesis of surfactin by Bacillus species. The negative positive regulation is indicated closed circles and close-head arrows indicate positive regulation [Adopted from Roongsawang et al.]
[78]
Kinetics of Biosurfactant Production
The BioS production kinetics has substantial variance among diverse systems. The different kinetic parameters to be considered are assembled below:
-
(a)
Growth-dependent;
-
(b)
Growth-limiting;
-
(c)
Synthesis by immobilized or resting cells; and
-
(d)
Synthesis with precursor supplements [80].
In production related to growth, there exists a parallel correlation between cellular growth, substrate usage, and increased BioS production. A heightened increase in BioS concentration due to the restraint of one or more medium constituents characterizes the synthesis under growth-limiting conditions. The synthesis by immobilized or resting cells is a type where the cells use carbon substrates continuously for BioS synthesis, with relatively no cell multiplication. The last kinetic parameter, as listed above, involves the addition of BioS precursors to the production medium. As revealed by researchers, precursor addition often results in qualitative and quantitative variations in BioS product yield [67].
Substrate Concentrations and Formulations used for Biosurfactant Production
In the last decade, many studies have been undertaken for media optimization, especially for most prominent BioS producers—Bacillus, Pseudomonas, Candida, and Acinetobacter species. Different parameters such as type and amount of carbon and nitrogen sources, the ratio of metal supplements, pH, temperature, aeration, dissolved oxygen, and cell density have been studied expansively and found to be fundamental for BioS production [81, 82]. Table 1 gives an overview of a few of the prominent BioS producers and concentrations of carbon sources, which showed the highest reported BioS yield when grown in shake flasks and large‐scale fermenter vessels. The output and productivity of BioS are significant in the range of culture conditions, medium compositions, primary substrate concentration, and current operating scales at relatively small-scale synthesis. However, techniques such as response surface methodology and statistical methods like Box–Behnken, Taguchi, and Plackett–Burman Design have been frequently used to handle multiple data and optimize BioS production processes [83,84,85].
Additionally, artificial intelligence‐based technique is another method that has been utilized for media optimization and yield improvement of BioS [86, 87]. In an enormous scope, the BioS synthesis costs are commonly revealed as being fundamentally subject to bioreactor volume (impacted by profitability and fairly by titer because of startup/closure time and batch scheduling), costs of raw materials (affected by the decision of crude materials and yield), and costs of separation and purification [88, 89]. Another key expense at industrial scale is the startup, cleansing, and costs of sterilization caused when running sequenced batch/fed-batch production campaigns, which are frequently neglected; however, these have been distinguished by mechanical accomplices as a vital objective for the decrease in costs. The frequency of production campaigns is inversely proportional to the product titer for a given quantity of BioS. As such, having the option to run a fermentation process for a more extended period while keeping up profitability and arriving at higher titers will decrease the entire production costs [90]. There is a need for cost-effectiveness through the utilization of renewable raw materials for most biotechnological product processes. This is why many products dependent on BioS and bioemulsifier are very costly and still in low amounts. Consequently, despite expanded interest in BioS, their bulk use has not been acknowledged because of huge costs, particularly when contrasted with surfactants of chemical origin [90, 91].
Genetic Engineering Strategies for Enhancing Biosurfactant Production
It has been hard to accomplish significant breakthroughs regarding BioS production with traditional techniques such as breeding, fermentation, optimization, and statistical optimization [23]. Thus, current genetic engineering approaches can be used to satisfy the need for new, competitive, and environmentally friendly BioS products as the improvement of microbial strains offers a great prospect in reducing the cost of production [92, 93]. The enhanced strains compared with parent strains also use the same quantity of raw materials thereby synthesizing a higher amount of the desired products [93]. The genetics of the producer organism is an essential factor that affects the yield of all biotechnological products since the ability to produce a metabolite is conferred by organism genes [94]. Genetic engineering is a strategy that includes the modification of genetically engineered microorganisms, naked genes, or plasmid-containing BioS genes using biotechnological techniques. This method helps in the manipulation of single organism's genes or operons, including heritable and non-heritable DNA, constructing biosynthetic pathways, and sequence modification of existing BioS synthetic genes. Therefore, it is of great significance to discuss the genetic methods utilized for improved synthesis of BioSs.
Recombinant DNA Technology
Recombinant microorganisms can be obtained by introducing an exogenous nucleic acid encoding BioS in a host microorganism. Here, the host microorganism can be chosen and utilized subjectively with the ability to produce the diverse BioS compounds. For example, the incorporation of a vector suitable for nucleic acid can be introduced into the host cell in any manner. A vector is a nucleic acid molecule incorporated and transported to the cell (carrier) with its capability in replication and expression for a host cell. Scientists have been successful in the usage of recombinant DNA technology to increase BioS production yield. For instance, metabolic engineering strategy led to an increased rhamnolipid production by recombinant E. coli expressing rhlAB compared to the parent strain (Pseudomonas aeruginosa) and other E. coli strains [95].
Similarly, the rate of fatty acid synthesis in recombinant E. coli increases 1.3-fold when RhlA is expressed, confirming that RhlA is required and sufficient in the formation of the rhamnolipid acyl moiety [96]. Anburajan et al. [97] also reported the heterologous expression of the surfactin synthetase gene from Bacillus licheniformis NIOT-06 into E. coli M15. Phosphopantetheinyl transferases are fundamental in activating polyketide, fatty acid, and non-ribosomal peptide synthetase enzymes. Therefore, it was proposed that Sfp phosphopantetheinyl transferase represents an important component of peptide synthesis systems and indispensable in the biosynthesis of lipopeptide BioS [13]. Porob et al. [60] reported that an sfp gene cloned from Bacillus tequilensis encoded 224 amino acids and the role of surfactin genes in BioS synthesis from 6 different Bacillus species. Additionally, the genes sfp, sfp0, and srfA were cloned into recombinant microbial strains BioSa, BioSb, and BioS, leading to improved BioS activity. The outcomes also discovered conserved family characteristics between BioS and esterase genes [98]. The production of lipopeptides with improved properties was facilitated through the engineering (cloning and sequencing) of the arthrofactin synthetase gene cluster, thus exploiting its usage industrially [51].
Mutagenesis
Mutagenesis is one of the methods by which the gene sequence of the microbial strain can be transformed in order to induce its activity [93]. This method is also used extensively as a mechanism for the development of hyper-producing microbial strains tailored for improved BioS synthesis. Different mutation techniques such as site-directed mutagenesis, treatment with DNA ligase, substitutions, deletions, and insertions have been proposed by researchers. Additionally, it is conceivable to get a mutation variant by other techniques, such as ultraviolet irradiation, physical and/or chemical mutagenesis methyl-N’-nitro-N-nitrosoguanidine, or by selection based on resistance to ionic detergents such as CTAB [93, 99]. One of the strategies to cope with economic constrains associated with BioS production is the development of a hyper-producing mutants. From the literature search, there has been limitation of the hyper-producing mutants for improved BioS synthesis to genera of Acinetobacter, Pseudomonas, and Bacillus, which are the producers of emulsan, rhamnolipid, and, respectively [94]. The overproduction of BioS by mutagenesis technique has been shown by previous reports. For instance, there was higher production of surfactants from a B. subtilis E8 mutant after being induced with ion beam [100].
Similarly, the use of gamma irradiation enabled a Pseudomonas aeruginosa MR01 mutant with a more than 1.5-fold BioS production [101]. The introduction of gamma-ray on P. aeruginosa S8 enabled the formation of mutant strain with 2–3 times BioS production than the wild-type cells [102]. In a report, the RhlA and RhlB mutants showed that swarming requires the expression of the rhlA gene but does not necessitate rhamnolipid production. It was also demonstrated that ammonium used instead of nitrate as a nitrogen source along with excess available iron decreases RhlA expression and swarming motility [103]. The genes responsible for the biosynthesis and control of emulsan heteropolysaccharides BioS were also targeted in a different report. The mutants deleted for several of the genes were defective in emulsification, indicating that polysaccharide is essential for extracellular emulsifying activity [50]. The mutated proteins flawed during the catalytic activity performed better in enhancing apoemulsan-mediated emulsifying activity [104]. Acinetobacter venetianus RAG1 also forms emulsan as reported in another study. The removal of the protein fraction yield apoemulsan shows a lower emulsifying activity on hydrophobic pollutants such as n-hexadecane [48]. In another report, the arfB gene disruption mutant did not produce arthrofactin but exhibited low swarming activity while enhancing biofilm formation and extracellular fiber production. These results suggest that the arthrofactin synthetase gene may have multiple functions [51].
Overexpression of Extracellular Peptides
The overexpression of small signaling molecules that activate and regulate the biosynthetic cluster of an antibiotic revealed the possibility of enhancing biosurfactant production [19]. As reported, the overexpression of extracellular peptides, ComX, and PhrC is a crucial resource for improving surfactant production as cells grow at low-level cell density. The production of surfactant in the strain, B. subtilis (pHT43-comXphrC) after 48 h cultivation was 6.4-fold higher than in the wild strain. Both extracellular signaling factors and different response pathways in B. subtilis were responsible for improving surfactin production through the development of genetic competence and quorum response trigger at low cell density [19]. Table 2 shows different microbial strains utilized for the improved synthesis of BioS, including the different genetic engineering techniques and BioS yields.
Substitution, Replacement, and Modifications of Amino Acids
The substitution or modification of amino acid refers to the replacement of an amino acid residue with a side chain with another amino residue with similar properties. This method is dependent on the various side chains which could be acidic side chains (e.g., aspartic acid, glutamic acid), basic side chains (e.g., lysine, arginine, histidine), non-polar side chains (e.g., alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine, tryptophan), beta-branched side chains (e.g., threonine, valine, isoleucine), uncharged polar side chains (e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine), and aromatic side chains (e.g., tyrosine, phenylalanine, tryptophan, such as the histidine). The intensity of the promoter affects the transcription level of a target gene directly, which in turn impacts the way the intensity is expressed. Also, one of the most direct and effective ways to regulate the expression of key genes is through promoter substitution [18]. Two earlier studies investigated surfactin yields after promoter exchange in the presence of the srfA operon. The studies were, however, conducted with different surfactin producer strains and substitute promoter sequences, which provided conflicting results [20, 105]. Coutte et al. [105] described the replacement of the srfA native promoter by the constitutive promoter PrepU in B. subtilis 168 after the integration of a functional sfp gene. Replacement of surfactin operon promoter by a constitutive one prevented lipopeptide synthetase expression thereby revealing only a small enhancement of surfactin production that suggests that the precursor supply is the problem for the surfactin overproduction in B. subtilis 168 derivative strains and not the synthetase expression [105].
In contrast, Sun et al. [20] reported tenfold enhanced surfactin yields after replacement of PsrfA with Pspac, an IPTG-inducible hybrid promoter originating from B. subtilis bacteriophage SP01 and E. coli lac operon. Another study reported a similar output as the substitution of the native srfA promoter for the constitutive Pveg could significantly increase surfactin production in a strain with only the compound's low native production [8]. Similarly, when the promoter of the iturin operon was replaced by the repU promoter of the plasmid pUB110 replication protein, a threefold increase in the production of iturin A was observed [62].
Gene/Gene Cluster Knock-Out
This is a genetic technique that requires that one of an organism's genes is made inactive (“knocked out” of the organism) for enhanced BioS synthesis. The best approach in this technique is homologous recombination and through which a single gene gets deleted without effecting all other genes in an organism. Therefore, it could be a double knock-out, triple knock-out, or quadruple knock-outs depending on the number of genes knocked out in a specific microorganism [106]. The biosynthesis of different lipopeptides and polyketides would compete with biosurfactant production for energy, NADPH, and direct precursors unavoidably. Therefore, eliminating these large gene clusters could enhance the synthesis of BioSs. Table 2 shows that the deletion of competitive extracellular matrix formation pathways and other NRPS/PKS pathways improved surfactin production significantly by 3.3-fold compared to 168S1, which may be possibly due to the elimination of precursor competition [21].
Bioremediation Applications of Genetically Engineered Microorganisms with Biosurfactant Properties
Biosurfactant-facilitated bioremediation (BFB) is an approach that involves the inoculation of BioS producers or BioS monomers into the contaminated environments. BFB has attracted increasing attention recently, as more researches have focused on this innovative strategy. This methodology is adopted to overcome the constraints of bioavailability encountered during the transformation of contaminants in complex and harsh environments [107,108,109,110,111,112,113,114]. Microorganisms are proficient in producing different kinds of BioSs, which range from the low molecular weight to the high molecular weight BioS. These BioS-producing microorganisms belong to several genera such as Pseudomonas, Penicillium, Clostridium, Acinetobacter, Bacillus, Aeromonas, Brevibacterium, Lactobacillus, Arthrobacter, Citrobacter, Candida, Corynebacterium, Yarrowia, Ustilago, Aspergillus, Torulopsis, Ochrobactrum, Pseudozyma, Saccharomyces, Gordonia, Enterobacter, Rhodococcus, Halomonas, Serratia, Leuconostoc, and Thiobacillus [44, 115]. BioSs are amphiphilic surface-active agents that are known to have both polar and non-polar groups, and they decrease surface tension at the interface between two liquids incapable of forming a homogeneous substance, similar to water and oil [116]. BioSs synthesized by microorganisms are environmentally compatible, biodegradable, non-toxic, effective at extreme environmental conditions, and have higher foaming capacity, making them more suitable over their synthetic counterparts [23]. In different environments, bioremediation can be less efficient due to the low bioavailability of petroleum hydrocarbons. Thus, introducing BioS-producing microorganisms that emulsify hydrocarbons would make remediation more efficient and reliable [23, 117]. The amphiphilic nature of BioSs enables the solubilization of water-insoluble hydrophobic pollutants through emulsification and surface-area reduction [117, 118]. BioS bioaugmentation is an essential strategy to enhance the degradation rate in contaminated environments. However, for a safe and successful introduction of BioS, the toxicity and effectiveness of BioSs must be assessed before inoculation of the BioS-producing organism. Furthermore, BioS compounds and degradative enzymes termed biomaterials can be directly introduced into the contaminated environment. This strategy may minimize the regulatory burden that is imposed with the direct inoculation of foreign organisms [119].
There is increased demand for effective BioSs because of its biotechnological, industrial, and environmental applications [120]. There was an increase in the extracellular anionic polysaccharide, biodispersan produced by A. calcoaceticus A2, a mutant strain defective in protein secretion. The reduction in secreted proteins presented on the extracellular fluid reduced problems in the purification and application of biodispersan. Moreover, recombinant strains often give rise to better product characteristics [120]. The engineered E. coli M15 strain has potential for biotechnological application since it produces BioS at high rates and can avoid the complex downstream process associated with the conventional bioprocess [97]. Besides, enhanced expression of the rhlAB operon in wild-type strain PG201 resulted in increased rhamnolipid production. The expression of the rhlAB operon on a plasmid led to rhamnolipid BioS and/or polyhydroxyalkanoate hyper-production that can be used for the synthesis of biodegradable plastics [121]. In a different research, recombinant E. coli pSKA cloned with olive as sole carbon source containing the BioS gene srfA showed higher esterase and BioS activity in comparison to Bacillus sp. SK320 [122]. In another study, data revealed that the BioS genes were successfully cloned, expressed, and overexpressed in BioSa, BioSb, and BioSc, showing a twofold increase in the activity than the parent strain. It was further reported in the study that cloning of the BioS genes from Bacillus subtilis SK320 into E. coli resulted in the expression of the BioS activity and conferred enhanced esterase production in the recombinant cells BioSa, BioSb, and BioSc as compared to Bacillus subtilis SK320 [98]. Subsequently, it was discovered that emulsion formed in the presence of olive oil by the recombinant cells of Bacillus subtilis SK320 emphasizing that the cloning of the BioS gene conferred on the E. coli cells enhances its ability to utilize olive oil as a sole carbon source [98]. The gene encoding AlnB was also cloned, sequenced, and overexpressed in E. coli. Recombinant AlnB had no emulsifying activity, but it stabilizes oil-in-water emulsion generated by AlnA [66]. To further justify the finding, the apoemulsan recombinant-esterase mixture was investigated for emulsification of a wide range of pure and crude oil products from various sources, comparing the activities with that of fully proteinated emulsan. The results revealed that the esterase–apoemulsan complex was more effective as it emulsifies various hydrophobic substrates that are typically not likely to be emulsified by crude emulsan itself [10].
In this context, rhIB is a gene that has been reported to be a BioS-producing gene. In a study, BioS-producing gene was amplified from Pseudomonas aeruginosa, and the gene regulated by regulatory gene reduced the surface tension, thereby emulsifying the oils (petrol, diesel, and kerosene) [123]. The sfp nucleotide sequence was expressed in E. coli, and its putative product was purified for use in antibody production and the analysis of the amino acid sequence. Overproduction of sfp in Bacillus subtilis did not cause the production of an increased amount of surfactin, but it resulted in the repression of a lacZ transcriptional fusion of srfA operon, which encodes enzymes that catalyze surfactin synthesis [59]. There was formation of stable oil–water emulsions with hydrophobic substrates such as hexadecane from the mixture of apoemulsan that included the catalytically active soluble form of the recombinant esterase isolated from cell extracts or the solubilized inert form of the enzyme recovered from the inclusion bodies [104]. The first time investigation of Rhodococcus strain accounted for genetic enhancement in the synthesis of BioS and the role played in environmental remediation [124]. In another instance, the possibility of B. subtilis mutant BioS for biotechnological uses was shown by its stability to environmental factors such as pH and temperature and its applicability in more than 90% of the oil recovery process motor oil adsorbed to a sand sample [93].
Further researches are essential to improve the environmental scale applications with consideration on numerous ecological complexes and factors that limit BioS synthesis and utilization. To encourage field uses of these BioS innovations, substantial tests are foreseen to consolidate heterogeneities in topographical/hydrological features and bioremediation of contaminated sites. With the new improvement in this field and the spotlight on interdisciplinary research joined with advancements of metabolic and genetic engineering, the prospects of BioSs will be financially practical. The exploration in this field is progressing quickly, and it envelops areas as diverse as textiles, pharmaceutics, cosmetics, petroleum, wastewater treatment, agriculture, natural science, and molecular biology.
Conclusion and Future Perspectives
The inclusion of genetic engineering can improve not only BioS surface activity but also its production yield. Genetically engineered hyper-producing organisms can bring breakthroughs in the production process and give high yields if the microbial surfactant production's genetics is known in detail. Therefore, it is desirable and recommended that future research on BioSs be focused on the use of genetic engineering to develop hyper-producing microbial strains. New and emerging concepts in genetic engineering should be employed to produce organisms that can enhance production with better product characteristics. Furthermore, the ability of mutant and engineered hyper-producing microbial strains to grow on a wide range of economical and renewable substrates could produce high yield BioSs and a cost-effective bioprocess. This approach will benefit from the recent advancements in strain engineering, structural elucidation, and characterization focused on producing novel BioS compounds with unique properties. Consequently, improving the quality and intensity of research in this field will help increase yield production and produce novel types of BioSs that can enhance their utilization in hydrocarbon bioremediation, antimicrobials, microbial enhance oil recovery, environmental and industrial processes.
References
Gautam, G., Mishra, V., Verma, P., Pandey, A. K., & Negi, S. (2014). A cost effective strategy for production of biosurfactant from locally isolated Penicillium chrysogenum SNP5 and its applications. Journal of Bioprocessing & Biotechniques, 4(6), 1.
Cai, Q., Zhang, B., Chen, B., Zhu, Z., Lin, W., & Cao, T. (2014). Screening of biosurfactant producers from petroleum hydrocarbon contaminated sources in cold marine environments. Marine Pollution Bulletin, 86, 402–410.
Al-Sulaimani, H., Al-Wahaibi, Y., Al-Bahry, S., Elshafie, A., Al-Bemani, A., Joshi, S., & Zargari, S. (2011). Optimization and partial characterization of biosurfactants produced by Bacillus species and their potential for ex-situ enhanced oil recovery. SPE Journal, 16, 672–682.
Elshafie, A. E., Joshi, S. J., Al-Wahaibi, Y. M., Al-Bemani, A. S., Al-Bahry, S. N., Al-Maqbali, D., & Banat, I. M. (2015). Sophorolipids production by Candida bombicola ATCC 22214 and its potential application in microbial enhanced oil recovery. Frontiers in Microbiology, 6, 1324.
Khan, A., & Butt, A. (2016). Biosurfactants and their potential applications for microbes and mankind: an overview. Middle East Journal of Business, 55, 1–10.
Martins, P. C., & Martins, V. G. (2018). Biosurfactant production from industrial wastes with potential remove of insoluble paint. International Biodeterioration & Biodegradation, 127, 10–16.
Zhang, R., & Somasundaran, P. (2006). Advances in adsorption of surfactants and their mixtures at solid/solution interfaces. Advances in Colloid and Interface Science, 123, 213–229.
Willenbacher, J., Mohr, T., Henkel, M., Gebhard, S., Mascher, T., Syldatk, C., & Hausmann, R. (2016). Substitution of the native srfA promoter by constitutive Pveg in two B. subtilis strains and evaluation of the effect on surfactin production. Journal of Biotechnology, 224, 14–17.
Makkar, R. S., Cameotra, S. S., & Banat, I. M. (2011). Advances in utilization of renewable substrates for biosurfactant production. AMB Express, 1(1), 5.
Sekhon, K. K., Khanna, S., & Cameotra, S. S. (2012). Biosurfactant production and potential correlation with esterase activity. Journal of Petroleum & Environmental Biotechnology, 3(133), 10–4172.
Piccolo, L. L., De Pasquale, C., Fodale, R., Puglia, A. M., & Quatrini, P. (2011). Involvement of an alkane hydroxylase system of Gordonia sp. strain SoCg in degradation of solid n-alkanes. Applied and Environmental Microbiology, 77, 1204–1213.
Aliakbari, E., Tebyanian, H., Hassanshahian, M., & Kariminik, A. (2014). Degradation of alkanes in contaminated sites. International Journal of Advanced Biological and Biomedical Research, 2, 1620–1637.
Jimoh, A. A., & Lin, J. (2019). Heterologous Expression of Sfp-Type phosphopantetheinyl transferase is indispensable in the biosynthesis of lipopeptide biosurfactant. Molecular Biotechnology, 61(11), 836–851.
Satpute, S. K., Bhuyan, S. S., Pardesi, K. R., Mujumdar, S. S., Dhakephalkar, P. K., Shete, A. M. and Chopade, B. A. (2010), in Biosurfactants, Springer, pp. 14–41.
Thakur, A., Putatunda, C., Sharma, R., Mehta, R., Solanki, P., & Bhatia, K. (2020). in Microbial Diversity (pp. 157–184). Interventions and Scope: Springer.
Wasilkowski, D., Swedziol, Z., & Mrozik, A. (2012). The applicability of genetically modified microorganisms in bioremediation of contaminated environments. Chemik, 66, 817–826.
De Almeida, D. G., Soares Da Silva, R., Luna, J. M., Rufino, R. D., Santos, V. A., Banat, I. M., & Sarubbo, L. A. (2016). Biosurfactants: promising molecules for petroleum biotechnology advances. Frontiers in Microbiology, 7, 1718.
Jiao, S., Li, X., Yu, H., Yang, H., Li, X., & Shen, Z. (2017). In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnology and Bioengineering, 114, 832–842.
Jung, J., Yu, K. O., Ramzi, A. B., Choe, S. H., Kim, S. W., & Han, S. O. (2012). Improvement of surfactin production in Bacillus subtilis using synthetic wastewater by overexpression of specific extracellular signaling peptides, comX and phrC. Biotechnology and Bioengineering, 109, 2349–2356.
Sun, H., Bie, X., Lu, F., Lu, Y., Wu, Y., & Lu, Z. (2009). Enhancement of surfactin production of Bacillus subtilis fmbR by replacement of the native promoter with the Pspac promoter. Canadian Journal of Microbiology, 55, 1003–1006.
Wu, Q., Zhi, Y., & Xu, Y. (2019). Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis 168. Metabolic Engineering, 52, 87–97.
Hu, F., Liu, Y., & Li, S. (2019). Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microbial Cell Factories, 18(1), 42.
Jimoh, A. A., & Lin, J. (2019). Biosurfactant: A new frontier for greener technology and environmental sustainability. Ecotoxicology and Environmental Safety, 184, 109607.
de França, Í. W. L., Lima, A. P., Lemos, J. A. M., Lemos, C. G. F., Melo, V. M. M., de Sant’ana, H. B., & Gonçalves, L. R. B. (2015). Production of a biosurfactant by Bacillus subtilis ICA56 aiming bioremediation of impacted soils. Catalysis Today, 255, 10–15.
Uzoigwe, C., Burgess, J. G., Ennis, C. J., & Rahman, P. K. (2015). Bioemulsifiers are not biosurfactants and require different screening approaches. Frontiers in Microbiology, 6, 245.
de Jesus Cortes-Sanchez, A., Hernández-Sánchez, H., & Jaramillo-Flores, M. E. (2013). Biological activity of glycolipids produced by microorganisms: new trends and possible therapeutic alternatives. Microbiological Research, 168, 22–32.
Bodour, A. A., Drees, K. P., & Maier, R. M. (2003). Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Applied and Environmental Microbiology, 69, 3280–3287.
Christova, N., Tuleva, B., Lalchev, Z., Jordanova, A., & Jordanov, B. (2004). Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane. Zeitschrift für Naturforschung C, 59, 70–74.
Nalini, S., & Parthasarathi, R. (2013). Biosurfactant production by Serratia rubidaea SNAU02 isolated from hydrocarbon contaminated soil and its physico-chemical characterization. Bioresource Technology, 147, 619–622.
Dubeau, D., Deziel, E., Woods, D. E., & Lepine, F. (2009). Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiology, 9(1), 263.
Chen, J., Huang, P. T., Zhang, K. Y., & Ding, F. R. (2012). Isolation of biosurfactant producers, optimization and properties of biosurfactant produced by Acinetobacter sp. from petroleum-contaminated soil. Journal of Applied Microbiology, 112, 660–671.
Edwards, J. R., & Hayashi, J. A. (1965). Structure of a rhamnolipid from Pseudomonas aeruginosa. Archives of Biochemistry and Biophysics, 111, 415–421.
Abdel-Mawgoud, A. M., Hausmann, R., Lépine, F., Müller, M. M. and Déziel, E. (2011), in Biosurfactants, Springer, pp. 13–55.
Van Bogaert, I. N., Saerens, K., De Muynck, C., Develter, D., Soetaert, W., & Vandamme, E. J. (2007). Microbial production and application of sophorolipids. Applied Microbiology and Biotechnology, 76, 23–34.
Morita, T., Konishi, M., Fukuoka, T., Imura, T., Kitamoto, H. K., & Kitamoto, D. (2006). Characterization of the genus Pseudozyma by the formation of glycolipid biosurfactants, mannosylerythritol lipids. FEMS Yeast Research, 7, 286–292.
Morita, T., Ishibashi, Y., Fukuoka, T., Imura, T., Sakai, H., Abe, M., & Kitamoto, D. (2011). Production of glycolipid biosurfactants, cellobiose lipids, by Cryptococcus humicola JCM 1461 and their interfacial properties. Bioscience Biotechnology and Biochemistry, 75, 1597–1599.
Saravanakumari, P., & Mani, K. (2010). Structural characterization of a novel xylolipid biosurfactant from Lactococcus lactis and analysis of antibacterial activity against multi-drug resistant pathogens. Bioresource Technology, 101, 8851–8854.
Adetunji, J. (2012). Microbial Enhanced Oil Recovery. PhD thesis, University of Aalborg, Denmark.
Ndlovu, T., Rautenbach, M., Vosloo, J. A., Khan, S., & Khan, W. (2017). Characterisation and antimicrobial activity of biosurfactant extracts produced by Bacillus amyloliquefaciens and Pseudomonas aeruginosa isolated from a wastewater treatment plant. AMB Express, 7, 108.
Ohadi, M., Dehghannoudeh, G., Forootanfar, H., Shakibaie, M., & Rajaee, M. (2018). Investigation of the structural, physicochemical properties, and aggregation behavior of lipopeptide biosurfactant produced by Acinetobacter junii B6. International Journal of Biological Macromolecules, 112, 712–719.
Mnif, I., & Ghribi, D. (2015). Review lipopeptides biosurfactants: mean classes and new insights for industrial, biomedical, and environmental applications. Peptide Science, 104, 129–147.
Jacques, P. (2011), in Biosurfactants, Springer, pp. 57–91.
Bodour, A. A., Guerrero-Barajas, C., Jiorle, B. V., Malcomson, M. E., Paull, A. K., Somogyi, A., et al. (2004). Structure and characterization of flavolipids, a novel class of biosurfactants produced by Flavobacterium sp. strain MTN11. Applied and Environmental Microbiology, 70, 114–120.
Shekhar, S., Sundaramanickam, A., & Balasubramanian, T. (2015). Biosurfactant producing microbes and their potential applications: A review. Critical Reviews in Environmental Science and Technology, 45, 1522–1554.
Park, S.-B., Lih, E., Park, K.-S., Joung, Y. K., & Han, D. K. (2017). Biopolymer-based functional composites for medical applications. Progress in Polymer Science, 68, 77–105.
Vater, J., Wilde, C., & Kell, H. (2009). In situ detection of the intermediates in the biosynthesis of surfactin, a lipoheptapeptide from Bacillus subtilis OKB 105, by whole-cell cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in combination with mutant analysis. Rapid Communications in Mass Spectrometry, 23, 1493–1498.
Abdel-Mawgoud, A. M., Lépine, F., & Déziel, E. (2014). A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chemistry & Biology, 21, 156–164.
Das, P., Mukherjee, S., & Sen, R. (2008). Genetic regulations of the biosynthesis of microbial surfactants: an overview. Biotechnology and Genetic Engineering Reviews, 25, 165–186.
Anuradha S, N. (2010). Structural and molecular characteristics of lichenysin and its relationship with surface activity. Biosurfactants, 304–315.
Nakar, D., & Gutnick, D. L. (2001). Analysis of the wee gene cluster responsible for the biosynthesis of the polymeric bioemulsifier from the oil-degrading strain Acinetobacter iwoffii RAG-1. Microbiology, 147, 1937–1946.
Roongsawang, N., Hase, K.-I., Haruki, M., Imanaka, T., Morikawa, M., & Kanaya, S. (2003). Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chemistry & Biology, 10, 869–880.
Das, P., Mukherjee, S., & Sen, R. (2008). Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. Journal of Applied Microbiology, 104, 1675–1684.
Ndlovu, T. (2017). Bioprospecting for novel biosurfactants and biosurfactant producing bacteria in wastewater. Stellenbosch University.
Madslien, E., Rønning, H., Lindbäck, T., Hassel, B., Andersson, M., & Granum, P. (2013). Lichenysin is produced by most Bacillus licheniformis strains. Journal of Applied Microbiology, 115, 1068–1080.
Qiu, Y., Xiao, F., Wei, X., Wen, Z., & Chen, S. (2014). Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Applied Microbiology and Biotechnology, 98, 8895–8903.
Finking, R., & Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides. Annual Review of Microbiology, 58, 453–488.
Wilkinson, B., & Micklefield, J. (2009). Biosynthesis of nonribosomal peptide precursors. Methods in Enzymology, 458, 353–378.
Schwarzer, D., Finking, R., & Marahiel, M. A. (2003). Nonribosomal peptides: From genes to products. Natural Product Reports, 20, 275–287.
Nakano, M. M., Corbell, N., Besson, J., & Zuber, P. (1992). Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Molecular and General Genetics MGG, 232, 313–321.
Porob, S., Nayak, S., Fernandes, A., Padmanabhan, P., Patil, B. A., Meena, R. M., & Ramaiah, N. (2013). PCR screening for the surfactin (sfp) gene in marine Bacillus strains and its molecular characterization from Bacillus tequilensis NIOS11. Turkish Journal of Biology, 37, 212–221.
Koumoutsi, A., Chen, X.-H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. Journal of Bacteriology, 186, 1084–1096.
Tsuge, K., Akiyama, T., & Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin A operon. Journal of Bacteriology, 183, 6265–6273.
Nakar, D., & Gutnick, D. L. (2003). Involvement of a protein tyrosine kinase in production of the polymeric bioemulsifier emulsan from the oil-degrading strain Acinetobacter iwoffii RAG-1. Journal of Bacteriology, 185, 1001–1009.
Coyne, M., Russell, K. S., Coyle, C. L., & Goldberg, J. B. (1994). The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. Journal of Bacteriology, 176, 3500–3507.
Toren, A., Segal, G., Ron, E. Z., & Rosenberg, E. (2002). Structure–function studies of the recombinant protein bioemulsifier AlnA. Environmental Microbiology, 4, 257–261.
Bekerman, R., Segal, G., Ron, E., & Rosenberg, E. (2005). The AlnB protein of the bioemulsan alasan is a peroxiredoxin. Applied Microbiology and Biotechnology, 66, 536–541.
Santos, D. K. F., Rufino, R. D., Luna, J. M., Santos, V. A., & Sarubbo, L. A. (2016). Biosurfactants: multifunctional biomolecules of the 21st century. International Journal of Molecular Sciences, 17, 401.
Bezza, F. A., & Chirwa, E. M. (2015). Biosurfactant from Paenibacillus dendritiformis and its application in assisting polycyclic aromatic hydrocarbon (PAH) and motor oil sludge removal from contaminated soil and sand media. Process Safety and Environmental Protection, 98, 354–364.
Perfumo, A., Smyth, T., Marchant, R. and Banat, I. (2010), in Handbook of hydrocarbon and lipid microbiology, Springer, pp. 1501–1512.
Satpute, S. K., Banpurkar, A. G., Dhakephalkar, P. K., Banat, I. M., & Chopade, B. A. (2010). Methods for investigating biosurfactants and bioemulsifiers: a review. Critical Reviews in Biotechnology, 30, 127–144.
Joshi-Navare, K. (2013). Biosynthesis of novel glycolipids: basic and applied aspects. Pune: CSIR-National Chemical Laboratory.
Van Bogaert, I. (2008). Microbial synthesis of sophorolipids by the yeast Candida bombicola. ed. Ghent University.
Morita, T., Kitagawa, M., Yamamoto, S., Sogabe, A., Imura, T., Fukuoka, T., & Kitamoto, D. (2010). Glycolipid biosurfactants, mannosylerythritol lipids, repair the damaged hair. Journal of Oleo Science, 59, 267–272.
Hewald, S., Linne, U., Scherer, M., Marahiel, M. A., Kämper, J., & Bölker, M. (2006). Identification of a gene cluster for biosynthesis of mannosylerythritol lipids in the basidiomycetous fungus Ustilago maydis. Applied and Environmental Microbiology, 72, 5469–5477.
Rao, K. N., Kumaran, D., Seetharaman, J., Bonanno, J. B., Burley, S. K., & Swaminathan, S. (2006). Crystal structure of trehalose-6-phosphate phosphatase–related protein: Biochemical and biological implications. Protein Science, 15, 1735–1744.
Lodish, H., Berk, A., Kaiser, C. A., Krieger, M., Scott, M. P., Bretscher, A., & Matsudaira, P. (2008). Molecular cell biology. Macmillan.
Eyéghé-Bickong, H. A. (2011). Role of surfactin from Bacillus subtilis in protection against antimicrobial peptides produced by Bacillus species. (Doctoral dissertation, Stellenbosch: University of Stellenbosch).
Roongsawang, N., Washio, K., & Morikawa, M. (2010). Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. International Journal of Molecular Science, 12, 141–172.
Sullivan, E. R. (1998). Molecular genetics of biosurfactant production. Current Opinion in Biotechnology, 9, 263–269.
Desai, J. D., & Banat, I. M. (1997). Microbial production of surfactants and their commercial potential. Microbiology and Molecular Biology Reviews, 61, 47–64.
Singh, P., Patil, Y., & Rale, V. (2019). Biosurfactant production: emerging trends and promising strategies. Journal of Applied Microbiologyl, 126, 2–13.
Eslami, P., Hajfarajollah, H., & Bazsefidpar, S. (2020). Recent advancements in the production of rhamnolipid biosurfactants by Pseudomonas aeruginosa. RSC Advances, 10, 34014–34032.
Eswari, J. S., Anand, M., & Venkateswarlu, C. (2016). Optimum culture medium composition for lipopeptide production by Bacillus subtilis using response surface model-based ant colony optimization. Sadhana, 41, 55–65.
Hassan, M., Essam, T., Yassin, A. S., & Salama, A. (2016). Optimization of rhamnolipid production by biodegrading bacterial isolates using Plackett-Burman design. International Journal of Biological Macromolecules, 82, 573–579.
Jimoh, A. A., & Lin, J. (2020). Bioremediation of contaminated diesel and motor oil through the optimization of biosurfactant produced by Paenibacillus sp. D9 on waste canola oil. Bioremediation Journal, 24, 21–40.
Sivapathasekaran, C., & Sen, R. (2013). Performance evaluation of an ANN–GA aided experimental modeling and optimization procedure for enhanced synthesis of marine biosurfactant in a stirred tank reactor. Journal of Chemical Technology & Biotechnology, 88, 794–799.
Ahmad, Z., Crowley, D., Marina, N., & Jha, S. K. (2016). Estimation of biosurfactant yield produced by Klebseilla sp. FKOD36 bacteria using artificial neural network approach. Measurement, 81, 163–173.
Roelants, S., Van Renterghem, L., Maes, K., Everaert, B., Redant, E., Vanlerberghe, B., Demaeseneire, S. L. and Soetaert, W. (2019). Microbial biosurfactants: from lab to market. Microbial Biosurfactants and their Environmental and Industrial Applications, 341–363.
Sastoque-Cala, L., Cotes-Prado, A. M., & Pedroza-Rodríguez, A. M. (2010). Effect of nutrients and fermentation conditions on the production of biosurfactants using rhizobacteria isolated from fique plants. Universitas Scientiarum, 15, 251–264.
Dolman, B. M., Wang, F., & Winterburn, J. B. (2019). Integrated production and separation of biosurfactants. Process Biochemistry, 83, 1–8.
Banat, I. M., Satpute, S. K., Cameotra, S. S., Patil, R., & Nyayanit, N. V. (2014). Cost effective technologies and renewable substrates for biosurfactants’ production. Frontiers in Microbiology, 5, 697.
Kuhn, D., Blank, L. M., Schmid, A., & Bühler, B. (2010). Systems biotechnology–Rational whole-cell biocatalyst and bioprocess design. Engineering in Life Sciences, 10, 384–397.
Bouassida, M., Ghazala, I., Ellouze-Chaabouni, S., & Ghribi, D. (2018). Improved biosurfactant production by Bacillus subtilis SPB1 mutant obtained by random mutagenesis and its application in enhanced oil recovery in a sand system. Journal of Microbiology and Biotechnology, 28, 95–104.
Mukherjee, S., Das, P., & Sen, R. (2006). Towards commercial production of microbial surfactants. Trends in biotechnology, 24, 509–515.
Cabrera-Valladares, N., Richardson, A.-P., Olvera, C., Treviño, L. G., Déziel, E., Lépine, F., & Soberón-Chávez, G. (2006). Monorhamnolipids and 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Applied Microbiology and Biotechnology, 73, 187–194.
Zhu, K., & Rock, C. O. (2008). RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. Journal of Bacteriology, 190, 3147–3154.
Anburajan, L., Meena, B., Raghavan, R. V., Shridhar, D., Joseph, T. C., Vinithkumar, N. V., et al. (2015). Heterologous expression, purification, and phylogenetic analysis of oil-degrading biosurfactant biosynthesis genes from the marine sponge-associated Bacillus licheniformis NIOT-06. Bioprocess and Biosystems Engineering, 38, 1009–1018.
Sekhon, K. K., Khanna, S., & Cameotra, S. S. (2011). Enhanced biosurfactant production through cloning of three genes and role of esterase in biosurfactant release. Microbial Cell Factories, 10, 49.
Xu, J. Z., & Zhang, W. G. (2016). Strategies used for genetically modifying bacterial genome: site-directed mutagenesis, gene inactivation, and gene over-expression. Journal of Zhejiang University Science-B, 17, 83–99.
Gong, G., Zheng, Z., Chen, H., Yuan, C., Wang, P., Yao, L., & Yu, Z. (2009). Enhanced production of surfactin by Bacillus subtilis E8 mutant obtained by ion beam implantation. Food Technology and Biotechnology, 47, 27–31.
Lotfabad, T. B., Abassi, H., Ahmadkhaniha, R., Roostaazad, R., Masoomi, F., Zahiri, H. S., et al. (2010). Structural characterization of a rhamnolipid-type biosurfactant produced by Pseudomonas aeruginosa MR01: enhancement of di-rhamnolipid proportion using gamma irradiation. Colloids Surface B: Biointerfaces, 81, 397–405.
Iqbal, S., Khalid, Z., & Malik, K. (1995). Enhanced biodegradation and emulsification of crude oil and hyperproduction of biosurfactants by a gamma ray-induced mutant of Pseudomonas aeruginosa. Letters in Applied Microbiology, 21, 176–179.
Deziel, E., Lepine, F., Milot, S., & Villemur, R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology, 149, 2005–2013.
Bach, H., Berdichevsky, Y., & Gutnick, D. (2003). An exocellular protein from the oil-degrading microbe Acinetobacter venetianus RAG-1 enhances the emulsifying activity of the polymeric bioemulsifier emulsan. Applied and Environmental Microbiology, 69, 2608–2615.
Coutte, F., Leclere, V., Bechet, M., Guez, J. S., Lecouturier, D., Chollet-Imbert, M., et al. (2010). Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of Bacillus subtilis 168 derivatives. Journal of Applied Microbiology, 109, 480–491.
Baranwal, D. K., Singh, P., Singh, R. K. and Shekhar, S. (2003). Gene knockout technology and its application. BIOLOGIX, 55.
Zhou, H., Huang, X., Liang, Y., Li, Y., Xie, Q., Zhang, C., & You, S. (2020). Enhanced bioremediation of hydraulic fracturing flowback and produced water using an indigenous biosurfactant-producing bacteria Acinetobacter sp. Y2. Chemical Engineering Journal, 397, 125348.
Huang, Y., Zhou, H., Zheng, G., Li, Y., Xie, Q., You, S., & Zhang, C. (2020). Isolation and characterization of biosurfactant-producing Serratia marcescens ZCF25 from oil sludge and application to bioremediation. Environmental Science and Pollution Research International, 27, 27762–27772.
Huang, X., Zhou, H., Ni, Q., Dai, C., Chen, C., Li, Y., & Zhang, C. (2020). Biosurfactant-facilitated biodegradation of hydrophobic organic compounds in hydraulic fracturing flowback wastewater: A dose–effect analysis. Environmental Technology & Innovation, 19, 100889.
Wei, Z., Wang, J. J., Gaston, L. A., Li, J., Fultz, L. M., DeLaune, R. D., & Dodla, S. K. (2020). Remediation of crude oil-contaminated coastal marsh soil: Integrated effect of biochar, rhamnolipid biosurfactant and nitrogen application. Journal of Hazardous Materials, 396, 122595.
Machado, T. S., Decesaro, A., Cappellaro, Â. C., Machado, B. S., van Schaik Reginato, K., Reinehr, C. O., et al. (2020). Effects of homemade biosurfactant from Bacillus methylotrophicus on bioremediation efficiency of a clay soil contaminated with diesel oil. Ecotoxicology and Environmental Safety, 201, 110798.
Vaishnavi, J., Devanesan, S., AlSalhi, M. S., Rajasekar, A., Selvi, A., Srinivasan, P., & Govarthanan, M. (2020). Biosurfactant mediated bioelectrokinetic remediation of diesel contaminated environment. Chemosphere, 264, 128377.
Durval, I. J. B., Mendonça, A. H. R., Rocha, I. V., Luna, J. M., Rufino, R. D., Converti, A., & Sarubbo, L. (2020). Production, characterization, evaluation and toxicity assessment of a Bacillus cereus UCP 1615 biosurfactant for marine oil spills bioremediation. Marine Pollution Bulletin, 157, 111357.
Chirwa, E. M. N., Mampholo, C. T., Fayemiwo, O. M., & Bezza, F. A. (2017). Biosurfactant assisted recovery of the C5–C11 hydrocarbon fraction from oily sludge using biosurfactant producing consortium culture of bacteria. Journal of Environmental Management, 196, 261–269.
Li, J., Deng, M., Wang, Y., & Chen, W. (2016). Production and characteristics of biosurfactant produced by Bacillus pseudomycoides BS6 utilizing soybean oil waste. International Biodeterioration & Biodegradation, 112, 72–79.
Banat, I. M., Franzetti, A., Gandolfi, I., Bestetti, G., Martinotti, M. G., Fracchia, L., et al. (2010). Microbial biosurfactants production, applications and future potential. Applied Microbiology and Biotechnology, 87, 427–444.
Jimoh, A. A., & Lin, J. (2019). Enhancement of Paenibacillus sp. D9 lipopeptide biosurfactant production through the optimization of medium composition and its application for biodegradation of hydrophobic pollutants. Applied Biochemistry and Biotechnology, 187, 724–743.
Jimoh, A. A., & Lin, J. (2019). Production and characterization of lipopeptide biosurfactant producing Paenibacillus sp. D9 and its biodegradation of diesel fuel. International Journal of Environmental Science and Technology, 16(8), 4143–4158.
Chaudhary, D. K., & Kim, J. (2019). New insights into bioremediation strategies for oil-contaminated soil in cold environments. International Biodeterioration & Biodegradation, 142, 58–72.
Calvo, C., Manzanera, M., Silva-Castro, G., Uad, I., & González-López, J. (2009). Application of bioemulsifiers in soil oil bioremediation processes. Future prospects. Science of the Total Environment, 407, 3634–3640.
Soberón-Chávez, G., Lépine, F., & Déziel, E. (2005). Production of rhamnolipids by Pseudomonas aeruginosa. Applied Microbiology and Biotechnology, 68, 718–725.
Khanna, S., Sekhon, K., & Prakash, N. (2009). Cloning and expression of a biosurfactant gene from endosulfan degrading Bacillus sp: correlation between esterase activity and biosurfactant production. Biotechnology, 8, 235–241.
Mathiyazhagan, N., Danashekar, K. and Natarajan, D. (2011). Amplification of biosurfactant producing gene (rhlB) from Pseudomonas aeruginosa isolated from oil contaminated soil. International Journal of Pharma and Bio Sciences, 2(1).
Cai, Q., Zhang, B., Chen, B., Cao, T., & Lv, Z. (2016). Biosurfactant produced by a Rhodococcus erythropolis mutant as an oil spill response agent. Water Quality Research Journal, 51, 97–105.
Morita, T., Fukuoka, T., Imura, T., & Kitamoto, D. (2013). Production of mannosylerythritol lipids and their application in cosmetics. Applied Microbiology and Biotechnology, 97, 4691–4700.
Ongena, M., & Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends in Microbiology, 16, 115–125.
Aparna, A., Srinikethan, G. and Hegde, S. (2012). Isolation, screening and production of biosurfactant by Bacillus clausii 5B. Research in Biotechnology, 3(2).
Gudina, E. J., Fernandes, E. C., Rodrigues, A. I., Teixeira, J. A., & Rodrigues, L. R. (2015). Biosurfactant production by Bacillus subtilis using corn steep liquor as culture medium. Frontiers in Microbiology, 6, 59.
Xia, W.-J., Luo, Z.-B., Dong, H.-P., Yu, L., Cui, Q.-F., & Bi, Y.-Q. (2012). Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous Pseudomonas aeruginosa WJ-1 using waste vegetable oils. Applied Biochemistry and Biotechnology, 166, 1148–1166.
Silva, S. N., Farias, C. B., Rufino, R. D., Luna, J. M., & Sarubbo, L. A. (2010). Glycerol as substrate for the production of biosurfactant by Pseudomonas aeruginosa UCP0992. Colloids Surface B: Biointerfaces, 79, 174–183.
Santos, E., Teixeira, M., Converti, A., Porto, A., & Sarubbo, L. (2018). Production of a new lipoprotein biosurfactant by Streptomyces sp. DPUA1566 isolated from lichens collected in the Brazilian Amazon using agroindustry wastes. Biocatalysis and Agricultural Biotechnology, 17, 142–150.
Jimoh, A. A., & Lin, J. (2020). Biotechnological applications of Paenibacillus sp. D9 lipopeptide biosurfactant produced in low-cost substrates. Applied Biochemistry and Biotechnology, 191(3), 921–941.
Luna, J., Rufino, R., Campos, G., & Sarubbo, L. (2012). Properties of the biosurfactant produced by Candida sphaerica cultivated in low-cost substrates. Chemical Engineering, 27, 67–72.
Andrade, R. F. S., Silva, T. A. L., Ribeaux, D. R., Rodriguez, D. M., Souza, A. F., Lima, M. A. B., et al. (2018). Promising biosurfactant produced by Cunninghamella echinulata UCP 1299 Using renewable resources and its application in cotton fabric cleaning process. Advances in Materials Science and Engineering, 2018, 1–12.
Parthipan, P., Preetham, E., Machuca, L. L., Rahman, P. K., Murugan, K., & Rajasekar, A. (2017). Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Frontiers in Microbiology, 8, 193.
Rane, A. N., Baikar, V. V., Ravi Kumar, V., & Deopurkar, R. L. (2017). Agro-industrial wastes for production of biosurfactant by Bacillus subtilis ANR 88 and its application in synthesis of silver and gold nanoparticles. Frontiers in Microbiology, 8, 492.
Bao, M., Pi, Y., Wang, L., Sun, P., Li, Y., & Cao, L. (2014). Lipopeptide biosurfactant production bacteria Acinetobacter sp. D3–2 and its biodegradation of crude oil. Environmental Science and Process Impacts, 16, 897–903.
Darvishi, P., Ayatollahi, S., Mowla, D., & Niazi, A. (2011). Biosurfactant production under extreme environmental conditions by an efficient microbial consortium, ERCPPI-2. Colloids Surface B: Biointerfaces, 84, 292–300.
Das, P., Mukherjee, S., & Sen, R. (2009). Substrate dependent production of extracellular biosurfactant by a marine bacterium. Bioresource Technology, 100, 1015–1019.
Deng, M. C., Li, J., Hong, Y. H., Xu, X. M., Chen, W. X., Yuan, J. P., et al. (2016). Characterization of a novel biosurfactant produced by marine hydrocarbon-degrading bacterium Achromobacter sp. HZ01. Journal of Applied Microbiology, 120, 889–899.
Freitas, B. G., Brito, J. M., Brasileiro, P. P., Rufino, R. D., Luna, J. M., Santos, V. A., & Sarubbo, L. A. (2016). Formulation of a commercial biosurfactant for application as a dispersant of petroleum and by-products spilled in oceans. Frontiers in Microbiology, 7, 1646.
Gaur, V. K., Bajaj, A., Regar, R. K., Kamthan, M., Jha, R. R., Srivastava, J. K., & Manickam, N. (2019). Rhamnolipid from a Lysinibacillus sphaericus strain IITR51 and its potential application for dissolution of hydrophobic pesticides. Bioresource Technology, 272, 19–25.
George, S., & Jayachandran, K. (2009). Analysis of rhamnolipid biosurfactants produced through submerged fermentation using orange fruit peelings as sole carbon source. Applied Biochemistry and Biotechnology, 158, 694–705.
George, S., & Jayachandran, K. (2013). Production and characterization of rhamnolipid biosurfactant from waste frying coconut oil using a novel Pseudomonas aeruginosa D. Journal of Applied Microbiology, 114, 373–383.
Ghribi, D., & Ellouze-Chaabouni, S. (2011). Enhancement of Bacillus subtilis lipopeptide biosurfactants production through optimization of medium composition and adequate control of aeration. Biotechnology Research International, 2011, 653654.
Hassen, W., Neifar, M., Cherif, H., Najjari, A., Chouchane, H., Driouich, R. C., et al. (2018). Pseudomonas rhizophila S211, a new plant growth-promoting rhizobacterium with potential in pesticide-bioremediation. Frontiers in Microbiology, 9, 34.
Hentati, D., Chebbi, A., Hadrich, F., Frikha, I., Rabanal, F., Sayadi, S., et al. (2019). Production, characterization and biotechnological potential of lipopeptide biosurfactants from a novel marine Bacillus stratosphericus strain FLU5. Ecotoxicology and Environmental Safety, 167, 441–449.
Jain, R. M., Mody, K., Joshi, N., Mishra, A., & Jha, B. (2013). Effect of unconventional carbon sources on biosurfactant production and its application in bioremediation. International Journal of Biologicaal Macromolecules, 62, 52–58.
Joshi, S. J., Geetha, S., & Desai, A. J. (2015). Characterization and application of biosurfactant produced by Bacillus licheniformis R2. Applied Biochemistry and Biotechnology, 177, 346–361.
Joshi-Navare, K., Khanvilkar, P., & Prabhune, A. (2013). Jatropha oil derived sophorolipids: production and characterization as laundry detergent additive. Biochemistry Research International, 2013, 169797.
Coutte, F., Niehren, J., Dhali, D., John, M., Versari, C., & Jacques, P. (2015). Modeling leucine’s metabolic pathway and knockout prediction improving the production of surfactin, a biosurfactant from Bacillus subtilis. Biotechnology Journal, 10, 1216–1234.
Fickers, P., Guez, J. S., Damblon, C., Leclere, V., Bechet, M., Jacques, P., & Joris, B. (2009). High-level biosynthesis of the anteiso-C(17) isoform of the antibiotic mycosubtilin in Bacillus subtilis and characterization of its candidacidal activity. Applied Environmental Microbiology, 75, 4636–4640.
Bechet, M., Castera-Guy, J., Guez, J. S., Chihib, N. E., Coucheney, F., Coutte, F., et al. (2013). Production of a novel mixture of mycosubtilins by mutants of Bacillus subtilis. Bioresource Technology, 145, 264–270.
Li, X., Yang, H., Zhang, D., Li, X., Yu, H., & Shen, Z. (2015). Overexpression of specific proton motive force-dependent transporters facilitate the export of surfactin in Bacillus subtilis. Journal of Ind Microbiology and Biotechnology, 42, 93–103.
Acknowledgements
The authors would like to acknowledge Dr. Wesley Ng'etich of the Department of Chemistry, University of KwaZulu-Natal (Westville campus), South Africa for his assistance in the drawing of the chemical structures provided.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
AAJ was responsible for conceptualization, design, drafting, illustrations, editing, and finalizing the manuscript. TS contributed to the writing and editing of the manuscript. RA and JL participated in the revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jimoh, A.A., Senbadejo, T.Y., Adeleke, R. et al. Development and Genetic Engineering of Hyper-Producing Microbial Strains for Improved Synthesis of Biosurfactants. Mol Biotechnol 63, 267–288 (2021). https://doi.org/10.1007/s12033-021-00302-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00302-1