Abstract
Purpose
Experiments were conducted to examine the effects of mercury (Hg) on soil nitrification activities and the microbial communities of ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA).
Methods
The soil samples spiked with different Hg concentrations were incubated for a period of 1, 2, 4, and 8 weeks in triplicate and the potential nitrification rate (PNR) of the samples was determined. The abundance of AOB and AOA was measured after an 8-week incubation by real-time polymerase chain reaction (PCR) assay of the amoA genes, while the community compositions by cloning and sequencing approaches.
Results
The soil PNR differed with different incubation periods. It tended to decrease with increasing soil Hg concentrations at week 1, basing on which the half-maximal effective concentration (EC50) was 1.59 mg kg−1. There was no significant difference in the abundance of AOB or AOA among the treatments. The AOB community was dominated by Nitrosospira-like sequences and more than 70% of the obtained clones were affiliated with the cluster 3a.2. The percentage of cluster 3a.1 in AOB community appeared to a consistent trend of decreasing with ascending soil Hg concentrations. While all the AOA sequences in the clone libraries were grouped into cluster S (soil and sediment origin).
Conclusions
This study revealed that Hg could inhibit soil potential nitrification and the extent varied with incubation periods. Soil Hg pollution changed the composition of soil AOB to some extent. These findings will be helpful to recognize the effects of Hg on the activity and community composition of soil ammonia oxidizers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Background, aim, and scope
Heavy metals are known to have a considerable effect on the activities of microorganisms and their mediated biogeochemical processes (Ehrlich 1997; Li et al. 2009). Mercury (Hg), as one of the most toxic heavy metals, has been widely studied, especially for its effects on soil microbes (Ranjard et al. 1997; Müller et al. 2002; Hayyis-Hellal et al. 2009). For example, the effect of Hg on the bacterial diversity has been demonstrated based on total community DNA, which indicated that microbial community in mercury-polluted soil diverged from the diversity in the control soil (Rasmussen and Sorensen 2001). However, it was also reported that an increase of mercury-resistant colonies was found in the Hg-treated soil (Ranjard et al. 1997). Some studies have also indicated that Hg could influence the soil particular functions which the microbes mediate, such as soil respiration, enzymatic activities, and nitrification through a combination of physiological and soil microcosm experiments (Rasmussen and Sorensen 2001; Müller et al. 2002). Nitrification has been considered to be an important process in the nitrogen cycle and one of the most sensitive soil microbial processes with regard to heavy metal stress (Broos et al. 2005; Xu and Chen 2006; Xu et al. 2009). Therefore, the potential nitrification rate (PNR) has been used as an endpoint to evaluate the risk of contaminated soil (Smolders et al. 2001). However, little information is available for the effects of Hg on the PNR of soil.
Ammonia-oxidizing bacteria (AOB) are a group of autotrophic microbes which have a key position in the rate-limiting step of nitrification (Purkhold et al. 2000; Di et al. 2009). A number of studies have shown that the community structure of AOB would shift in response to different environmental conditions, such as temperature, fertilization, and pH (Phillips et al. 2000; He et al. 2007, 2009; Nicol et al. 2008). Some groups belonging to the AOB appeared to be particularly sensitive to heavy metals though it is still unclear how the contaminants affect these Proteobacteria (β-subgroup ammonia oxidizers; Frey et al. 2008). It was reported that the AOB were completely inhibited in the heavy metal-contaminated soil according to the measurement of the potential ammonia oxidation (Gremion et al. 2004). However, AOB populations from long-term contaminated soil samples were able to tolerate higher Zn concentrations than those from uncontaminated soil samples (Mertens et al. 2006). Recent study also found that the ammonia-oxidizing population had a trend of resilience after the relief of heavy metal stress although the soil remained depressed microbial activities (Frey et al. 2008). Both metal-sensitive and metal-tolerant AOB populations could be observed in agricultural soil amended with heavy metals (Stephen et al. 1999). There were markedly different responses of AOB community to the different pollution stress. However, little information is available regarding the effects of Hg pollution on the abundance and composition of AOB community in soil.
Non-thermophilic crenarchaea have a potential role in soil ammonia oxidization (Nicol et al. 2008). Some studies had showed that there was a higher transcriptional activity of archaeal over bacterial ammonia oxidizers in soils (Chen et al. 2008; Nicol et al. 2008), which provides increasingly convincing evidences for the importance of ammonia-oxidizing archaea (AOA) in the global nitrogen cycle. Recently, the impacts on AOA community by soil factors, such as fertilization, pH, rhizosphere, and addition of antibiotics have been reported (He et al. 2007; Shen et al. 2008; Chen et al. 2008; Schauss et al. 2009), which presented considerable information recognizing the community characteristics of AOA in soils.
The aim of this study was to investigate the responses of ammonia-oxidizing prokaryotes to the toxicity of Hg in soils during an 8-week incubation period. To achieve this goal, the soil PNRs were determined in the soils with series of Hg concentrations at different incubation periods. Then, the real-time polymerase chain reaction (PCR) and clone-library sequencing were used to characterize the abundance and community composition of AOB and AOA in the treatments.
2 Materials and methods
2.1 Soil samples and experimental design
The soil sample was collected from a vegetable field in a suburb of Beijing, China, and its characteristics are listed in Table 1. All the samples were air-dried and ground to a size finer than 2.0 mm. The samples were spiked with HgCl2 solutions to give the final Hg concentrations of 0, 4, 8, 12, 15, and 30 mg kg−1 dry soil. A total of 60 g of each sample was placed in a 300-ml-plastic jar microcosm in triplicate for each Hg concentration and each harvest time. The microcosms were incubated at 22 ± 1°C for 8 weeks with 55% of soil water holding capacity and destructively harvested at week 1, 2, 4, and 8 in triplicate for soil analyses. Soil samples harvested at week 8 were stored at −80°C for subsequent DNA extraction and molecular analysis.
2.2 Soil chemical analysis and potential nitrification rates
Soil pH was determined with a soil to water ratio of 1:2.5, and soil organic matter was determined by K2Cr2O7 oxidation method (Walkley and Black 1934). Available P was analyzed according to Olsen P method. Soil samples were digested with HNO3 + HCl (10 ml, 1:1 v/v) for analysis of Hg, while soil samples were digested with HNO3 and H2O2 for analysis of other elements. The concentrations of Mn, Fe, Cu, Zn, and Cr were analyzed by an inductively coupled plasma optical emission spectrometer (ICP-OES, PerkinElmer Co., America), and Hg was analyzed by an atomic fluorescence spectrometer (AFS-2202E Haiguang Analytical Instrument Co., Beijing, China). Two standard reference materials, GBW-07401 (GSS-1) and GBW-07405 (GSS-5), obtained from the Center for National Standard Reference Materials of China, were inserted to the analytical process for quality assurance/quality control. PNR was measured using the chlorate inhibition method (Kurola et al. 2005). Briefly, 5.0 g of fresh soil was added to 50 ml centrifuge tube containing 20 ml of phosphate buffer solution (g l−1: NaCl, 8.0; KCl, 0.2; Na2HPO4, 0.2; NaH2PO4, 0.2; pH 7.4) with 1 mM (NH4)2SO4. Potassium chlorate with a final concentration of 10 mM was added to inhibit nitrite oxidation. The suspension was incubated in a dark incubator at 25°C for 24 h. And then nitrite was extracted with 5 ml of 2 M KCl and determined spectro-photometrically at 540 nm with N-(1-naphthyl) ethylenediamine dihydrochloride.
2.3 DNA extraction
DNA was extracted from 0.5 g fresh soil samples using Ultra-cleanTM soil DNA Isolation Kits (MoBio Laboratory, USA) according to the manufacturer’s protocol. The extracted DNA was evaluated on a 1% agarose gel and the concentration and purity of the extracts were estimated by spectrophotometry (NanoDrop, USA).
2.4 Quantification of amoA genes by real-time PCR assays
Abundances of AOB and AOA were determined by quantitative real-time PCR method targeting on amoA gene with the iCycler iQ5 thermocycler (Bio-Rad, USA; He et al. 2007). The 25 μl reaction mixture included 12.5 μl Premix Ex TaqTM (Takara Biotechnology, Japan), 1 μl of BSA, 0.5 μl of each primer (10 μM), and 2 μl of tenfold diluted extracted DNA as template. Bacterial amoA gene was quantified using the primers amoA1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′; Rotthauwe et al. 1997), and the primers for archaeal amoA gene were arch-amoAF (5′-STAATGGCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′; Francis et al. 2005).
2.5 Construction of the amoA gene clone library and restriction fragment length polymorphism analysis
In order to investigate the effects of Hg on the community composition of soil AOB and AOA, three soil treatments spiked with 0, 8, and 30 mg kg−1 Hg were selected and the DNA extracts from three replicated samples were mixed. The products of bacterial and archaeal amoA genes amplified with the primers for the real-time PCR assays were purified and cloned, which produces a 490 bp fragment for bacterial amoA gene and 635 bp for archaeal amoA gene. Clones were randomly chosen for PCR reamplification using the primers T7 and SP6. For restriction fragment length polymorphism (RFLP) and sequencing, the inserted fragments were digested with restriction endonuclease HhaI (Takara Biotechnology, Japan) and incubated at 37°C for 2 h. The restricted fragments were separated by gel electrophoresis on 3% agarose gel with ethidium bromide and observed under UV illumination. The fingerprint images of the RFLP band types in each replicate of three HgCl2 concentrations treatments were analyzed by the software Quantity One (Bio-Rad, USA) and grouped based on different band patterns. Evaluation of the clone libraries was performed by a rarefaction analysis using software aRarefactWin Version 1.3 (Steven Holland, Stratigraphy Lab, University of Georgia [http://www.uga.edu/strata/software/Software.html]).
2.6 Sequencing and phylogenetic analysis
The clones of unique RFLP band types in each library were sequenced. All amoA gene sequences were deposited in the GenBank nucleotide sequence database under accession no. FJ940180 to FJ940192, FJ999545 to FJ999552 for AOB and FJ940130 to FJ940179 for AOA. The amoA nucleotide sequences were trimmed and assembled using SeqMan (DNAStar, Madison, WI, USA). Homology analysis of the amoA sequences was performed using the software DNAMAN version 4.0 (Lynnon Biosoft, USA). Sequences displaying more than 98% identity with each other were grouped into the same operational taxonomic units (OTUs). Phylogenetic analysis was performed using MEGA version 4.0 and the neighbor-joining trees were constructed using the Poisson correction model with bootstrap value of 1,000.
2.7 Statistical analysis
Copy numbers for the amoA gene analysis were log-transformed as needed to normalize the distributions prior to statistical analysis. All statistical analyses were performed using SPSS version 11.5 and one-way analysis of variance followed by S-N-K-test was used to check for quantitative differences between treatments. P < 0.05 was considered to be statistically significant. Effects on the PNR were plotted against added Hg concentration in the soil samples. A logistic response model was fitted to the data (Vanewijk and Hoekstra 1993):
Where y = response of measured endpoint (PNR), k = response of measured endpoint at x = 0, s = slope parameter, x = concentration (on a liner scale), and x 50 = median effective concentration (EC50).
3 Results
3.1 Potential nitrification rates in the Hg-spiked soils
Soil PNRs were measured at week 1, 2, 4, and 8 after addition of HgCl2 (Fig. 1). The PNRs in all Hg treatments were significantly lower than those in control in the respective incubation period, and the PNRs at week 1 were much lower than those at week 2, 4, and 8. No significant differences were found between the treatments with soil Hg concentrations higher than 8 mg kg−1. Only the PNR curve of week 1 could be fitted well with logistic dose-response model (P < 0.05), from which the EC50 of PNRs was 1.59 mg kg−1.
3.2 Abundance of AOB and AOA in the soils
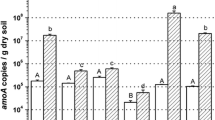
There was no significant difference for the bacterial or archaeal amoA gene copy numbers among all the treatments, respectively (Fig. 2). The average bacterial amoA gene copy numbers of 1.63 × 107 copies per gram of dry soil was found in all the tested soils, in which the average archaeal amoA gene copy numbers was 7.73 × 106 copies.
3.3 Analysis of amoA gene clone libraries
Each of the bacterial amoA gene clone libraries was clustered into seven to eight RFLP groups based on the band patterns, while all the three archaeal amoA gene clone libraries were clustered into six RFLP groups (Table 2). Rarefaction curves were constructed for each amoA gene library. All of the amoA gene libraries were sampled adequately as the rarefaction curves approached a plateau (Fig. 3). To determine the effects of Hg addition on soil AOB and AOA populations, the clones from amoA gene clone libraries that representing different RFLP patterns were partially sequenced. The obtained sequences from clone libraries were grouped into 18 bacterial amoA gene OTUs and 19 archaeal amoA gene OTUs by the Blast comparisons of nucleotide sequences in the NCBI database.
Rarefaction analysis of the amoA clones obtained from the different treatments of Hg concentrations using software aRarefactWin Version 1.3. Treatment: bacterial amoA genes without Hg (AOB0), bacterial amoA genes with 8 (AOB8) and 30 (AOB30) mg kg−1 soil Hg, archaeal amoA genes without Hg (AOA0), archaeal amoA genes with 8 (AOA8) and 30 (AOA30) mg kg−1 soil Hg
3.4 Phylogenetic analysis of the bacterial and archaeal amoA sequences
Phylogenetic trees of the bacterial amoA gene sequences and related NCBI sequences are showed in Fig. 4. All AOB sequences were affiliated with Nitrosospira amoA cluster 3a.1, 3a.2 and Nitrosomonsa amoA cluster 7 (Avrahami and Conrad 2003). Sequences in cluster 3a fell within two sub-cluster 3a.1 and 3a.2 with bootstrap value at 90%. The percentage of the sequences with similarity less than 98% in all 50 tested clones was calculated (Table 3). The sequences affiliated with 3a.2 had high proportions (more than 70%) in the three clone libraries. The proportion of cluster 3a.1sequence in the control was 28.0%, while it reduced to 9.5% in the treatment of 8 mg kg−1. No sequence affiliated with cluster 3a.1 was detected in the highest Hg concentration treatment (30 mg kg−1). In addition, no sequence affiliated with cluster 7 in the control but there was in the treatment of 8 or 30 mg kg−1 soil Hg. These data indicated substantial differences in AOB community composition among the soil samples with different Hg concentrations.
Phylogenetic relationships among bacterial amoA sequences retrieved from the three Hg levels soil samples. Designation of the clones in bold includes the following information: accession number in the Genbank with Hg treatments in the parentheses. Bootstrap values (>50%) are indicated at branch points. The scale bar represents 1% estimated sequence divergence. Treatment: bacterial amoA genes without Hg (AOB0), with 8 mg kg−1 Hg (AOB8), and with 30 mg kg−1 Hg (AOB30)
All the detected AOA sequences fell within the previous characterized soil and sediment and were grouped into two distinct clades (data not shown). These were designated as soil and sediment cluster 1 and 2 according to the latest classification (Zhang et al. 2009). Soil and sediment cluster 1 and 2 represented all the AOA clones, presenting in all soils at slightly different proportions (Table 3). Eighteen AOA OTUs belonged to soil and sediment cluster 2 while only one OTU were grouped into soil and sediment cluster 1.
4 Discussion
The soil PNR curve of week 1 could be fitted well with the logistic model, suggesting that the inhibition of soil Hg to the oxidation of ammonia could be predicted with dose-response relation in toxicology. The EC50 value calculated from the model was 1.59 mg kg−1, which was lower than another EC50 (3.26 mg kg−1) based on the reproduction of soil animal Folsomia candida stressed by soil Hg (Lock and Janssen 2001). According to the previous report (Fountain and Hopkin 2005), the F. candida was very sensitive to the soil pollution and used as indicator to assess soil quality. Therefore, PNRs could be a sensitive parameter in the soil Hg pollution diagnostics as well, especially in the early diagnostics since there were no dose-response relations found in the treatments over 1 week incubation in this study. Mertens et al. (2006) mentioned the same phenomena in the study using Zn as a stressor, stating that the long-term Zn pollution could hardly change the PNRs in the soil.
No difference of AOB abundance was observed between treatments in this study, which was in agreement with the previous report suggesting that heavy metal contaminated soil did not decrease the abundance of AOB (Stephen et al. 1999; Frey et al. 2008). It was also reported that there was no significant difference in the number of 16S rRNA gene fragment of AOB using most probable number method (Mertens et al. 2006). Phylogenetic analysis of the bacterial amoA gene sequences indicated that Nitrosospira cluster 3 was the dominant AOB group and the community composition changed with different soil Hg concentrations. The sequence affiliated with cluster 3a.1 decreased when soil Hg was 8 mg kg−1, and no cluster 3a.1 sequence was found when soil Hg was increased to 30 mg kg−1. On the other hand, cluster 7, which was absent in the soil without addition of Hg, could be detected in the other two soils with Hg spiked. Intriguingly, based on the current phylogenic analysis, it might be possible to reveal some dose-response relation in some populations from cluster 3a.1 or to do some screening in cluster 7 for possible discovery of resistant strains or genes of AOB. As for the sequences affiliated with cluster 3a.2, which had a high percentage in community composition and only slight difference between the treatments, they could hardly be affected by the addition of Hg. Although it was unclear whether these populations were intrinsically tolerant to Hg or tolerance was only induced by the extrinsic addition of Hg, this resistance could account for minimal difference of AOB abundance in the different treatments.
Meanwhile, the addition of Hg did not change the abundance of AOA. And there was no remarkable shift of AOA community between the treatments. The information about the effect of heavy metal on AOA was still limited up to now. Although it was reported that AOA might be in general less susceptible than AOB to inhibitor because of the remarkably different physiological characteristics of archaea compared with bacteria (Schauss et al. 2009), the mechanistic information about the response of AOA to environmental stress remains to be unclear due to limited cultivated AOA cluster so far (Konneke et al. 2005; de la Torre et al. 2008).
According to the results in this study, it was not an appropriate option to use the amoA gene copy number of AOB or AOA for the purpose of soil pollution assessment. No significant correlation was found between PNRs and the abundance of AOB or AOA, which also proved the difficulties to build quantity relations between the abundance of AOB or AOA and the ammonia-oxidizing function of the soil. Then some new techniques or concepts should be introduced to assess the effects of pollutants on the soil nitrification function, for example exploration on the species level of microbes as mentioned above. How to link the soil function and the ecotoxicology still needs a lot of studies.
5 Conclusions
Our results revealed that the PNR in the soil incubated for 1 week was very sensitive to Hg stress and accorded with logistic dose-response model. The addition of Hg distinctly changed the composition of soil AOB, whereas the AOA presented minimal shift. Most of AOB sequences belonged to the cluster 3a.2, and the percentage of cluster 3a.1 in AOB community appeared to a consistent trend of decreasing with ascending soil Hg concentrations. Therefore, the addition of Hg impacted the soil potential nitrification activity and community composition of AOB in this study.
6 Recommendations and perspectives
These findings demonstrated the effects of Hg on the activity and community composition of soil ammonia oxidizers.
Further studies should be performed to determine how those sensitive markers respond to soil heavy metal stress at molecular level. Thus, studies of transcriptional activity of ammonia oxidizers in the contaminated soils may be especially valuable in further exploring this issue. Furthermore, more detailed studies on the effects of contaminants on soil ammonia oxidizers should be conducted and thus to collect more toxicological data for evaluating their ecological effects.
References
Avrahami S, Conrad R (2003) Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl Environ Microbiol 69:6152–6164
Broos K, Mertens J, Smolders E (2005) Toxicity of heavy metals in soil assessed with various soil microbial and plant growth assays: As comparative study. Environ Toxicol Chem 24:634–640
Chen XP, Zhu YG, Xia Y, Shen JP, He JZ (2008) Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol 10:1978–1987
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nature Geoscience 2:621–624
Ehrlich HL (1997) Microbes and metals. Appl Microbiol Biotechnol 48:687–692
Fountain MT, Hopkin SP (2005) Folsomia candida (Collembola): a "standard" soil arthropod. Annu Rev Entomol 50:201–222
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102:14683–14688
Frey B, Pesaro M, Rudt A, Widmer F (2008) Resilience of the rhizosphere pseudomonas and ammonia-oxidizing bacterial populations during phytoextraction of heavy metal polluted soil with poplar. Environ Microbiol 10:1433–1449
Gremion F, Chatzinotas A, Kaufmann K, Von Sigler W, Harms H (2004) Impacts of heavy metal contamination and phytoremediation on a microbial community during a twelve-month microcosm experiment. FEMS Microbiol Ecol 48:273–283
Hayyis-Hellal J, Vallaeys T, Garnier-Zarli E, Bousserrhine N (2009) Effects of mercury on soil microbial communities in tropical soils of French Guyana. Appl Soil Ecol 41:59–68
He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:3152–3152
He JZ, Ge Y, Xu ZH, Chen CR (2009) Linking soil bacterial diversity to ecosystem multifunctionality using backward-elimination boosted tree analysis. J Soils Sediments 9:547–554
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M (2005) Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol Lett 250:33–38
Li F, Zheng YM, He JZ (2009) Microbes influence the fractionation of arsenic in paddy soils with different fertilization regimes. Sci Total Environ 407:2631–2640
Lock K, Janssen CR (2001) Ecotoxicity of mercury to Eisenia fetida, Enchytraeus albidus and Folsomia candida. Biol Fertil Soils 34:219–221
Mertens J, Springael D, De Troyer I, Cheyns K, Wattiau P, Smolders E (2006) Long-term exposure to elevated zinc concentrations induced structural changes and zinc tolerance of the nitrifying community in soil. Environ Microbiol 8:2170–2178
Müller AK, Westergaard K, Christensen S, Sorensen SJ (2002) The diversity and function of soil microbial communities exposed to different disturbances. Microb Ecol 44:49–58
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Phillips CJ, Harris D, Dollhopf SL, Gross KL, Prosser JI, Paul EA (2000) Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl Environ Microbiol 66:5410–5418
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16 S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Ranjard L, Richaume A, Jocteur-Monrozier L, Nazaret S (1997) Response of soil bacteria to Hg(II) in relation to soil characteristics and cell location. FEMS Microbiol Ecol 24:321–331
Rasmussen LD, Sorensen SJ (2001) Effects of mercury contamination on the culturable heterotrophic, functional and genetic diversity of the bacterial community in soil. FEMS Microbiol Ecol 36:1–9
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schauss K, Focks A, Leininger S, Kotzerke A, Heuer H, Thiele-Bruhn S, Sharma S, Wilke BM, Matthies M, Smalla K, Munch JC, Amelung W, Kaupenjohann M, Schloter M, Schleper C (2009) Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11:446–456
Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ (2008) Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10:1601–1611
Smolders E, Brans K, Coppens F, Merckx R (2001) Potential nitrification rate as a tool for screening toxicity in metal-contaminated soils. Environ Toxicol Chem 20:2469–2474
Stephen JR, Chang YJ, Macnaughton SJ, Kowalchuk GA, Leung KT, Flemming CA, White DC (1999) Effect of toxic metals on indigenous soil p-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol 65:95–101
Vanewijk PH, Hoekstra JA (1993) Calculation of the EC50 and its confidence interval when subtoxic stimulus is present. Ecotox Environ Saf 25:25–32
Walkley A, Black IA (1934) An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci 37:29–38
Xu ZH, Chen CR (2006) Fingerprinting global climate change and forest management within rhizosphere carbon and nutrient cycling processes. Environ Sci Pollut Res 13:293–298
Xu ZH, Chen CR, He JZ, Liu JX (2009) Trends and challenges in soil research 2009: linking global climate change to local long-term forest productivity. J Soils Sediments 9:83–88
Zhang LM, Wang M, Prosser JI, Zheng YM, He JZ (2009) Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everset. FEMS Microbiol Ecol 70:208–217
Acknowledgments
This work was supported by the Chinese Academy of Sciences (grant no. KZCX1-YW-0603) and the Ministry of Sciences and Technology (grant no. 2009AA06Z402).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Zhihong Xu
Rights and permissions
About this article
Cite this article
Liu, YR., Zheng, YM., Shen, JP. et al. Effects of mercury on the activity and community composition of soil ammonia oxidizers. Environ Sci Pollut Res 17, 1237–1244 (2010). https://doi.org/10.1007/s11356-010-0302-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-010-0302-6