Abstract
Despite the advances in transgenesis, transformation technologies still rely on the introduction of a selectable marker gene to identify cells and tissues that have integrated the gene of interest in their genome. The continuous presence of the marker genes in the transgenics is often controversial as it can potentially have multiple undesirable impacts. The present study employed the self-excising Cre-loxP system to generate marker-free Arabidopsis thaliana expressing the agronomically important glyoxalase I (glyI) gene from Brassica juncea to confer salt stress tolerance. A binary vector was constructed wherein the salt-inducible rd29A promoter was used to drive the expression of the glyI gene and the transformants of A. thaliana were recovered using kanamycin resistance as the selectable marker. The neomycin phosphotransferase II (nptII) gene was flanked by the loxP sites followed by the introduction of a heat-inducible Cre-recombinase in between the loxP sites. The kanamycin-resistant transgenic lines of A. thaliana using this vector showed an ability to withstand stress imposed by 150 mM NaCl. The exposure of the T2 transgenic lines to a mild heat shock (37°C) resulted in the recovery of salt-tolerant, kanamycin-sensitive T3 progeny. Molecular analyses of the T3 transgenic lines following the heat shock treatment confirmed the excision of the nptII gene and the completion of their life cycle in the presence of 150 mM NaCl-induced stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glyoxalase I (EC 4.4.1.5, lactoylglutathione lyase) and glyoxalase II (EC 3.1.2.6, hydroxacylglutathione hydrolase) constitute the glyoxalase system. In a two step reaction, these enzymes act using glutathione as a cofactor to coordinately convert cytotoxic methylglyoxal and other 2-oxoaldehydes to their 2-hydroxyacids (Thornalley 1990) as follows
Glyoxalase I first catalyses the formation of S-D-lactoylglutathione from hemithioacetal, which is formed by a non-enzymatic reaction between reduced glutathione (GSH) and methylglyoxal. Glyoxalase II catalyzes the hydrolysis of this S-D-lactoylglutathione to D-lactate with the regeneration of GSH in the second step (Uotila 1989). The primary physiological function of the glyoxalase system appears to be the detoxification of methylglyoxal, which is mainly synthesized as a byproduct of carbohydrate metabolism. Besides this, the glyoxalase system also increases the level of free reduced glutathione which is essential for scavenging of toxic reactive oxygen species (such as H2O2) and organic peroxides) that are increased in plants under stress conditions and in the maintenance of other antioxidants such as ascorbates and tocopherols (Alscher 1989). The involvement of the glyoxalase system in plants under stress conditions was first observed by Esparteo et al. (1995) where the glyoxalase I activity was shown to be upregulated under abiotic stresses. Overexpression of the glyoxalase I (gly1) gene under the control of the constitutive CaMV35S promoter in tobacco and rice has been shown to impart tolerance to salt, drought, and heavy metal stress (Veena et al. 1999; Singla-Pareek et al. 2003, 2006). Similarly, CaMV35S–mediated constitutive overexpression of glyoxalase II, the other gene of this system, has been shown to confer salt tolerance in tobacco (Singla-Pareek et al. 2003) and rice (Singla-Pareek et al. 2008). Since both the enzymes act in the same pathway, overexpression of either of these enzymes automatically shifts the enzyme catalyzed reaction in the forward direction that ultimately leads to increased release of reduced glutathione that ultimately detoxifies reactive oxygen species. This might result in a similar stress tolerant phenotype in the plants overexpressing either of the enzymes. However, constitutive overexpression of the transgene may compete for the building blocks and machinery needed for RNA and protein synthesis under stress-free conditions. Hence we constructed and analyzed plants with transgene expression driven by a stress-inducible promoter, rd29A, so that the specific mRNA and proteins required for stress alleviation are only produced under stress conditions. It has been reported that the rd29A and rd29B genes are induced under conditions of high temperature, high salt or upon treatment with exogenous abscisic acid (Yamaguchi-Shinozaki and Shinozaki 1993a, 1994). The sequence analysis of rd29A promoter showed the presence of drought-responsive element (DRE) and ABA-responsive element (ABRE). The 9 bp DRE element is involved in the first rapid response of rd29A to conditions of dehydration or high salt. ABA has also been found to be produced in plants under stress.
Selectable marker genes (SMGs) are used in nearly all plant transformation experiments and do not serve any purpose after the gene of interest is established. Besides precluding its reuse for future transformation experiments, the continuous presence of the SMG also raises issues of ecological concerns (Hill and Sendashonga 2006). It is, therefore, desirable to remove the selectable marker gene after it has served its crucial role in selection. There are several approaches for the removal of the marker gene like the simultaneous delivery of two T-DNA elements, one having the marker gene and the other having the gene of interest, as used by (Park et al. 2004; Matthews et al. 2001; Chen et al. 2005), transposition mediated repositioning and subsequent elimination of marker genes (Goldsbrough et al. 1993), use of homologous recombination (Iamtham and Day 2000) and the Cre-loxP recombination system (Dale and Ow 1991; Russell et al. 1992; Hoa et al. 2002). The Cre-loxP recombination system is often used due to its simplicity because apart from the 38 kDa Cre recombinase and the 34 bp loxP sites, no other factor is required for recombination to occur (Sternberg and Hamilton 1981; Stemberg et al. 1986). The cre gene for producing the Cre recombinase can be introduced into the loxP background by crossing plants harboring the loxP sequences with the plants expressing the recombinase gene or by sequential transformation of a plant with loxP and cre bearing constructs, respectively. Recently, various approaches were investigated to reproducibly obtain optimum CRE activity (Marjanac et al. 2008). An exciting area that deserves attention is the use of site-specific recombinases under the control of inducible promoters to excise SMGs after transgenic plants have been recovered (Hoff et al. 2001; Cuellar et al. 2006). Based on this premise, the cre gene under a heat shock inducible promoter (hsp) was cloned in a binary vector containing the glyI gene so that the cre gene, the loxP sequences and the glyI gene can be transferred to the target species. Arabidopsis thaliana was transformed with this novel construct to obtain salt tolerant kanamycin resistant transformants which upon exposure to a mild heat shock resulted in the recovery of marker-free salt tolerant transgenics. This could be used subsequently for obtaining abiotic stress tolerant marker-free transgenics of agronomically important crops.
Materials and methods
Construction of self-excising plant transformation vector
Escherichia coli strain DH5α was used as the host for recombinant vector constructions. The binary vector pCAMBIA 2301 (pC2301) which had the nptII (neomycin phosphotransferase) gene coding for kanamycin resistance as the plant selectable marker and gus (β-glucuronidase) as the reporter gene, both driven by the CaMV35S promoter, was chosen as the basic vector for this study. Two pairs of complementary single-stranded oligonucleotides (containing the loxP sites) were chemically synthesized (Qiagen Inc.) in such a manner that each pair, when annealed, would result in double stranded loxP oligonucleotides having EcoRI and XhoI overhangs and the restriction sites used for cloning would be preserved after their ligation into the parent vector. These loxP oligos were independently cloned into pC2301 after digestion with EcoRI and XhoI. Thus, the 34 bp loxP sequence was introduced both upstream of the 35S promoter (at EcoRI restriction site) and also between the nptII coding region and the 35S terminator (at XhoI restriction site). The positive clones were confirmed by restriction digestion and sequencing. The resultant vector had the nptII gene flanked on both the sides by the loxP sites in direct orientation and was denoted as pnpt-lox.
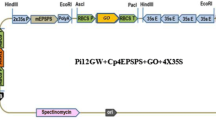
A 950 bp fragment of rd29A promoter (Yamaguchi-Shinozaki and Shinozaki 1993b) was amplified from A. thaliana cv. Columbia genomic DNA with primers having XbaI and NcoI overhangs (5′-GAGCTCTAGATGCAATTCAATCAAACTG-3′ and 5′-TGATCCATGGTCCAAAGATTTTTTTCTTTCCAATAG-3′). The Brassica juncea glyI cDNA, cloned in pBI-SI (Veena et al. 1999), was amplified with primers having NcoI and BstEII overhangs (TTCTCCATGGCGTCGGAAGCGAAGGAATC-3′ and 5′-TTTTGGTCACCGATAACAACTTATTTAACTCAACTC-3′). The binary vector pnpt-lox was digested with XbaI and BstEII, which led to the removal of the gus reporter gene. A three-fragment ligation was done with the XbaI-BstEII fragment of the vector, XbaI-NcoI fragment of rd29A promoter and NcoI-BstEII fragment of the glyI gene. This resulted in the construct pnpt-lox + rd29A-glyI, wherein the gene of interest, glyI was placed under the control of the rd29A promoter. Finally, a DNA fragment comprising the cre-recombinase (hsp-cre) driven by the heat-shock inducible promoter was obtained as a PCR product using primers with EcoRI and BamHI overhangs (5′-GCCAGAATTCATCGGTTTGAAGATGGCAAGTGTT-3′ and 5′-AATTGGATCCTAATCGCCATCTTCCAGCA-3′) and the pCrox 18 vector (Hoff et al. 2001) as the template. The CaMV35S terminator was obtained as a BamHI-EcoRI digest from dsProA (Pooggin et al. 2003). The hsp-cre fragment and CaMV35S terminator were introduced as a three-fragment ligation into pnpt-lox + rd29A-glyI, which was linearized using EcoRI. The integrity and orientation of the double insert was confirmed by restriction analysis and later by sequencing. This resulted in the vector phsp-cre + npt-lox + rd29A-glyI wherein both the nptII and hsp-cre were flanked by the loxP sites. The resulting vector, phsp-cre + npt-lox + rd29A-glyI, contained the glyI gene driven by the rd29A promoter and both the nptII and hsp-cre were flanked by the loxP sites (Fig. 1).
Schematic representation of construction of phsp-cre + rd29A-glyI binary vector used for A. thaliana transformation. In the first step two lox sites were introduced in pC2301 to get pnpt-lox vector (a) and then the glyI gene was introduced under the control of rd29A promoter at XbaI and BstEII sites of this vector to develop pnpt-lox + rd29A-glyI vector (b) and finally cre gene under the control of hsp was introduced in this vector to generate the final phsp-cre + rd29A-glyI vector (c)
The vector phsp-cre + npt-lox + rd29A-glyI was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. A. tumefaciens was grown on YEB (1.0 g/l yeast extract, beef extract 5 g/l, peptone 5 g/l, sucrose 5 g/l, 0.2 g/l MgSO4, 15 g/l agar) semi-solid medium containing 50 mg/l rifampicin, 25 mg/l gentamycin and 50 mg/l kanamycin. A single bacterial colony was inoculated into 5 ml of liquid YEB containing the same antibiotics and grown overnight at 28°C on a shaker at 200 rpm. A 200 μl aliquot of bacterial suspension was added to 20 ml of YEB liquid medium supplemented with the same antibiotics and grown overnight before using the culture for transformation of A. thaliana.
Plant material and growth conditions
The seeds of A. thaliana ecotype Columbia were surface-sterilized by incubation for 1 min in 70% ethanol, 10 min in 2% hypochlorite/0.01% Tween-20 and rinsed four times with sterile water. Seeds were imbibed for 2 days at 4°C before germination in a growth chamber (22°C, 16 h light/8 h dark, 100 μmol m−2 s−1, 60% relative humidity) on agarified half-strength Murashige–Skoog (MS) medium. Flowering Arabidopsis plants were transformed with GV 3101 (phsp-cre + npt-lox + rd29A-glyI) using the floral-dip method (Bechtold et al. 1993).
The T1 seeds from primary transformants were treated as before and selected on half-strength MS medium supplemented with 50 mg l−1 kanamycin in magenta boxes. The seedlings were transferred to soil in pots, checked for the presence of the glyI and the nptII gene by PCR analysis, allowed to self-fertilize and form T2 seeds. The T2 seeds were collected and germinated on selection medium as described above to obtain the T3 progeny.
PCR and Northern analysis
Total genomic DNA was isolated from the transgenic as well as untransformed control plants by a refined protocol of Murray and Thompson (1980). The 780 bp region of glyI gene was amplified using the primers (5′-CGGGGTACCATGGCGTCGGAAGCGAAGG-3′ and 5′-TGCTCTAGCGCTCTCAAGCTGCGTTTCCGGCTG-3′) and the 700 bp nptII gene coding region was amplified using the primers (5′-GGAGCGGCGATACCGTAAAGC-3′ and 5′-GAGGCTATTCGGCTATGACTG-3′). The amplification reaction was carried out using a thermal cycler (Techne Inc.) under the following conditions: for glyI gene: one cycle of 94°C for 5 min; 29 cycles of 94°C for 30 s (denaturation), 55°C for 30 s (annealing), 72°C for 90 s (extension); a final extension at 72°C for 5 min (one cycle); for nptII gene: one cycle of 94°C for 5 min; 29 cycles of 94°C for 60 s (denaturation), 58°C for 30 s (annealing), and 72°C for 45 s (extension); a final extension at 72°C for 5 min (one cycle). The PCR was performed using ca. 100 ng of purified genomic DNA and Taq polymerase (NEB). The amplified products were separated by electrophoresis on a 1% agarose gel (Sambrook et al. 1989) and visualized by ethidium bromide staining.
For Northern blot analysis, the standard protocol of Sambrook et al. (1989) was followed. Total RNA was isolated (Chomczynski and Sachi 1987) separated by formaldehyde gel electrophoresis and probed with 32P-dCTP-labelled glyI cDNA.
Glyoxalase I assay
Leaves of A. thaliana were crushed using liquid nitrogen and mixed thoroughly with the extraction buffer (SPB pH 7.0 also containing 50% glycerol, 16 mM MgSO4, 0.2 mM PMSF, and 0.2% PVPP). The crushed tissue extract was centrifuged twice at 13,000 rpm at 4°C for 30 min so as to obtain the crude protein extract as a clear supernatant. The gly I activity was assayed according to the protocol described by Ramaswamy et al. (1983). The standard enzyme assay mixture comprised 0.1 M SPB pH7.5, 3.5 mM methylglyoxal, 1.7 mM GSH and 16 mM MgSO4 in a final volume of 1 ml. This assay mixture was incubated for 7 min at room temperature prior to addition of crude protein extract (to allow non-enzymatic formation of hemithioacetal from methylglyoxal and GSH). After addition of the protein extract, the gly I activity was measured spectrophotometrically as a function of thioester formation (S-D lactoylglutathione) by measuring the rate of change in absorbance at 240 nm. The molar absorption coefficient of the thioester (SLG) at 240 nm is 3,370 m−1 cm−1. Three different enzyme extractions were performed per sample for three independent plants of the five T2 generation transformants. The specific activity of enzyme was expressed in units per mg−1 of protein.
Heat induction experiments
The T2 seedlings grown on kanamycin-supplemented MS medium were used for heat induction experiments. In the first phase, the 2-week-old seedlings were incubated at 37°C for 16 h, after which they were allowed to recover at 20°C for 30 h. This was followed by the second phase of heat treatment where the seedlings were re-incubated at 37°C for 16 h and immediately transferred to half-strength MS medium without kanamycin and maintained at 20°C. After one week they were transferred to soil in the pots, allowed to grow and set seed. The T3 progenies obtained from the heat shocked T2 lines were checked for the marker excision by PCR and Southern blot analyses.
Results
Construct designing, transformation and selection of A. thaliana
The vector pC2301 was modified such that the nptII gene was flanked by the loxP sites and the glyI gene was cloned downstream of the stress inducible rd29A promoter. The hsp-cre fragment from the pCrox18 vector (Hoff et al. 2001) was also introduced in the same vector within the loxP sites (see Materials and methods and Fig. 1). The binary vector used in the present investigation was constructed in such a manner that the Cre recombinase induced by heat shock would act at the loxP sites excising the hsp-cre fragment along with the expression cassette of the marker gene from the transgenic plants which would then contain only the desired gene (glyI) of interest. The glyI gene driven by the rd29A promoter was presumed to express only when the plants experienced salt stress.
Arabidopsis thaliana cv. Columbia was transformed with this modified pC2301 vector and the seeds (T1) were collected and germinated on ½ MS medium containing kanamycin (50 mg l−1). The putative transgenic seedlings growing on the selection medium were screened for the presence of the glyI and the nptII gene, respectively by PCR using the gene specific primers. Forty percent of the putative transgenic plants from different lines gave the expected bands corresponding to ~780 bp for the glyI and ~700 bp for the nptII gene. No corresponding bands were obtained in the case of untransformed control line. The transgenic lines grew normally and did not show any deleterious effect due to the presence of the cre driven by a CaMV35S promoter gene as reported earlier by Coppoolse et al. (2003). When transferred to the soil, they flowered and set seed. The T2 seeds from five of these lines were used for further experiments on salinity stress tolerance and marker excision. Figure 2a shows PCR analysis of the untransformed control plants as well as plants from the five representative lines. The expected bands corresponding to ~780 bp for the glyI and ~700 bp for the nptII gene are seen in the transgenic lines while they are absent in the untransformed control plant.
Analyses of T2 transgenic lines of A. thaliana before the heat shock treatment (a) PCR analyses of T2 transgenic lines for the presence of nptII gene and the glyI gene. (b) Glyoxalase I enzyme activities of five independent transgenic lines (1–5) and wild type untransformed control (Wt) plants. The error bars in the graph indicate standard deviation. (c) Enhanced germination of T2 transgenic (line 3) vs. untransformed control seeds on ½ MS medium supplemented with 150 mM NaCl. Ten μg of RNA was denatured and electrophoresed through a 1.5% agarose gel containing formaldehyde (7%). Transfer on nylon membrane and blotting was performed according to Sambrook et al. (1989). (d) Induction of glyI gene expression in transgenic (T2) A. thaliana (line 3) exposed to different concentrations of salt (NaCl). The numbers 1, 2, 3, 4, and 5 represent the different transgenic lines, wt represents untransformed control
Comparison of salt stress tolerance in transgenic vs. the untransformed control lines
Different transgenic lines were tested for the glyoxalase I enzyme activity (Fig. 2b) and the transgenic line showing maximum enhancement of the activity (Line 3) when compared to untransformed control plant, was checked for salinity tolerance test. Seeds of this transgenic line as well as the untransformed control plants were inoculated in ½ MS medium with 150 mM and 200 mM NaCl, respectively. No germination was observed on 200 mM NaCl in either control or transformed seeds. At 150 mM NaCl only 15% of the untransformed control seeds germinated as against 75% of the T2 transgenic seeds (Fig. 2c). The transgenic plants appeared healthy and normal in morphology, whereas the control plants were stunted and showed slow growth with yellowish leaves.
Effect of induction of the gly I transgene in A. thaliana in response to salt stress
The T2 seeds from the transgenic as well as the untransformed control plants were germinated on ½ MS medium. After one week, 10–15 seedlings from Line 3 were transferred to ½ MS medium containing 50 mM, 100 mM, 150 mM, and 200 mM NaCl, respectively. It was observed that the untransformed control plants grown on 100 and 150 mM NaCl were shorter as compared to the transgenic plants. In ½ MS medium (without the addition of NaCl) all plants flowered at the same time. However, delayed flowering (ca. 10 days) was observed in the untransformed control plants in the presence of even low (50 mM) NaCl concentration in ½ MS medium, in which the flowering in transgenic plants remained unaffected. It was only in the presence of a higher concentration (150 mM) of NaCl in the medium that delayed flowering (ca. 7 days) in the transgenic plants was observed as compared to the untransformed control plants.
Total RNA was isolated from the transgenic as well as the untransformed control plants grown at 0 mM, 50 mM, 100 mM, 150 mM, and 200 mM NaCl, blotted and probed with the glyI cDNA. No expression of the transgene was seen in the untransformed controls at any of the salt concentrations used (Fig. 2d). Though, hardly any expression was observed in the transgenic plants growing on salt free medium, very good expression of the glyI gene was observed on exposure of these plants to salinity stress (Fig. 2d).
Recovery of marker free salt tolerant transgenic plants
The seeds of the five selected PCR positive T2 transgenic lines were germinated on ½ MS medium with kanamycin (50 mg l−1). Fully mature transgenic plants were given heat shock treatment (as described in Material and methods). The heat shock treatment was repeated and the plants were transferred to fresh medium (½ MS without kanamycin). These plants were later transferred to soil where they flowered, self fertilized and set seed. The T3 transgenic seeds, thus obtained, were germinated to obtain the T3 transgenic lines. The T3 transgenic as well as the untransformed control lines were subjected to PCR analysis using the nptII and the glyI primers, respectively, to check for the excision of the antibiotic resistance marker, nptII, gene. Almost, all the transgenic lines which were subjected to heat shock treatment showed amplification with the glyI primers but did not show any amplification with the nptII primers (Fig. 3a). This showed that the antibiotic resistance marker gene (nptII) had been excised after the heat shock treatment.
Analyses of T3 transgenic lines of A. thaliana after the heat shock treatment (a) PCR analyses of T3 transgenic lines for the presence of nptII gene and the glyI gene. (b) A representative (T3) transgenic line and the untransformed control line (wt) grown in ½ MS medium supplemented with 150 mM NaCl. (c) Northern blot of total RNA of untransformed control and transgenic (T3) A. thaliana (line 3) grown in ½ MS medium supplemented with 150 mM NaCl. Ten μg of RNA was denatured and electrophoresed through a 1.5% agarose gel containing formaldehyde (7%). Transfer onto nylon membrane and blotting was performed according to Sambrook et al. (1989). The blot was probed with the glyI cDNA. The numbers 1, 2, 3, 4, and 5 represent the different transgenic lines, wt represents untransformed control
To check the expression of the gene of interest, the glyI, after the excision of the marker gene, the T3 seeds from the transgenic (Line 3) and untransformed control lines were germinated on ½ MS medium. After one week, the seedlings were transferred to ½ MS medium supplemented with 150 mM NaCl. While the transgenic lines grew normally, flowered and formed seeds, the untransformed control lines showed stunted growth (Fig. 3b). The expression of the glyI transgene was checked by northern analysis of the T3 transgenic lines vs. the untransformed control line which confirmed the expression of the gene of interest, the glyI, which remained unaffected by the excision of the fragment within the loxP sites in response to heat shock (Fig. 3c). Thus, marker free salt tolerant transgenic lines of A. thaliana were successfully developed.
Discussion
Excision of the antibiotic resistance marker gene is desirable for the genetically modified plants to be acceptable to the consumers. Many recombination approaches have been used for the successful deletion of DNA from transgenic plants but the Cre-loxP system is one of the best characterized and widely used (Dale and Ow 1991; Russell et al. 1992). Recently, Arumugam et al. (2006) reported the use of Cre-loxP system where a passage through in vitro culture of F1 leaf explants resulted in efficient development of marker-free transgenics in F2 generation in Brassica juncea. However, no gene of interest was used. In this study, the commonly used binary vector pC2301 was modified such that the plant selection marker gene (neomycin phosphotransferase II) and its promoter were flanked by the loxP sites. The final vector had the cre recombinase gene under the heat shock promoter which was also inserted between the loxP sites, thus circumventing the need to co-transform the cre gene or wait for another round of transformation. The hsp81-1 promoter was chosen for this study as it has been reported to be tightly regulated (Hoff et al. 2001) and because of the ease of heat shock treatments to the plants. The gene of interest, the glyI gene, driven by the salt inducible promoter rd29A, was present outside the loxP sites. This system of auto-excision of the marker gene has several advantages over the crossing strategy. Expression of the cre gene for a shorter period of time might overcome the unexpected effects which might arise due to expression for longer duration. Since the selectable marker gene and the cre gene can be removed simultaneously, in a single step, while retaining the gene of interest, this methodology offers another advantage in saving time and additional efforts. In this investigation, no deleterious effect of the cre gene expression, were observed in the plants. Since the excision of the nptII gene occurred after the heat shock treatments while the glyI gene was retained, it proves that the approach used in this study was successful for developing agronomically important marker free transgenic plants.
Significant progress has been made towards developing salt stress tolerant plants using various genes (Blumwald et al. 2004). The reports suggest that although abiotic stress is a multigenic trait, salinity stress tolerant plants can be produced by the transfer of a single gene utilizing the transgenic approaches. Overexpression of the glyoxalase I gene under the constitutive CaMV35S promoter was shown to impart salt and heavy metal tolerance in transgenic tobacco plants (Veena et al. 1999; Singla-Pareek et al. 2003, 2006. However, in the present investigation, the glyI gene driven by a salt inducible promoter has been shown to impart tolerance in transgenic A. thaliana plants exposed to salt stress. The glyI transcript was observed only in those transgenic lines which were exposed to salinity stress and probed with the transgene.
The transgenic plants were able to cope up better with salt stress, appeared healthier and grew faster as compared to the untransformed control plants under salinity stress. The fact that the salt tolerant transgenic plants developed during this study were also free of the antibiotic resistance marker gene is significant. This strategy if used for the transformation of crop plants will offer at least two major advantages, viz., the expression of the gene of interest under an inducible promoter under salt stress; and a simplified usage of the Cre-loxP system where no crossing of the plants having the cre gene and the antibiotic resistance gene flanked by loxP sites, respectively is required. This not only circumvents the controversies related to marker genes conferring antibiotic resistance, which have practically no use after the transformed plants are established, but also avoids the use of any viral constitutive promoter. Moreover, the foreign DNA (the glyI gene) that ultimately remained in the transgenic plants was of plant origin (B. juncea). The strategy used in this investigation is similar to that used by Cuellar et al. (2006) for developing antibiotic marker free transgenic potato and by Wang et al. (2005) for tobacco. However, no transgene of agronomic importance was introduced in the transgenic plants. Recently an embryo specific promoter driving the cre gene has been used for generating marker excision in soybean (Li et al. 2007). Other workers (Zuo et al. 2001; Sreekala et al. 2005; Zhang et al. 2003) have used chemically induced autoexcision of selectable markers. The efficiency of the use of heat shock, chemical and tissue specific promoter for the autoexcision of marker gene may vary in different plant systems and needs to be tested.
Abbreviations
- ABA:
-
Abscisic acid
- ABRE:
-
ABA- responsive element
- Cre:
-
Cre-recombinase
- DRE:
-
Drought responsive element
- gly I:
-
Glyoxalase I
- hsp:
-
Heat shock promoter
- MS:
-
Murashige and Skoog
- SPB:
-
Sodium Phosphate Buffer
- GSH:
-
Reduced glutathione
- PMSF:
-
Phenylmethylsulfonylfluoride
- PVPP:
-
Polyvinyl polypyrrolidone
References
Alscher RG (1989) Biosynthesis and antioxidant function of glutathione in plants. Physiol Plant 77:457–464. doi:10.1111/j.1399-3054.1989.tb05667.x
Arumugam N, Gupta V, Jagannath A, Mukhopadhyay, Pradhan AK, Burma PK and Pental D (2006) A passage through in vitro culture leads to efficient production of marker-free transgenic plants in Brassica juncea using the Cre-loxP system. Trans Res 16:703–712. doi:10.1007/s11248-006-9058-7
Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Life Sci 316:1194–1199
Blumwald E, Grover A, Good AG (2004) Breeding for abiotic stress tolerance: challenges and opportunities. Proc. 4th Int. Crop Science Congress
Chen S, Li X, Liu X, Xu H, Meng K, Xiao G et al (2005) Green fluorescent protein as a vital elimination marker to easily screen marker-free transgenic progeny derived from plants co-transformed with double T-DNA binary vector system. Plant Cell Rep 23:625–631. doi:10.1007/s00299-004-0853-4
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi:10.1016/0003-2697(87)90021-2
Coppoolse ER, de Vroomen MJ, Roelofs D, Smit J, van Gennip F, Hersmus BJ et al (2003) Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol 51:263–279. doi:10.1023/A:1021174726070
Cuellar W, Gaudin A, Solorzano D, Casas A, Nopo L, Chudalayandi P et al (2006) Self-excision of the antibiotic resistance gene nptII using a heat inducible Cre-loxP system for transgenic potato. Plant Mol Biol 62:71–82. doi:10.1007/s11103-006-9004-3
Dale EC, Ow DW (1991) Gene transfer with subsequent removal of the selection gene from the host genome. Proc Natl Acad Sci USA 88:10558–10562. doi:10.1073/pnas.88.23.10558
Esparteo J, Sanchez-Aguayo I, Pardo JM (1995) Molecular characterization of glyoxalase I from a higher plant; upregulated by stress. Plant Mol Biol 29:1223–1233. doi:10.1007/BF00020464
Goldsbrough AP, Lastrella CN, Yoder JI (1993) Transposition mediated re-positioning and subsequent elimination of marker genes from transgenic tomato. Bio/technology 11:1286–1292
Hill R, Sendashonga C (2006) Conservation biology, genetically modified organism, and the biosafety protocol. Conserv Biol 20(6):1620–1625. doi:10.1111/j.1523-1739.2006.00534.x
Hoa TT, Bong BB, Huq E, Hodges TK (2002) Cre-lox site specific recombination controls the excision of a transgene from the rice genome. Theor Appl Genet 104:518–525. doi:10.1007/s001220100748
Hoff T, Schnorr KM, Mundy J (2001) A recombinase–mediated transcriptional induction system in transgenic plants. Plant Mol Biol 45:41–49. doi:10.1023/A:1006402308365
Iamtham S, Day A (2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol 18:1172–1176
Li Z, Xing A, Moon BP, Burgoyne SA, Guida AD, Liang H et al (2007) A Cre/loxP- mediated self-activating gene excision system to produce marker gene free transgenic soyabean plants. Plant Mol Biol 65:329–341. doi:10.1007/s11103-007-9223-2
Marjanac G, De Paepe A, Peck I, Jacobs A, De Buck S, Depicker A (2008) Evaluation of CRE-mediated excision approaches in Arabidopsis thaliana. Transgenic Res 17:239–250
Matthews PR, Wang MB, Waterhouse PM, Thornton S, Fieg SJ, Gubler F et al (2001) Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNA’ on a standard Agrobacterium transformation vector. Mol Breed 7:195–202. doi:10.1023/A:1011333321893
Murray GC, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucl Acid Res 8:4321–4325
Park J, Lee YK, Kang BK, Chung WH (2004) Co-transformation using a negative selectable marker gene for the production of selectable marker gene-free transgenic plants. Theor Appl Genet 109:1562–1567. doi:10.1007/s00122-004-1790-x
Pooggin M, Shivaprasad PV, Veluthambi K, Hohn T (2003) RNAi targeting of DNA virus in plants. Nat Biotechnol 21:131–132. doi:10.1038/nbt0203-131b
Ramaswamy O, Guha-Mukherjee S, Sopory SK (1983) Presence of glyoxalase I in pea. Biochem Int 7:307–318
Russell SH, Hoopes JL, Odell JT (1992) Directed excision of a transgene from the plant genome. Mol Gen Genet 234:49–59
Sambrook J, Fritsch EF, Maniatis T (1989) In Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Singla-Pareek SL, Reddy M, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA 100(25):14672–14677. doi:10.1073/pnas.2034667100
Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK (2006) Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc spiked soils. Plant Physiol 140:613–623. doi:10.1104/pp.105.073734
Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK (2008) Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Trans. Res. 17:171–180. doi:10.1007/s11248-007-9082-2
Sreekala C, Wu L, Gu K, Wang D, Tian D, Yin Z (2005) Excision of selectable marker in transgenic rice (Oryza sativa L.) using a chemically regulated Cre/loxP system. Plant Cell Rep 24:86–94. doi:10.1007/s00299-004-0909-5
Stemberg N, Sauer B, Hoess R, Abremski K (1986) Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and regulation by DNA methylation. J Mol Biol 187:197–212. doi:10.1016/0022-2836(86)90228-7
Sternberg N, Hamilton D (1981) Bacteriophage P1 site-specific recombination between lox P sites. J Mol Biol 150:467–486. doi:10.1016/0022-2836(81)90375-2
Thornalley PJ (1990) Glyoxalase system: new developments towards functional characterization of metabolic pathways fundamental to biological life. Biochem J Kasu 269:1–11
Uotila L (1989) Glutathione thiol esterases. In: Dolphin D, Poulson R, Avramovic O (eds) Glutahione: Chemical, Biochemical and Medical Aspects, Coenzymes and Cofactors; Vol III. Part A. Wiley-Interscience, New York, pp 767–804
Veena, Reddy VS, Sopory SK (1999) Glyoxalase I from Brassica juncea: molecular cloning, regulation and its overexpression confer tolerance in transgenic tobacco under stress. Plant J 17:385–395
Wang Y, Chen B, Hu Y, Li J, Lin Z (2005) Inducible excision of selectable marker gene from transgenic plants by the cre/lox site-specific recombination system. Transgenic Res 14:605–614
Yamaguchi-Shinozaki K, Shinozaki K (1993a) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340. doi:10.1007/BF00277130
Yamaguchi-Shinozaki K, Shinozaki K (1993b) Arabidopsis DNA encoding two dessication–responsive rd29 genes. Plant Physiol 101:1119–1120. doi:10.1104/pp.101.3.1119
Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature or high salt stress. Plant Cell 6:251–254
Zhang W, Subbarao S, Addae P, Shen A, Armstrong C, Peschke V et al (2003) Cre/lox-mediated marker gene excision in transgenic maize (Zea mays L.) plants. Theor Appl Genet 107:1157–1168. doi:10.1007/s00122-003-1368-z
Zuo J, Niu QW, Moller SG, Chua NH (2001) Chemical-regulated, site specific DNA excision in transgenic plants. Nat Biotechnol 19:157–161. doi:10.1038/84428
Acknowledgements
We thank Prof. S. K. Sopory I.C.G.E.B., New Delhi, for the gift of the glyI cDNA, Prof. John Mundy, University of Copenhagen, Denmark, for the pCrox vector, Prof. Barbara Hohn, F.M.I., Switzerland for discussions and suggestions and Dr. Mohd. Aslam Yusuf and Mr. Ravi Rajwanshi, Jawaharlal Nehru University, New Delhi for their valuable inputs. Thanks are due to Prof. Anna Depicker for her help with advice and vectors. This research project has been implemented with financial contributions from the Swiss Agency for Development and Cooperation, Government of Switzerland and the Department Biotechnology, Government of India under the Indo-Swiss Collaboration of Biotechnology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roy, S.D., Saxena, M., Bhomkar, P.S. et al. Generation of marker free salt tolerant transgenic plants of Arabidopsis thaliana using the gly I gene and cre gene under inducible promoters. Plant Cell Tiss Organ Cult 95, 1–11 (2008). https://doi.org/10.1007/s11240-008-9402-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-008-9402-0