Abstract
In this paper, we consider the process of pattern formation induced by nonlinear diffusion in the gene network with cross-diffusion. We present a theoretical analysis of the pattern formation and show how cross-diffusion is able to destabilize the uniform equilibrium, being therefore liable for the emergence of spatial patterns. Through the linear stability analysis, we analytically derive a set of sufficient conditions which guarantee that the system generates turing instability, indicating that the competition and cross-diffusion between protein and mir-17–92 can lead to the turing pattern formation. Furthermore, we also obtain the Turing regions in which Turing patterns are generated.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Turing instability was proposed by Turing [24] in 1952; it is the phenomenon that initially stable steady state of a dynamical system can become unstable if we additionally consider diffusion in the system. This is surprising and unexpected phenomenon because diffusion usually makes things more smooth and uniform. Loss of stability due to diffusion is called Turing instability. Since the seminal paper of Turing [21], more and more attention is paid on the theoretical models to explain self-organized pattern formation in many different areas of physics, chemistry, biology, geology, and so on, especially biology. Recently, the entrainment and modulation of time-evolutional patterns are investigated numerically in one dimension [26]. Lee and Cho [15] find that the shape and type of Turing patterns depend on dynamical parameters and external periodic forcing. Moreover, Pena and Perez [18] show that slightly squeezed hexagons are locally stable in a full range of distortion angles. The domain coarsening process is strongly affected by the spatial separation among groups created by the Turing pattern formation process [19]. And the robustness problem is also investigated in [11, 17]. The dynamics of spontaneous pattern formation was first introduced to biology by Turing. Several groups have proposed possible candidates generating patterns [4, 10].
In the reaction–diffusion system, diffusion can induce the instability of a uniform equilibrium which is stable with respect to a constant perturbation, as shown by Turing in 1950s. The equilibrium of the nonlinear system is asymptotically stable in the absence of diffusion but unstable in the presence of diffusion [6]. Spatial patterns in reaction–diffusion systems have attracted the interest of experimentalists and theorists during the last decades. Until now, however, there has been no general analysis of the possible role of cross-diffusion in dissipative pattern formation [33]. Shi [20] show that cross-diffusion can destabilize a uniform equilibrium which is stable for the kinetic and self-diffusion reaction systems. On the other hand, cross-diffusion can also stabilize a uniform equilibrium which is stable for the kinetic system but unstable for the self-diffusion reaction system [11, 20]. Cross-diffusion which can lead to Turing pattern formation in biological systems has been reported [3, 9]. The effects of cross-diffusion on pattern formation in reaction–diffusion systems have been discussed in many theoretical papers in which the gradient in the concentration of one species lead to the diffusion in other species [25]. Other two relevant examples are on chemotaxis in E. coli bacteria [8, 25], a phenomenon where cells direct their motion toward or away from higher concentrations of other chemical species.
The above results in the turing instability were focused on the chemistry, ecology, and seldom on gene network. However, synthetic gene network and its dynamics were more and more concerned [27–31, 35–37], and we know that diffusion process is ubiquitous when the gene information was transported from cytoplasm to cell nucleus. In the following, we will investigate the character of diffusion process involving cancer network regulated by microRNA (mir-17–92). MicroRNAs (miRNAs) are an abundant class of small non-coding RNA that functions to regulate the activity and stability of specific mRNA targets through posttranscriptional regulatory mechanism and play a role of repressing translation of mRNA or degrading mRNAs [1, 2, 12, 21]. Recent studies show that miRNAs play a central role in many biological (cellular) processes, including developmental timing, cell proliferation, apoptosis, metabolism, cell differentiation, somitogenesis, and tumor genesis [2]. The induction of specific E2F activities is an essential component in the Myc pathways that control cell proliferation and cell fate decisions [7, 16, 32]. In normal cells, E2F-1 exhibits key negative feedback regulation on c-Myc-induced TERT expression, which is critical to control the transmission of c-Myc-mediated oncogenic signals [14, 22, 34]. The protooncogene Pim-1 is a part of the network that regulates transcription of the human miR-17–92 cluster [5, 23].
In recent years, many scientists deemed that mathematical modeling could be used to investigate the differences at the dynamical level between healthy and pathologic configurations of biological pathways [13]. Using the mathematical model, the researchers can detect the key points regulating main properties of biological system and find the methods to solve the different diseases. In order to understand further the miR-17–92 network involving in Myc and E2F, we also make use the mathematical model to investigate above biological network involving miR-17–92 and look further into the turing instability induced by cross-diffusion, and then understand how the turing pattern inside cells provides the checkpoint that combines mechanical and biochemical information to trigger events during the cell division process.
The paper is organized as follow. In Sect. 2, we give the gene network represented by mathematical model. In Sect. 3, we analyze the stability of pattern formation and selection. In Sect. 4, we show the numerical analysis of the network. Finally, we summarize our results.
2 The model
In this section, we consider a cancer network, where the reaction kinetics describes the interaction between microRNA and protein model. As we all know that molecular diffusion is ubiquitous when molecules interaction or pass though the cytomembrane, we should consider the effect of diffusion on the network. In this paper, we modified the system (1) and (2) given by Aguda et al. [1] and add the diffusion term to the system, obtain the reaction–diffusion system as follows:
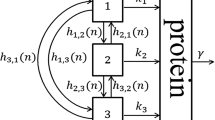
where \(u\) is a vector \(u_{i}(x, t), i = 1, 2\) of species densities, in which \(u_1\) represents transcription factor \(p\) in Fig. 1c and \(u_2\) represents microRNA in Fig. 1c; \(D\) is a \(2\times 2\) matrix of the diffusion coefficients, where the diagonal element is called the self-diffusion coefficient and the non-diagonals are called cross-diffusion coefficients; and \(G\) is the reaction term indicating the interaction between the involved species. The nonlinear diffusion coefficients matrix of our model as follows:
And the interaction contribution is included in the reaction term \(G\). Specifically, the underlying model consists as follows:
where \(\Omega _{T}=\Omega \times (0,T),\Sigma _{T}=\partial \Sigma \times (0,T)\) for a fixed \(T>0\). In biological terms, the homogeneous Neumann boundary condition indicates that there is no population flux across the boundary. Here, the presence of \(u_{2}\) in the denominator accounting for the miR-dependent down-regulation of protein expression is determined by the term (\(a+bu_{2}\)) in the denominator. The value of the parameter \(b\) is a measure of the efficiency of miRNA inhibition of protein expression, and lumps all factors that could affect the binding of the members of the miR-17–92 cluster to their targets and the inhibition of protein translation. The constant term \(\alpha \) stands for constitutive protein expression due to signal transduction pathways stimulated by growth factors present in the extracellular medium. The parameter \(\alpha \), therefore, corresponds to an experimentally controllable conditions such as the concentration of nutrients in the cell culture medium. \(\delta \) is the rate of the protein degradation. The constant term \(\beta \) represents p-independent constitutive transcription of \(m\). And the last term is a degradation term with rate coefficient \(\gamma \). The presence of the nonlinear diffusion term means basically that the disperse direction of \(u_{2}\) not only contains the self-diffusion (in which way the species move from a region of high density to a region of low density), but also contains cross-diffusion. More specifically, species \(u_{2}\) diffuses with a flux
Notice that, as \(-d_{2}d_{3}u_{2}<0\), the part \(-d_{2}d_{3}u_{2}\nabla u_{2}\) of the corresponding flux is directed toward the decreasing population density of the species \(u_{1}\).
a A cancer network. This network is part of the mammalian G1-S regulatory network involving miR-17–92, arrowhead represents the activation, hammerhead represents the inhibition, b first stage in the reduction of the model, c the final network model that abstracts the essential structure of the network in a, \(p\) presents the transcription factor (Myc and E2F), \(m\) represents microRNA (miR-17–92) cluster
3 Cross-diffusion-driven spatial patterns
In this section, we derive some sufficient conditions for spatial patterns. In particular, we not only show that the patterns formation without cross-diffusion, but also the formation of spatial patterns with cross-diffusion.
3.1 Linear stability analysis
We assume that the system (3.1) is true.
where
And we can know from [1] that there is a unique positive equilibrium point to system (2.1), denoted by \(u^{*}=(u^{*}_{1}, u^{*}_{2})\).
In order to study the locally asymptotic stability of the system (2.1), we give the notation as follows.
Notation 3.1
Let \(0=\mu _{1}<\mu _{2}<\cdots \rightarrow \infty \) be the eigenvalues of \(-\triangle \) on \(\Omega \) under no-flux boundary conditions,and \(E(\mu _{i})\) be the space of eigenfunctions corresponding to \(\mu _{i}\). We define the space decomposition as follows:
-
(i)
\(\mathbf{X}_{ij}\,{:=}\,\{\mathbf{c}\cdot \phi _{ij}:\mathbf{c}\in R^{2}\}\), where \({\phi _{ij}}\) is an orthonormal basis of \(E(\mu _{i})\) for \(j=1,\ldots , \mathrm{{dim}} E(\mu _{i})\).
-
(ii)
\(\mathbf{X}\,{:=}\,\{\mathbf{u} \in [C^1(\overline{\Omega })]^{2}:\frac{\partial u_{1}}{\partial \eta }=\frac{\partial u_{2}}{\partial \eta }=0\) on \(\partial \Omega \}\), and thus \( \mathbf X =\oplus ^{\infty }_{i=1}\mathbf X _{i}\), where \(\mathbf X _{i}=\oplus ^{\mathrm{{dim}}E(\mu _{i})}_{j=1}\mathbf X _{ij}\).
Theorem 3.1
The positive equilibrium point \(u^{*}\) of (2.1) without cross-diffusion is locally asymptotically stable when (3.1) is true.
Proof
First, for the sake of simplicity, we will denote in this paper
The linearization of (2.1) around the state \(u^{*}\) can be expressed by
where \(D=\mathrm{{diag}}(d_{1}, d_{2})\), \(u=(u_{1}, u_{2})\) and
According to Notation 3.1, the space \(\mathbf X _{i}\) is invariant under the operator \(D\Delta +G_{u}(u^{*})\), and \(\lambda \) is an eigenvalue of this operator on \(\mathbf X _{i}\), if and only if it is an eigenvalue of the matrix \(-\mu _{i}D+G_{u}(u^{*})\). The characteristic polynomial of \(-\mu _{i}D+G_{u}(u^{*})\) is given by
where
Recalling condition (3.1), it is easy to verify that \(B_{i}\) and \(-C_{i}\) are negative. Thus, for each \(i\ge 1\), the two roots \(\lambda _{i,1}\) and \(\lambda _{i,2}\) of \(\psi _{i}(\lambda )=0\) have negative real parts, and this completes the proof.
Theorem 3.2
Assume that the system (3.1) holds. If \(\mu _{2}<\widetilde{\mu }\), then there exists a positive constant \(d^{*}_{3}\) such that the equilibrium point \(u^{*}\) of (2.1) is unstable provided that \(d_{2}>d^{*}_{3}\), where \(\mu _{2}\) is given in Notation 3.1 and \(\widetilde{\mu }\) will be given in (3.2).
Proof
For simplicity, we denote \(\Phi (u)=(d_{1}u_{1},d_{2}(u_{2}+d_{3}u_{1}u_{2}))^T\). Linearizing (2.1) around the state \(u^{*}\) yields
where
We can obtain that the characteristic polynomial of \(-\mu _{i}\Phi _{u}+G_{u}(u^{*})\) is
where
We denote \(\lambda _{1}(\mu _{i})\) and \(\lambda _{2}(\mu _{i})\) be the roots of \(\psi _{i}(\lambda )=0\), and then have
In order to get that \(Re\lambda _{1}(\mu _{i})<0\) and \(Re\lambda _{2}(\mu _{i})>0\), a sufficient condition is \(E_{i}<0\) (based on the fact that \(D_{i}<0\)).
In the following, we look for conditions that make \(E_{i}<0\) hold. First, since \(E_{i}=\mathrm{{det}}(\mu _{i}\Phi _{u}-G_{u}(u^{*}))\), it is easy to deduce that
where
We denote \(Q(\mu )=Q_{2}\mu ^{2}_{i}+Q_{1}\mu _{i}+\mathrm{{det}}(G_{u})\), and \(\widetilde{\mu _{1}}\) and \(\widetilde{\mu _{2}}\) be the roots of \(Q(\mu )=0\) with \(Re(\widetilde{\mu _{1}})<Re(\widetilde{\mu _{2}})\). Condition (3.1) implies that
Furthermore,we have
We see that \(\lim _{d_{3}\rightarrow \infty }\frac{Q}{d_{3}}=0\) has two real roots, one being zero and the other being positive. A continuity argument allows us to show that \(\widetilde{\mu _{1}}\) and \(\widetilde{\mu _{2}}\) are real and positive when \(d_{3}\) is large enough, and in addition
Hence, there exists a positive number \(d^{*}_{3}\) such that the inequalities \(\widetilde{\mu }<\mu _{2}\) when \(d_{3}>d^{*}_{3}\), and \(Q(\mu )<0\) when \(\mu \in (0,\widetilde{\mu })\), are valid. Since \(0<\mu _{2}<\widetilde{\mu }\), then \(u_{2}\in (\widetilde{u_{1}}, \widetilde{u_{2}})\). It follows that \(Q(\mu )<0\), which finally implies that \(E_{i}<0\), which completes the proof.
The above theorems reveal that the cross-diffusion effect is able to destabilize the positive equilibrium point, and thereby the solution results in spatial patterns.
3.2 Turing parameter space
In view of Theorem 3.2, the fulfillment of the following conditions is sufficient for the positive equilibrium point \(u^{*}\) being linearly unstable with respect to the particular case of system (2.1):
-
(i)
\(A-\delta <\gamma \) and \(4d_{1}d_{2}k_{2}B>(d_{2}(A-\delta )+d_{1} \gamma )^{2}\).
-
(ii)
\(\mu _{2}<\frac{u^{*}_{1}+(A-\delta )() u^{*}_{1}+Bd_{2}u^{*}_{2}}{d_{1}u^{*}_{1}}\).
In this paper, the values satisfying the parameter spaces are taken as (3.3)
For this particular choice, the positive stationary uniform solution is given by
In Fig. 2, we depict the parameter spaces where instabilities are expected to appear according to conditions (i) and (ii) above. These graphics are obtained by fixing all parameters in (3.3) except for \(a\) and \(b\) (Fig. 2a), \(\gamma \) and \(k_{1}\) (Fig. 2b).
Turing parameter spaces for model (2.1). In the A domain, the solutions remain stable, while in the B, C, D domain, instabilities are expected to appear
If no diffusion is considered, then the system (2.1) boils down to the dynamical system
whose phase diagram is presented in Fig. 3a. We show trajectories for different initial values of \(u_{1,0}\) and \(u_{2,0}\) in which the trajectories converge to the equilibrium state (3.4), shown by the intersection of the (nontrivial) nullclines
We are able to calculate the wavenumber explicitly and determine the pattern selection by linearizing cross-diffusion around the stationary uniform solution and taking \(d_{3}\) as the Turing bifurcation parameter. From the mathematical viewpoint, the Turing bifurcation occurs when \(Im(\lambda _{k})= 0\), \(Re(\lambda _{k})=0\) at \( k\not \equiv 0\), where \(k_{c}\) is the critical wavenumber and \(|k|^{2}\) is equivalent to the eigenvalue \(\mu _{i}\) in Notation 3.1.
a Phase plane for model (2.1). Four trajectories are represented, starting from the initial states \(A,B,C,D\) and reaching the intersection of the nullclines, b plot of the largest of the eigenvalues \(Re(\lambda )\), there is a range of wave numbers which are linearly unstable
Now, replacing \(\mu _{i}\) with \(|k|^{2}\), the problem can obtain from the linearization of (2.1) around \(u^{*}\) as follows:
And the characteristic equation is as follows:
In order to find out the critical wavenumber, we only need to confirm that
which is a quadratic polynomial of \(|k|^{2}\). In this way, the Turing bifurcation threshold is given by \( d^{c}_{3}\) which satisfies the following equation:
and the critical wavenumber \(k_c\) is given by
4 Numerical analysis
In this section, we will demonstrate Turing pattern according to the previous theoretical results. First, we introduce our numerical scheme using finite difference method. In this paper, we consider zero-flux boundary conditions with region defined as square region \([0, L] \times [0, L] \in \mathfrak {R}^{2}\), then discretize the space and time, in which the square space is divided as \(M\times N\) lattice sites domain with the \(h\) length of lattices, and time step is set as constant \(\tau \). The system (2.1) is performed using the standard five-point approximation for the spatial derivative and an explicit Euler method for the time integration. In this paper, we set \(h=1,\ \tau =0.02\), and select coefficients of self-diffusion \((d_{1},d_{2})=(1,1)\), each frame is \(100\times 100\) space units, we choose parameters \((a,b)=(1,1)\), \((k_{1},k_{2},\alpha ,\beta )=(0.5,1,1,1)\). For our first test, we simulate the section of different patterns depending on the value of the cross-diffusion coefficient \(d_{3}\), and the initial data are taken as a uniformly distributed random perturbation. More precisely, we choose
where \(\eta _{1}\in [0,1],\eta _{2}\in [0,1]\).
First, we simulate the section of different patterns based on the the value of cross-diffusion coefficient \(d_{3}\), and depict the real part of the eigenvalues \(Re(\lambda )\) in correspondence with the norm of the wave vector in Fig. 3b. In Fig. 4, we choose the parameter \(d_{3}\) as \(d_{3}=0\), and changed it to \(d_{3}=17, d_{3}=18, d_{3}=19, d_{3}=19.5\) for Figs. 5, 6, 7, and 8, respectively. In addition, we can know that the initial value paly an important role in the selection of patterns and that the modes of the pattern convert from the stripes to the spots as the cross-diffusion coefficient \(d_{3}\) increases. Finally, we also find that the appearance of spots’ pattern will be delayed (\(d_{3}\) will be larger when the spots pattern appear)when the coefficient \(d_{1}\) increases in the simulation, and the delay value is in proportion to \(d_{1}\). From above Figs. 4, 5, 6, 7 and 8, the patterns of protein concentration evolve from stripe to spot pattern, and the spots pattern shows the Stationary localized pulse (dissipative soliton) which corresponds periodical traveling wave. These patterns are starting from a spatially uniform state which turing proved that stable inhomogeneities in “morphogen” concentration could emerge through a diffusion-driven symmetry-breaking instability.
5 Conclusion
In this paper, we have developed a cancer network regulated by microRNA with cross-diffusion and analyze the spatiotemporal dynamics of the cancer network with diffusion under the zero-flux boundary conditions. It is found that the steady state of the system can be globally asymptotically stable under some condition, and Turing patterns can be formed under other conditions. Importantly, we have shown that cross-diffusion can lead to the Turing pattern formation and influence on the distribution of protein and miRNA; obviously, the cross-diffusion is one of drivers which drive the turing pattern in biological system. We also find that the appearance of spots pattern will be delayed (\(d_{3}\) will be larger when the spots pattern appear) when the coefficient \(d_{1}\) increases in the simulation, and the delay value is in proportion to \(d_{1}\). As we know that the formation of Turing patterns in biochemical pathway could then be related to organizing centers in eukaryotic cells playing a role during cell division, and the Turing pattern inside cells could provide a checkpoint that combine mechanical and biochemical information to trigger events during the cell division process, in addition, the cancer is uncheck cell growth, and the unregulated checkpoint can induce to cancer.
References
Aguda, B.D., Kim, Y., Piper-Hunter, M.G., Friedman, A., Marsh, C.B.: MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. In: Proceedings of the National Academy of Sciences, vol. 105(50), pp. 19678–19683 (2008)
Ambros, V.: The functions of animal microRNAs. Nature 431, 350–355 (2004)
Almirantis, Y., Papageorgiou, S.: Cross-diffusion effects on chemical and biological pattern formation. J. Theoret. Biol. 151, 289–311 (1991)
Bard, J.: Lauder, I.:How well does Turings theory of morphogenesis work. J. Theoret. Biol 45, 501–531 (1974)
Bartel, D.P.: MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116, 281–297 (2004)
Baier, R.R., Tian, C.: Mathematical analysis and numerical simulation of pattern formation under cross-diffusion. Nonlinear Anal. Real World Appl. 10, 1–12 (2012)
Brandman, O., Ferrell, J.E., Li, R., et al.: Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310, 496–498 (2005)
Budrene, E.O., Berg, H.C.: Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature 376, 49–53 (1995)
Chung, J.M., Peacock-Lpez, E.: Bifurcation diagrams and Turing patterns in a chemical self-replicating reaction-diffusion system with cross diffusion. J. Chem. Phys. 127, 174903 (2007)
Cohen, S.M., Brennecke, J., Stark, A.: Denoising feedback loops by thresholding—a new role for microRNAs. Genes. Dev 20, 2769–2772 (2006)
Fanelli, D., Cianci, C., Di Patti, F.: Turing instabilities in reaction-diffusion systems with cross diffusion. Eur. Phys. J. B 86, 1–8 (2013)
Gottesman, S.: Micros for microbes: Non-coding regulatory RNA in bacteria. Trands. Genet 21, 399–404 (2005)
Kitano, H.: Biological robustness. Nat. Rev. Genet 5, 826–837 (2004)
Lacerte, A., Korah, J., Roy, M., et al.: Transforming growth factor to be inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal 20, 50–59 (2008)
Lee, I., Cho, U.I.: Pattern formations with Turing and Hopf oscillating pattern. Bull. Korean. Chem. Soc. 21, 1213–1216 (2000)
Leone, G., Sears, R., Huang, E.: Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8, 105–113 (2001)
Maini, P.K., Woolley, T.E., Baker, R.E., et al.: Turings model for biological pattern formation and the robustness problem. Interface Focus 2, 487–496 (2012)
Pena, B., Perez, C.: Stability of Turing patterns in the Brusselator model. Phys. Rev. E 64, 056213 (2001)
Sayama, H., Marcus, A.M., Aguiar, D.E., et al.: Interplay between Turing pattern formation and domain coarsening in spatially extended population models. Forma 18, 19–36 (2003)
Shi, J., Xie, Z., Little, K.: Cross-diffusion induced instability and stability in reaction-diffusion systems. J. Appl. Anal. Comput. 1, 95–119 (2011)
Shoji, H.: Entrainment and modulation of Turing patterns under spatiotemporal forcing. Forma 24, 23–27 (2009)
Soucek, L., Evan, G.: The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 20, 91–95 (2010)
Thomas, M., Grnweller, K.L., et al.: Analysis of transcriptional regulation of the human miR-17-92 cluster. Int. J. Mol. Sci. 14, 12273–12296 (2013)
Turing, A.: The chemical basis of morphogenesis. Trans. R. Soc. B 237, 37–72 (1952)
Vanag, V.K., Epstein, I.R.: Cross-diffusion and pattern formation in reaction diffusion systems. Phys. Chem. Chem. Phys. 11, 897–912 (2008)
Wu, Y., Wang, P., et al.: Turing patterns in a reaction-diffusion system. Commun. Theor. Phys 45, 761–764 (2006)
Xu, Y., Feng, J., Li, J.J., Zhang, H.Q.: Lèvy noise induced switch in the gene transcriptional regulatory system. Chaos 23, 013110 (2013)
Xu, Y., Jin, X., et al.: Parallel logic gates in synthetic gene networks induced by non-Gaussian noise. Phys. Rev. E 88, 052721 (2013)
Xu, Y., Wu, J., Zhang, H.Q., Ma, S.J.: Stochastic resonance phenomenon in an underdamped bistable system driven by weak asymmetric dichotomous noise. Nonlinear Dyn. 70, 531–539 (2012)
Xu, Z.Y., Bagci, U., Kubler, A., et al.: Computer-aided detection and quantification of cavitary tuberculosis from CT scans. Med. Phys. 40, 113701 (2013)
Xu, Z.Y., Dasgupta, S.K., Saha, P.K.: Tensor scale: an analytic approach with efficient computation and applications. Comput. Vision Image Underst. 116, 1060–1075 (2012)
Yan, F., Liu, H., Hao, J., et al.: Dynamical behaviors of Rb-E2F pathway including negative feedback loops involving miR449. PLoS One 7, 43908 (2012)
Zemskov, E.P., Vanag, V.K., Epstein, I.R.: Amplitude equation for reaction-diffusion systems with cross diffusion. Phys. Rev. E 10, 1103 (2011)
Zhang, A., Zhang, Y.: Negative feedback regulation of E2F–1 on c-Myc-induced hTERT expression. Military Med. J. South China 26, 527–531 (2012)
Zheng, Q.Q., Shen, J.W.: Bifurcations and dynamics of cancer signaling network regulated by microRNA. Discret. Dyn. Nat. Soc. 2013, 176956 (2013)
Zhu, Y.N., Shen, J.W., Xu, Y.: Mechanism of stochastic resonance in a quorum sensing network regulated by small RNAs. Abstr. Appl. Anal. 2013, 105724 (2013)
Zhu, Y.N., Shen, J.W., Xu, Y.: Coherence resonance in a noise-driven gene network regulated by small RNA. Theoret. Appl. Mech. Lett. 4, 013008 (2014)
Acknowledgments
This work is supported by the National Natural Science Foundation of China (11272277), Program for New Century Excellent Talents in University (NCET-10-0238), the Key Project of Chinese Ministry of Education (211105) and Innovation Scientists and Technicians Troop Construction Projects of Henan Province (134100510013), Innovative Research Team in University of Henan Province (13IRTSTHN019).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, Q., Shen, J. Turing instability in a gene network with cross-diffusion. Nonlinear Dyn 78, 1301–1310 (2014). https://doi.org/10.1007/s11071-014-1516-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11071-014-1516-9