Abstract
Multiple phenotypic states of single cells often co-exist in the presence of positive feedbacks. Stochastic gene-state switchings and low copy numbers of proteins in single cells cause considerable fluctuations. The chemical master equation (CME) is a powerful tool that describes the dynamics of single cells, but it may be overly complicated. Among many simplified models, a fluctuating-rate (FR) model has been proposed recently to approximate the full CME model in the realistic intermediate region of gene-state switchings. However, only the scenario with two gene states has been carefully analysed. In this paper, we generalise the FR model to the case with multiple gene states, in which the mathematical derivation becomes more complicated. The leading order of fluctuations around each phenotypic state, as well as the transition rates between phenotypic states, in the intermediate gene-state switching region is characterized by the rate function of the stationary distribution of the FR model in the Freidlin–Wentzell-type large deviation principle (LDP). Under certain reasonable assumptions, we show that the derivative of the rate function is equal to the unique nontrivial solution of a dominant generalised eigenvalue problem, leading to a new numerical algorithm for obtaining the LDP rate function directly. Furthermore, we prove the Lyapunov property of the rate function for the corresponding deterministic mean-field dynamics. Finally, through a tristable example, we show that the local fluctuations (the asymptotic variance of the stationary distribution at each phenotypic state) in the intermediate and rapid regions of gene-state switchings are different. Finally, a tri-stable example is constructed to illustrate the validity of our theory.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Genes switch among different states due to the regulation of transcription factors and synthesise proteins at a state-dependent rate. This paper considers self-regulatory genes with positive feedback regulations, in which the transcription factors are synthesised by the regulated gene itself and reversely help the genes switch to a state with a relatively large synthesis rate. This may lead to a copy-number distribution with multiple modals, which, from a biological point of view, correspond to multiple phenotypic states of a living cell continuously exchanging materials and energy with its surroundings (Choi et al. 2008; Gupta et al. 2011; Ozbudak et al. 2004). Transitions among phenotypic states induced by intrinsic stochasticity are advantageous for cells to survive in fluctuating environments (Kussell and Leibler 2005; Acar et al. 2008).
Early mathematical works use the reaction rate equation to model the expression of a gene with two (Babloyantz and Sanglier 1972) or multiple (Santillán 2008) states, which neglects the randomness. The chemical-master-equation (CME) model, on the contrary, describes a random dynamics inside a single cell (Delbrück 1940; Gillespie 1977), and has been applied to study lots of gene-regulatory networks (Samad et al. 2005; Berg 1978; Thattai and van Oudenaarden 2001; Paulsson 2005; Jia et al. 2018; Newby and Chapman 2014; Newby 2015). Although the CME captures both types of randomness, i.e. stochastic gene-state switchings and low copy numbers of chemical species inside a single cell (Li and Xie 2011; Taniguchi et al. 2010; Eldar and Elowitz 2010), its mathematical computation is usually complex, and the exact solution is only available in simple cases (Hornos et al. 2005; Ramos et al. 2011).
Therefore, many algorithms have been proposed to numerically solve the probability distributions of CME accurately for problems of interests, without Monte Carlo simulations which are usually computationally expensive. For example, the Finite State Projection (FSP) algorithm (Munskya and Khammashb 2006) and its improved versions (Peleš et al. 2006; MacNamara et al. 2008; Kazeev et al. 2014; Hegland et al. 2008) utilize efficient projections in the vector-based state space of CME. Recently, the FSP is further developed to efficiently estimate the stationary distribution and the parameter sensitivities of the CME (Gupta et al. 2017; Dürrenberger et al. 2019). Other examples include the on-the-fly variant of the uniformisation technique, which improves the original algorithm at the cost of a small approximation error (Mateescu et al. 2010), the method of conditional moments (MCM), which employs a discrete stochastic description for low-copy number species and a moment-based description for medium/high-copy number species (Hasenauer et al. 2014), and so on.
Alternatively, various simplified mechanistic models rather than just numerical algorithms have also been proposed to approximate the CME and investigate the mechanism of single-cell dynamics. Their mathematical foundation is the limit behavior of general CME under different scales of reaction rate, species abundance, and time (Crudu et al. 2012; Kang and Kurtz 2013). Specifically, for gene-regulatory networks, simplified models are applicable under different parameter regions based on the relationship between the gene-state switchings and the birth–death kinetics of proteins. Most previous works assumed that gene-state switchings are extremely slow (Karmakar and Bose 2004; Artyomov et al. 2007; Qian et al. 2009; To and Maheshri 2010; Feng et al. 2011; Ochab-Marcinek and Tabaka 2010) or extremely rapid (Ge and Qian 2009; Wang et al. 2010; Zhou et al. 2012; Qian 2014; Lu et al. 2014; Hufton et al. 2019a, b) to avoid the mathematical difficulty in subsequent analyses. However, at least in bacteria, the single-cell gene-state switchings are neither extremely slow nor extremely rapid (Li and Xie 2011; Taniguchi et al. 2010; Choi et al. 2008; Gupta et al. 2011; Ozbudak et al. 2004). The relative stability of phenotypic states and the transition rates among them in such an intermediate region are far from quantitatively understood.
Recently, Ge et al. (2015) have proposed a so-called fluctuating-rate (FR) model for the more realistic intermediate region, which neglects the randomness caused by the low copy number of proteins but retains the randomness caused by gene-state switchings. The FR model is much more accessible for mathematical analyses than the full CME model because its mathematical prototype, i.e. the piecewise deterministic Markov processes (PDMP), has been well studied (Davis 1984, 1993). PDMP is a Markov process, whose randomness is only given by the jumps among different deterministic dynamics. PDMP has appeared in several previous studies of the gene-regulatory networks (Kepler and Elston 2001; Newby 2012; Hufton et al. 2016, 2018). These works are all restricted to the case of specific number (two or three) of gene states. In addition, Lin and Doering (2016) studied two-state cases theoretically, but only provided the numerical results for multiple-state ones.
Actually, it has already been proved that, after taking the limit of large active synthesis rates of proteins, the full CME model approaches the FR model, and if the switching rates between discrete gene states further tend to infinity, the FR model finally converges to the deterministic mean-field dynamics (DMFD) (Crudu et al. 2012; Faggionato et al. 2010). Studying the relative stability of phenotypic states and the transition rates among them should go one step further beyond this law of large number. That is the large deviation principle (LDP), especially the Freidlin–Wentzell-type one (Freidlin and Wentzell 2014). Although the general dynamic LDP theory of PDMP models has already been derived (Kifer 2009; Faggionato et al. 2009, 2010), explicit expressions of the LDP rate function of steady-state distribution as well as the proof for its Lyapunov property for the DMFD are still lacking for gene-regulatory FR models with multiple gene states. The case with only two gene states has been solved via the WKB expansion in Ge et al. (2015). The connection between LDP and WKB methods is justified in Bressloff and Faugeras (2017). People may believe that the scenario with multiple gene states should share the same mathematical results as the case with only two gene states, and the multiple-state FR model has indeed already been applied to investigate the stochastic kinetics of lac operon (Ge et al. 2018), but the mathematical methods in Ge et al. (2015) cannot be generalized straightforwardly.
Besides the rigorous LDP for PDMPs, there are a variety of application techniques in solving the first-passage time problems of stochastic hybrid systems, which include WKB approximations and matched asymptotics (Bressloff and Newby 2014b; Keener and Newby 2011; Newby 2012; Newby and Keener 2011; Newby et al. 2013), and path-integrals (Bressloff and Newby 2014a; Bressloff 2015). But the calculation of the LDP rate function by these techniques is still difficult for the multiple-state case even numerically. That is why most applications of the PDMP so far are still restricted to the case of two or three gene states.
We focus on the LDP rate function of the stationary distribution, which is a quasi-potential of the FR model and describes the leading order of the fluctuations of the protein abundance. We found out that its derivative with respect to the continuous variable can be formulated as the unique nontrivial solution of a dominant generalised eigenvalue problem in the case with arbitrary finite number of gene states, which generalises the results in Ge et al. (2015). Main mathematical tools are the famous Perron–Frobenius theorem (Frobenius 1912) and the convexity of the dominant eigenvalue of an essentially nonnegative matrix on diagonal elements (Deutsch and Neumann 1984). Such a detailed investigation of dominant generalised eigenvalue problems promotes a new numerical algorithm for obtaining the LDP rate function of steady-state distribution in the FR models. We further prove the Lyapunov property of the LDP rate function with respect to the DMFD, based on the above analysis. Whereas the results of Faggionato et al. (2009) are restricted to the case with unique fixed point, our result of the Lyapunov property is general. The prefactor of the FR model, which provides the next order of fluctuations, is also proved to be continuous and positive.
We use a tristable example to numerically show that the rate function of the FR model correctly predicts the transition rates between phenotypic states, i.e. different attractors of DMFD, in the intermediate region based on the Freidlin–Wentzell-type LDP. Moreover, the local fluctuations, i.e. asymptotic variance, of each phenotypic state in the intermediate region of gene-state switchings are highly different from those in the rapid region, even if their DMFDs are the same.
This paper is organised as follows. Heuristic derivation of the FR model with multiple gene states as well as the associated LDP rate function is given in Sect. 2. Mathematically rigorous proof of the LDP rate function as the unique nontrivial solution of a dominant generalised eigenvalue problem and the Lyapunov property with respect to DMFD are given in Sect. 3. In the same section, we propose a new numerical algorithm for the calculation of LDP rate function. In Sect. 4, a tristable example is analysed in detail to further justify the main results. The conclusions and remarks are presented in Sect. 5.
2 Approximate the full CME model with multiple gene states
We briefly describe the full CME model of protein syntheses in Sect. 2.1. In Sect. 2.2, we reduce the full CME model to the FR model through the rapid limit of protein synthesis. Then as the gene-state switching rates further approach infinity, the DMFD of the FR model and the LDP for its stationary distribution are given in Sects. 2.3 and 2.4, respectively. We briefly introduce the reduced CME model in Sect. 2.5, which approximates the full CME model through the rapid limit of gene-state switchings.
2.1 Full CME model
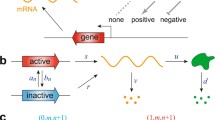
We assume the total number of gene states is G. A gene switches from state i to state j by the rate \(h_{i,j}\left( n\right) \), which depends on the protein copy number n. A gene in state i synthesises proteins by the rate \(k_i\), and a protein degrades by rate \(\gamma \). Without loss of generality, assume that \(k_1>k_2>\cdots >k_G\). Figure 1 is the diagram of an example of the full CME model with three gene states. The state of a single cell is characterized by the gene state and copy number of protein molecules. Define \(p_i\left( n,t\right) \) as the probability of the cell state in which the gene state is i and there are n protein molecules at the moment t. The CME is (Delbrück 1940; Grima et al. 2012; Gillespie 1976, 1977)
2.2 FR model
Let \(h_{i,j}\left( n\right) \) and \(\gamma \) be fixed, and define \(n_{\max }=\frac{k_1}{\gamma }\). Denote \(k_i=n_{\max }k_i^0\) with \(k_i^0\) being fixed, in which \(k_1^0=\gamma \). We give a heuristic derivation of the FR model which approximates the full CME model as \(n_{\max }\rightarrow \infty \). The FR model is actually a PDMP. Rigorous definition and proof of the convergence of the full CME model to PDMP as \(n_{\max }\rightarrow \infty \) are given by Theorem 3.1 in Crudu et al. (2012).
Define \(x:=n/n_{\max }\), \({\widetilde{p}}_i\left( x,t\right) :=p_i\left( n_{\max }\cdot x,t\right) \) and \({\widetilde{h}}_{i,j}\left( x\right) :=h_{i,j}\left( n_{\max }\cdot x\right) \). Then Eq. (1) becomes
Substitute
into Eq. (2), resulting in
It is exactly the Fokker–Planck equation of a PDMP process, called FR model for single-cell dynamics (Ge et al. 2015). The gene switches stochastically among different states, while at each gene state, the fluctuation of the protein kinetics is eliminated by taking the limit of the large active synthesis rate of proteins, leaving the rescaled protein number x to follow a deterministic dynamics.
2.3 DMFD of the FR model
Define the negative transition rate matrix as
where \({\widetilde{h}}_{i,\cdot }\left( x\right) :=\sum _{j\ne i}{\widetilde{h}}_{i,j}\left( x\right) \). In the FR model of Eq. (3), let \(k_i^0\) and \(\gamma \) be fixed, and \({\widetilde{H}}\left( x\right) ={\mathcal {H}}\widehat{{\widetilde{H}}}\left( x\right) \) (\({\widetilde{h}}_{i,j}\left( x\right) ={\mathcal {H}}\widehat{{\widetilde{h}}}_{i,j}\left( x\right) \)) with \(\widehat{{\widetilde{H}}}\left( x\right) \) being fixed and \({\mathcal {H}}\rightarrow \infty \). If \(\widehat{{\widetilde{H}}}\left( x\right) \) is irreducible, the Markov chain with the transition rate matrix \(-{\mathcal {H}}\widehat{{\widetilde{H}}}\left( x\right) \) has the unique stationary probability \({\widetilde{\zeta }}_i\left( x\right) \) for gene state i. We give a heuristic derivation of the DMFD of the FR model. The rigorous counterpart is the averaging principle of PDMP given by Theorem 2.2 in Faggionato et al. (2010).
As \({\mathcal {H}}\rightarrow \infty \), the characteristic time-scale of the gene switching becomes much faster than the dynamics of the protein abundance x in the FR model, and \({\widetilde{p}}_i\left( x,t\right) \propto {\widetilde{\zeta }}_i\left( x\right) \) approximately. Thus, we define
Let \({\widetilde{p}}\left( x,t\right) :=\sum _{i=1}^G{\widetilde{p}}_i\left( x,t\right) \). Then summing Eq. (3) over i, and substituting Eq. (4), we have
Equation (5) is exactly the Louville equation, describing the evolution of probability along the DMFD
2.4 LDP for the stationary distribution of the FR model
Except for the particular cases in Propositions 3.4 and 3.5 of Faggionato et al. (2009), obtaining the exact stationary solution of the FR model with more than two gene states is generally difficult. We alternatively give a heuristic derivation of the LDP rate function for the stationary distribution of the FR model.
Assume Eq. (3) has the unique stationary distribution \({\widetilde{p}}_i^{ss}\left( x\right) \), which is absolutely continuous w.r.t. the Lebesgue measure. Then
The pathwise LDP has been proved for many mesoscopic models approaching the macroscopic ones (including diffusion processes) (Freidlin and Wentzell 2014; Feng and Kurtz 2015; Olivieri and Vares 2005; Touchette 2009). For PDMP, the LDP of the path density is proved by Theorem 2.3 in Faggionato et al. (2010). Then by the same classic techniques used in Freidlin–Wentzell theory (Freidlin and Wentzell 2014), one can prove that the limit \(\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log \sum _i{\widetilde{p}}_i^{ss}\left( x\right) \) exists. We further assume that the limit \(\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log {\widetilde{p}}_i^{ss}\left( x\right) \) exists for each i and is independent of i, which is denoted by \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \).
In other words, the stationary distribution of the FR model satisfies \({\widetilde{p}}_i^{ss}\left( x\right) =C_i\left( x,{\mathcal {H}}\right) \exp \left( -{\mathcal {H}}\widehat{{\widetilde{\varPhi }}}\left( x\right) \right) \) scaled by the gene switching intensity \({\mathcal {H}}\), where \(C_i\left( x,{\mathcal {H}}\right) \) is the prefactor satisfying \(\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log C_i\left( x,{\mathcal {H}}\right) =0\).
We have assumed that \(\widehat{{\widetilde{\varPhi }}}_i\left( x\right) =\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log {\widetilde{p}}_i^{ss}\left( x\right) \) is independent of i. This is based on the following heuristic argument. Substitute \({\widetilde{p}}_i^{ss}\left( x\right) =C_i\left( x,{\mathcal {H}}\right) \exp \left( -{\mathcal {H}}\widehat{{\widetilde{\varPhi }}}_i\left( x\right) \right) \) into Eq. (7), and divide both sides by \(\exp \left( -{\mathcal {H}}\widehat{{\widetilde{\varPhi }}}_i\left( x\right) \right) \).
The third term on the right hand side of Eq. (8) will be exponentially large unless its exponential parts vanish. Thus,
Define \(C\left( x,{\mathcal {H}}\right) :=\left[ C_1\left( x,{\mathcal {H}}\right) ,C_2\left( x,{\mathcal {H}}\right) ,\cdots ,C_G\left( x,{\mathcal {H}}\right) \right] ^T\). Assume that
Then divide both sides of Eq. (8) by \({\mathcal {H}}\Vert C(x,{\mathcal {H}})\Vert _2\), neglect the first term in Eq. (8) as \({\mathcal {H}}\rightarrow \infty \), and substitute \(x_i^*:=k_i^0/\gamma \), we have
Define
then Eq. (9) can be rewritten in the matrix form:
\(\exists {\widetilde{C}}\left( x\right) \ne 0\) satisfying Eq. (11) iff
What we have used is the WKB method. The connections between LDP and less rigorous WKB methods for Markov chains have been established for a long time (Dykman et al. 1994; Hanggi et al. 1984; Knessl et al. 1985; Vellela and Qian 2008). Similar results for PDMP are recently proved in Bressloff and Faugeras (2017). Such a LDP rate function also provides the transition-rate formula between different attractors of DMFD, which takes the general Arrhenius/Kramers form (Keener and Newby 2011; Ge et al. 2015). In literature, people sometimes call the LDP rate function \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) the nonequilibrium landscape function (NLF) (Feng et al. 2011; Ge et al. 2015), which is an analog of the equilibrium landscape function established in the field of protein folding (Frauenfelder et al. 1991; Onuchic et al. 1997).
2.5 Reduced CME model
Let \(k_i\) and \(\gamma \) be fixed, and \(H\left( n\right) ={\mathcal {H}}{\widehat{H}}\left( n\right) \) with \({\widehat{H}}\left( n\right) \) being fixed and \({\mathcal {H}}\rightarrow \infty \). Resembling Ge et al. (2015), we give a heuristic derivation of the reduced CME model which averages the full CME model over the stationary distribution of the gene state as \({\mathcal {H}}\rightarrow \infty \). The rigorous averaging principle of the full CME model is given by Theorem 5.1 in Kang and Kurtz (2013).
Define \(p\left( n,t\right) :=\sum _{i=1}^Gp_i\left( n,t\right) \). Let \(\zeta _i\left( n\right) \) be the stationary probability for gene state i of the Markov chain with transition rate matrix \(-{\mathcal {H}}{\widehat{H}}\left( n\right) \). As \({\mathcal {H}}\rightarrow \infty \), \(p_i\left( n,t\right) \propto \zeta _i\left( n\right) \) approximately. Define \({\bar{k}}\left( n\right) :=\sum _{i=1}^G\zeta _i\left( n\right) k_i\). Sum Eq. (1) over i.
Equation (13) is called the reduced CME model.
Note that the reduced CME is actually a full CME with only one gene state. Thus, the FR approximation of it as in Sect. 2.2 directly leads to the DMFD. Use the same rate scales as in Sect. 2.2, and define \(\widetilde{{\bar{k}}}\left( x\right) :={\bar{k}}\left( n\right) \). By Eq. (4), \(\widetilde{{\bar{k}}}\left( x\right) =n_{\max }g\left( x\right) \). Then Eq. (13) becomes
Substitute
into Eq. (14), one arrives at
which is exactly Eq. (5). Therefore, the FR model and the reduced CME model share the same DMFD in Eq. (6).
Define \(p^{ss}\left( n\right) \) as the stationary distribution of \(p\left( n,t\right) \). By Eq. (13), for \(n\ge 1\),
\(0=\gamma p^{ss}\left( 1\right) -{\bar{k}}\left( 0\right) p^{ss}\left( 0\right) \) for \(n=0\). Therefore,
Define \(\widehat{{\widetilde{\varPhi }}}^R\left( x\right) :=\lim _{n_{\max }\rightarrow \infty }-\frac{1}{n_{\max }}\log \left[ p^{ss}\left( xn_{\max }\right) \right] =-\int _0^x\log \left( \frac{g\left( y\right) }{\gamma y}\right) dy\). Then
By Eqs. (6) and (16), we have the Lyapunov property of \(\widehat{{\widetilde{\varPhi }}}^R\left( x\right) \).
3 Rigorous analysis of the rate function and the prefactor of the FR model
Based on the heuristic arguments in the previous section, we propose several basic assumptions for the FR models:
Assumption 1
The stationary distribution \({\widetilde{p}}_i^{ss}\left( x\right) \) of the FR model can be expressed as
in which \(\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log C_i\left( x,{\mathcal {H}}\right) =0\) and \(\lim _{{\mathcal {H}}\rightarrow \infty }\frac{1}{{\mathcal {H}}}\partial _x \left[ \log C_i(x,{\mathcal {H}})\right] =0\).
Furthermore, \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) is continuously-differentiable, and \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \) satisfies Eq. (12).
Assumption 2
\(C(x,{\mathcal {H}})\) in Assumption 1 satisfies \(\lim _{{\mathcal {H}}\rightarrow +\infty }C(x,{\mathcal {H}})/\Vert C(x,{\mathcal {H}})\Vert _2={\widetilde{C}}(x)\), and \({\widetilde{C}}(x)\) satisfies Eq. (11).
Assumption 3
The negative transition matrixes \(\widehat{{\widetilde{H}}}\left( x\right) \) are irreducible and continuous in \(\left( x_G^*,x_1^*\right) \).
Generalised eigenvalue (GE) proposed in previous works (Chu 1987; Ikramov 1993) and dominant generalised eigenvalue (DGE) proposed in this work are the two most important concepts in this section. The classical eigenvalue problem of a matrix Z is to solve the equation \(\det \left( \lambda I-Z\right) =0\). Replacing the identity matrix I by another matrix D, roots of \(\det \left( \lambda D-Z\right) =0\) are called GEs of the matrix Z on D, which form the generilised spectrum. Assume Z is a Z-matrix, i.e. all the off-diagonal elements are nonpositive, then we can define the classical dominant eigenvalue problem for Z. For \(\lambda \) large enough, \(\lambda I-Z\) is a nonnegative matrix with a real positive dominant eigenvalue \(\lambda _0\) due to the Perron–Frobenius theorem. Then \(\lambda -\lambda _0\) is a real eigenvalue with the smallest real part among all eigenvalues of Z, which is called the classic dominant eigenvalue of Z and denoted by \(r\left( Z\right) \). Obviously, such a definition does not depend on the choice of \(\lambda \). Note that \(Z-r\left( Z\right) I\) is still a Z-matrix, and the eigenvalue of \(Z-r\left( Z\right) I\) with the smallest real part is exactly 0. Thus, \(r\left( Z-r\left( Z\right) I\right) =0\). Replacing I by another diagonal matrix D, any solution of \(r\left( Z-\lambda D\right) =0\) (see Definition A1) is called a DGE of Z on D.
By Assumption 1 and Definition A1, \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \) is a GE of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \). By Assumption 3, Corollary A1 and Theorem A1, \(\widehat{{\widetilde{H}}}\left( x\right) \) has at most two DGEs on \(A\left( x\right) \) for \(x\in \left( x_G^*,x_1^*\right) \). One is always 0. The other, if exists, is defined as the nontrivial DGE \(\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \). Otherwise, define \(\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) :=0\) (see Definition A4). For simplicity, define \(\mu \left( x\right) :=\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \) for \(x\in \left( x_G^*,x_1^*\right) \).
3.1 The relation between \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \) and \(\mu \left( x\right) \)
First, we justify the continuity of \(\mu (x)\), especially at those \(x_i^*\)’s with degenerated \(A\left( x\right) \). For \(x\ne x_i^*\), \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) is a \(\lambda \)-polynomial of order G and has G roots (GEs). However, \(\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] =0\) is of order \(G-1\) and has only \(G-1\) roots (GEs). The following theorem shows that one of the G GEs for \(x\ne x_i^*\), denoted by \(\lambda _{\infty }(x)\), diverges at \(x_i^*\), and the other \(G-1\) GEs for \(x\ne x_i^*\) tend to the \(G-1\) GEs at \(x_i^*\). Furthermore, for \(x\ne x_i^*\), the nontrival DGE is not \(\lambda _{\infty }(x)\) near \(x_i^*\), and converges at \(x_i^*\) to the nontrival DGE at \(x_i^*\).
Theorem 1
Under Assumption 3, we have for \(i\in [2,G-1]\)
-
1.
There exists a single root \(\lambda _{\infty }\left( x\right) \) of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) near \(x_i^*\) such that \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) =\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) \lambda _{\infty }\left( x\right) \).
-
2.
\(\forall \lambda \),
$$\begin{aligned} \lim _{x\rightarrow x_i^*}p\left( x\right) \left( \lambda \right) =\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] \Bigg /\left[ \widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) \prod _{i'\ne i}\gamma \left( x_i^*-x_{i'}^*\right) \right] , \end{aligned}$$where for \(x\in \bigcup _{i'=1}^{G-1}\left( x_{i'+1}^*,x_{i'}^*\right) \),
$$\begin{aligned} p\left( x\right) \left( \lambda \right) :=\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] \Bigg /\left\{ \left[ \lambda -\lambda _{\infty }\left( x\right) \right] \prod _{i'=1}^G\gamma \left( x-x_{i'}^*\right) \right\} . \end{aligned}$$(17) -
3.
\(\lim _{x\rightarrow x_i^*}\mu \left( x\right) =\mu \left( x_i^*\right) \).
Proof
For \(x\in \bigcup _{i'=1}^{G-1}\left( x_{i'+1}^*,x_{i'}^*\right) \), \(A\left( x\right) \) is nonsingular, thereby
and the GEs of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \) are the eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \). \(\forall \lambda \),
so as \(x\rightarrow x_i^*\), \(G-1\) eigenvalues (count multiplicity) of \(\gamma \left( x_i^*-x\right) \widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) tend to 0, and the remaining one tends to \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) \). The eigenvalues of \(\gamma \left( x_i^*-x\right) \widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) are just those of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) multiplying \(\gamma \left( x_i^*-x\right) \), so we have the statement 1.
Multiply both sides of Eq. (17) by \(\left[ \lambda -\lambda _{\infty }\left( x\right) \right] \prod _{i'=1}^G\gamma \left( x-x_{i'}^*\right) \).
Because \(\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) \lambda _{\infty }\left( x\right) =\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) \), for any fixed \(\lambda \),
This is the statement 2.
Because \(\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) \lambda _{\infty }\left( x\right) =\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) >0\), we have
Denote the number of positive and negative diagonal elements of \(A\left( x\right) \) by \(m\left[ A\left( x\right) \right] \) and \(n\left[ A\left( x\right) \right] \), respectively. Because \(m[A(x_i^*)],n[A(x_i^*)]>0\), some diagonal element of \(\widehat{{\widetilde{H}}}\left( x\right) -\lambda _{\infty }\left( x\right) A\left( x\right) \) tends to \(-\infty \) as \(x\rightarrow x_i^*\). By the Gershgorin circle theorem (Gershgorin 1931), \(\lim _{x\rightarrow x_i^*}r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda _{\infty }\left( x\right) A\left( x\right) \right] =-\infty \). Thus, \(\mu \left( x\right) \ne \lambda _{\infty }\left( x\right) \) for x sufficiently close to \(x_i^*\). By statement 2, the roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) except \(\lambda _\infty \left( x\right) \) tend to the \(G-1\) roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] =0\) as \(x\rightarrow x_i^*\), thereby continuous at \(x_i^*\) and bounded near \(x_i^*\); thus, \(-\infty<\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \le \varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) <+\infty \). Let \(\lim _{j\rightarrow +\infty }y_j=x_i^*\) be a sequence such that \(\lim _{j\rightarrow +\infty }\mu \left( y_j\right) =\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \). \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( y_j\right) >0\) because \(\widehat{{\widetilde{H}}}(x)\) is irreducible by Assumption 3. Then
Thus, \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \) is a DGE of \(\widehat{{\widetilde{H}}}\left( x_i^*\right) \) on \(A\left( x_i^*\right) \), and so is \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \).
Now we are going to prove that \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) =\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \) by contradiction. If not so, then \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \) and \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \) are the only two DGEs of \(\widehat{{\widetilde{H}}}\left( x_i^*\right) \) on \(A\left( x_i^*\right) \) by Corollary A1, and one must be 0 by Theorem A1. Assume without loss of generality that \(0=\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) >\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \). Let \({\bar{\mu }}:=\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) /2<0\). By Corollary A1, \(r\left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -{\bar{\mu }} A\left( x_i^*\right) \right] >0\) since \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right)<{\bar{\mu }}<\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \). For j large enough, \({\bar{\mu }}<\mu \left( y_j\right) \). Then \(r\left[ \widehat{{\widetilde{H}}}\left( y_j\right) -{\bar{\mu }}A\left( y_j\right) \right] <0\) by Corollary A1 since \({\bar{\mu }}<0\) and \({\bar{\mu }}<\mu (y_j)\). As \(j\rightarrow \infty \), \(r\left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -{\bar{\mu }} A\left( x_i^*\right) \right] \le 0\), conflicts. Therefore, \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) =\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \).
Now we show that \(\lim _{x\rightarrow x_i^*}\mu \left( x\right) =\mu \left( x_i^*\right) \).
-
1.
If \(\mu \left( x_i^*\right) =0\), then by Definition A4, 0 is the only DGE of \(\widehat{{\widetilde{H}}}\left( x_i^*\right) \) on \(A\left( x_i^*\right) \). Then \(\lim _{x\rightarrow x_i^*}\mu \left( x\right) =0=\mu \left( x_i^*\right) \) because \(\lim _{x\rightarrow x_i^*}\mu \left( x\right) \) is a DGE.
-
2.
If \(\mu \left( x_i^*\right) >0\), then following Definition A3, denote the number (count multiplicity) of GEs with positive (negative) real parts of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \) by \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \) (\(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \)). By symmetry, it is enough to prove \(\lim _{x\rightarrow x_i^*-}\mu \left( x\right) =\mu \left( x_i^*\right) \).
By Theorem A1, \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x_i^*\right) =m\left[ A\left( x_i^*\right) \right] =i-1\). By continuity, \(\exists \delta >0\) such that for \(x\in \left( x_i^*-\delta ,x_i^*\right) \), the \(i-1\) roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] =0\) with positive real parts at \(x_i^*\) still have positive real parts at x. By \(\lim _{x\rightarrow x_i^*-}\lambda _\infty \left( x\right) =+\infty \), it is possible to decrease \(\delta \) such that \(\lambda _\infty \left( x\right) >0\), thereby \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \ge i\) for \(x\in \left( x_i^*-\delta ,x_i^*\right) \). Conversely, for \(x\in \left( x_{i+1}^*,x_i^*\right) \), \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \le m\left[ A\left( x\right) \right] =i\) by Theorem A1. Therefore, \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =m\left[ A\left( x\right) \right] =i\) for \(x\in \left( x_i^*-\delta ,x_i^*\right) \). By Theorem A1, \(\mu \left( x\right) >0\) for \(x\in \left( x_i^*-\delta ,x_i^*\right) \). Denote the roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) other than \(\lambda _{\infty }(x)\) and \(\mu (x)\) by \(\{\lambda _j(x)\}_{j=1}^{G-2}\). Assume without loss of generality that \(\{\lambda _j(x)\}_{j=1}^{i-2}\) have positive real parts in \(\left( x_i^*-\delta ,x_i^*\right) \).
By statement 2, \(\mu (x)\) and \(\{\lambda _j(x)\}_{j=1}^{G-2}\) tend to the roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] =0\). If \(\lim _{x\rightarrow x_i^*-}\mu \left( x\right) =\mu \left( x_i^*\right) \) does not hold, then \(\lim _{x\rightarrow x_i^*-}\mu \left( x\right) =0\). Also, since \(\{\lambda _j\}_{j=i-1}^{G-2}\) have nonpositive real parts in \(\left( x_i^*-\delta ,x_i^*\right) \), \(\lim _{x\rightarrow x_i^*-}\mathfrak {R}[\lambda _j(x)]\le 0\) for \(j\in [i-1,G-2]\). Thus, \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x_i^*\right) \le i-2<i-1=m\left[ A\left( x_i^*\right) \right] \), which conflicts Theorem A1.
-
3.
The case for \(\mu \left( x_i^*\right) <0\) can be proved similarly as if \(\mu \left( x_i^*\right) >0\).
\(\square \)
Theorems A2 and 1 imply Corollary 1.
Corollary 1
Under Assumption 3, \(\mu \left( x\right) \) is continuous in \(\left( x_G^*,x_1^*\right) \).
Remark 1
A generalisation of Theorem 1 is given in Theorem A3 under a weaker assumption than Assumption 3 that for \(i\in \left[ 2,G-1\right] \), \(\widehat{{\widetilde{H}}}\left( x\right) \) is continuous at \(x_i^*\), \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }{|x-x_i^*|}=+\infty \), and \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,j}\left( x\right) }{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }\) exists \(\forall j\ne i\).
The following lemma is essential for the relation between \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \) and \(\mu \left( x\right) \).
Lemma 1
Under Assumption 3, any real continuous GE \(\lambda \left( x\right) \) in \(\left( x_G^*,x_1^*\right) \) satisfies \(\lambda \left( x\right) \in \left\{ 0,\mu \left( x\right) \right\} \), \(\forall x\in \left( x_G^*,x_1^*\right) \).
Proof
By Corollary 1, \(\mu \left( x\right) \) is continuous in \(\left( x_G^*,x_1^*\right) \). By Lemma A1, \(\mu \left( x\right) \) and 0 is not a single root of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) iff \(\mu \left( x\right) =0\). So \(\mu \left( x\right) \) and 0 do not intersect any other GE of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \). Thus, \(\exists x\in \left( x_G^*,x_1^*\right) \) such that \(\lambda \left( x\right) \in \left\{ 0,\mu \left( x\right) \right\} \) implies \(\lambda \left( x\right) \in \left\{ 0,\mu \left( x\right) \right\} \) \(\forall x\in \left( x_G^*,x_1^*\right) \). So it is enough to prove the former. By Lemma A6, \(0<\lambda \left( x\right) \) for \(x\in \left( x_G^*,x_{G-1}^*\right) \), and \(0>\lambda \left( x\right) \) for \(x\in \left( x_2^*,x_1^*\right) \). By continuity of \(\lambda \left( x\right) \), \(\exists x\in \left( x_G^*,x_1^*\right) \) such that \(\lambda \left( x\right) =0\in \left\{ 0,\mu \left( x\right) \right\} \). \(\square \)
By Assumption 1 and Lemma 1, \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \in \left\{ 0,\mu \left( x\right) \right\} \). However, we still can not uniquely determine \(\partial _x\widehat{{\widetilde{\varPhi }}}\). It is because if \(\mu \left( x\right) \) has two roots \(x_G^*<x'<x''<x_1^*\), one can let \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) =0\) in \(\left( x',x''\right) \) and \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) =\mu \left( x\right) \) in \(\left( x_G^*,x'\right) \cup \left( x'',x_1^*\right) \). Then \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \) is still continuous and does not violate Assumption 1 and Lemma 1. Therefore, we have to add another assumption in order to not consider those trivial scenarios.
Assumption 4
If \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \in \left\{ 0,\mu \left( x\right) \right\} \), then \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) =\mu \left( x\right) \).
3.2 The Lyapunov property of \(\widehat{{\widetilde{\varPhi }}}(x)\) for DMFD
Theorem 2 proves the Lyapunov property of \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \).
Theorem 2
Under Assumptions 1, 3 and 4, in \(\left( x_G^*,x_1^*\right) \):
-
1.
\(\mu (x)=0\) iff \(g(x)-\gamma x=0\);
-
2.
\(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) satisfies
$$\begin{aligned} \frac{d\widehat{{\widetilde{\varPhi }}}\left( x\right) }{dt}=\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) \frac{dx}{dt}=\mu \left( x\right) \left[ g\left( x\right) -\gamma x\right] \le 0, \end{aligned}$$(18)and the equality holds iff \(g(x)-\gamma x=0\).
Proof
\(\sum \limits _{j=1}^G\det \left[ \widehat{{\widetilde{H}}}^{{\bar{j}}}\left( x\right) \right] >0\) because \(\widehat{{\widetilde{H}}}\left( x\right) \) is irreducible. So
where \(\widehat{{\widetilde{H}}}^{{\bar{j}}}\left( x\right) \) is the submatrix of \(\widehat{{\widetilde{H}}}\left( x\right) \) deleting the jth row and column. For \(x\in \bigcup _{j=1}^{G-1}\left( x_{j+1}^*,x_j^*\right) \), denote the G roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] \) by \(\{\lambda _j\}_{j=1}^G\), and for \(x=x_i^*\) for some \(i\in [2,G-1]\), denote the \(G-1\) roots by \(\{\lambda _j\}_{j=1}^{G-1}\). Assume without loss of generality that \(\lambda _1\left( x\right) \equiv 0\). Observing the coefficients of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] \), we have for \(x\in \bigcup _{j=1}^{G-1}\left( x_{j+1}^*,x_j^*\right) \),
and for \(x=x_i^*\),
By Lemma A1, \(\mu \left( x\right) \) or 0 is not a single root of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) iff \(\mu \left( x\right) =0\). In other words, \(\prod _{j=2}^G\lambda _j\left( x\right) =0\) (or \(\prod _{j=2}^{G-1}\lambda _j\left( x\right) =0\) for \(x=x_i^*\)) is equivalent to \(\mu (x)=0\), thereby \(\sum _{j=1}^G\det \left[ \widehat{{\widetilde{H}}}^{{\bar{j}}}\left( x\right) \right] \gamma (x_j^*-x)=0\). This is the statement 1.
By Eqs. (19) and (21), and Assumption 4, for \(x\in \bigcup _{j=1}^{G-1}\left( x_{j+1}^*,x_j^*\right) \),
By Lemma A6, exactly one of the following occurs.
-
1.
\(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =m\left[ A\left( x\right) \right] \), \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =n\left[ A\left( x\right) \right] -1\), \(\mu \left( x\right) >0\).
-
2.
\(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =m\left[ A\left( x\right) \right] -1\), \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =n\left[ A\left( x\right) \right] \), \(\mu \left( x\right) <0\).
-
3.
\(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =m\left[ A\left( x\right) \right] -1\), \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =n\left[ A\left( x\right) \right] -1\), \(\mu \left( x\right) =0\).
The signs of \(\left[ \prod _{j=2}^G\lambda _j\left( x\right) \right] \) and \(\prod _{j=1}^G \gamma \left( x_j^*-x\right) \) are determined by \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \) and \(n\left[ A\left( x\right) \right] \), respectively. In case 1, they have opposite signs, and \(\mu (x)>0\). In case 2, they have the same sign, and \(\mu (x)<0\). In case 3, \(\mu (x)=0\). Since \(\sum _{i=1}^G\det \left[ \widehat{{\widetilde{H}}}^{{\bar{i}}}\left( x\right) \right] >0\), \(\frac{d\widehat{{\widetilde{\varPhi }}}\left( x\right) }{dt}\le 0\) in all cases. Because \(\mu \left( x\right) \) and \(g\left( x\right) \) are continuous in \(\left( x_G^*,x_1^*\right) \), \(\frac{d\widehat{{\widetilde{\varPhi }}}\left( x\right) }{dt}\) is also continuous by Eq. (18). Thus, \(\frac{d\widehat{{\widetilde{\varPhi }}}\left( x\right) }{dt}\le 0\) for \(x=x_i^*\) with \(i\in \left[ 2,G-1\right] \). The necessary and sufficient condition for the equality is implied by the statement 1. Hence the statement 2 is proved. \(\square \)
Proposition 1
Under Assumptions 2, 3 and 4, \({\widetilde{C}}_i\left( x\right) \) is continuous and positive in \(\left( x_G^*,x_1^*\right) \).
Proof
By definition, \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \right] =0\) for \(x\ne x_i^*\). By continuity, \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \right] =0\) \(\forall x\in \left( x_G^*,x_1^*\right) \). By Assumption 4, \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) =\mu (x)\). Then by Eq. (11), \({\widetilde{C}}\left( x\right) \) is the eigenvector of \(\widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \) corresponding to the dominant eigenvalue \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \right] =0\). Because \(\widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \) is irreducible, by the Perron–Frobenius theorem, \({\widetilde{C}}_i\left( x\right) >0\) \(\forall i\), and \({\widetilde{C}}_i\left( x\right) \) is continuous since \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\mu \left( x\right) A\left( x\right) \right] =0\) is a single eigenvalue. \(\square \)
Proposition 2
Under Assumptions 1, 3 and 4, for \(\epsilon >0\),
Proof
Statement 1 in Theorem 1 is also true for \(x_1^*\) and \(x_G^*\). So
By Assumption 4, \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x\right) =\mu (x)\). It is enough to prove that \(\mu \left( x\right) =\lambda _{\infty }\left( x\right) \) in \(\left( x_G^*,x_G^*+\epsilon \right) \) and \(\left( x_1^*-\epsilon ,x_1^*\right) \) for \(\epsilon \) small enough. Since \(\lim _{x\rightarrow x_G^*+}\lambda _{\infty }\left( x\right) =-\infty \), for \(\epsilon <x_{G-1}^*-x_G^*\) small enough, \(\lambda _{\infty }\left( x\right) <0\) in \(\left( x_G^*,x_G^*+\epsilon \right) \). Thus, \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \ge 1=n\left[ A\left( x\right) \right] \). By Theorem A1, \(n\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =n\left[ A\left( x\right) \right] \) (\(\lambda _{\infty }\left( x\right) \) is the only GE with negative real part) and \(\mu \left( x\right) <0\). Thus, \(\mu \left( x\right) =\lambda _{\infty }\left( x\right) \) in \(\left( x_G^*,x_G^*+\epsilon \right) \). The case for \(x_1^*\) can be proved similarly. \(\square \)
By Proposition 2,
It is appropriate to define \(\widehat{{\widetilde{\varPhi }}}\left( x\right) =+\infty \) for \(x\in \left[ 0,x_G^*\right] \cup \left[ x_1^*,+\infty \right) \) because the stationary distribution of the FR model vanishes outside \(\left( x_G^*,x_1^*\right) \). In fact, once x entering \(\left( x_G^*,x_1^*\right) \), it will never leave based on the piecewise-deterministic dynamics of the FR models.
3.3 The numerical algorithm for \(\widehat{{\widetilde{\varPhi }}}(x)\)
We need the following proposition to support our numerical algorithm.
Proposition 3
Under Assumption 3, if \(\mu \left( x\right) \ne 0\), then no real GE of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \) locates strictly between \(\mu \left( x\right) \) and 0.
Proof
Assume without loss of generality that \(\mu \left( x\right) >0\). By Corollary A1, \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] >0\) for \(0<\lambda <\mu \left( x\right) \). Recall that \(r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] \) is the eigenvalue of \(\widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \) with the smallest real part. Thus, all eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \) have positive real parts. Therefore, \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] >0\). So any \(\lambda \in \left( 0,\mu \left( x\right) \right) \) cannot be a GE.\(\square \)
Proposition 3 together with Theorem A1 can be used to calculate \(\mu \left( x\right) \) in \(\left( x_G^*,x_1^*\right) \) numerically, thereby \(\partial _{\left( x\right) }\widehat{{\widetilde{\varPhi }}}\left( x\right) \) under Assumption 4. Then based on the fact that the global minimum of the LDP rate function is always zero (Touchette 2009), one can numerically obtain \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \).
The rest of this subsection is the numerical algorithm for obtaining the nontrivial DGE at any fixed \(x\in \bigcup _{i=1}^{G-1}\left( x_{i+1}^*,x_i^*\right) \). Since A is nonsingular, the GEs of \(\widehat{{\widetilde{H}}}\) on A are the eigenvalues of \(\widehat{{\widetilde{H}}}A^{-1}\), which can be solved by existing numerical methods. We first assume the ideal case that the numerically solved spectrum is completely accurate. Then \(\mu \) can be obtained based on Theorem A1 and Proposition 3, following the four steps:
-
1.
Calculate \(m\left( \widehat{{\widetilde{H}}}|A\right) \) and \(n\left( \widehat{{\widetilde{H}}}|A\right) \) by the G eigenvalues of \(\widehat{{\widetilde{H}}}A^{-1}\).
-
2.
If \(m\left( \widehat{{\widetilde{H}}}|A\right) =m\left( A\right) \), let \(\mu \) be the smallest real positive eigenvalue in the spectrum.
-
3.
If \(n\left( \widehat{{\widetilde{H}}}|A\right) =n\left( A\right) \), let \(\mu \) be the largest real negative eigenvalue in the spectrum.
-
4.
If \(m\left( \widehat{{\widetilde{H}}}|A\right) =m\left( A\right) -1\) and \(n\left( \widehat{{\widetilde{H}}}|A\right) =n\left( A\right) -1\), let \(\mu =0\).
However, in real applications, the solved eigenvalues have round-off error. So we have to modify the above procedure as follows:
-
1.
Set the eigenvalue with the smallest absolute value as 0.
-
2.
Calculate \(m\left( \widehat{{\widetilde{H}}}|A\right) \) and \(n\left( \widehat{{\widetilde{H}}}|A\right) \) by the G eigenvalues.
-
3.
If \(m\left( \widehat{{\widetilde{H}}}|A\right) \ge m\left( A\right) \), then
-
find the eigenvalues with the smallest absolute imaginary part among all the eigenvalues with positive real parts;
-
let \(\mu \) be the smallest real part among the eigenvalues found in the above step.
-
-
4.
If \(n\left( \widehat{{\widetilde{H}}}|A\right) \ge n\left( A\right) \), do symmetrically as the case when \(m\left( \widehat{{\widetilde{H}}}|A\right) \ge m\left( A\right) \).
-
5.
Otherwise, let \(\mu =0\).
One may expect that the above modification is able to obtain an accurate \(\mu \) if the round-off error is small. Nevertheless, no theory promises its correctness. Thus, one still needs to check the obtained \(\mu \). Corollary A1 provides a possible method, which implies that
-
1.
If \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \ge 0\), then both \((-\infty ,\lambda ]\) and \([\lambda ,+\infty )\) contain DGE.
-
2.
If \(\lambda >0\) and \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \le 0\), then \((\lambda ,+\infty )\) contains no DGE.
-
3.
If \(\lambda <0\) and \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \le 0\), then \((-\infty ,\lambda )\) contains no DGE.
We assume that the above numerical algorithm is accurate enough such that the following statements are valid for some small error tolerances \(\tau>\epsilon >0\):
-
1.
If \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \ge 0\), then both \((-\infty ,\lambda +\epsilon ]\) and \([\lambda -\epsilon ,+\infty )\) contain DGE.
-
2.
If \(\lambda \ge \tau \) and \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \le 0\), then \((\lambda -\epsilon ,+\infty )\) contains no DGE.
-
3.
If \(\lambda \le \tau \) and \(r\left( \widehat{{\widetilde{H}}}-\lambda A\right) \le 0\), then \((-\infty ,\lambda +\epsilon )\) contains no DGE.
Denote the accurate value of \(\mu \) by \(\mu ^*\). The following procedure checks whether \(\mu \) is a good approximation of \(\mu ^*\). If not so, such a check procedure still can further narrow the possible range for the searching of \(\mu ^*\).
-
1.
If \(|\mu |<2\tau \), then
-
If \(r\left( \widehat{{\widetilde{H}}}+2\tau A\right) \le 0\) and \(r\left( \widehat{{\widetilde{H}}}-2\tau A\right) \le 0\), then \(\mu ^*\in [-2\tau -\epsilon ,2\tau +\epsilon ]\), thereby \(\mu \) is a good approximation of \(\mu ^*\).
-
If \(r\left( \widehat{{\widetilde{H}}}+2\tau A\right) > 0\) and \(r\left( \widehat{{\widetilde{H}}}-2\tau A\right) \le 0\), then \((-\infty ,-2\tau +\epsilon ]\) contains DGE. Since \(0\notin (-\infty ,-2\tau +\epsilon ]\), \(\mu ^*\in (-\infty ,-2\tau +\epsilon ]\).
-
If \(r\left( \widehat{{\widetilde{H}}}+2\tau A\right) \le 0\) and \(r\left( \widehat{{\widetilde{H}}}-2\tau A\right) > 0\), then \(\mu ^*\in [2\tau -\epsilon ,+\infty )\) symmetrically.
-
If \(r\left( \widehat{{\widetilde{H}}}+2\tau A\right) > 0\) and \(r\left( \widehat{{\widetilde{H}}}-2\tau A\right) > 0\), then \(\mu ^*\in (-\infty ,-2\tau +\epsilon ]\cap [2\tau -\epsilon ,+\infty )=\emptyset \), conflicts.
-
-
2.
If \(\mu \ge 2\tau \), then
-
If \(r\left( \widehat{{\widetilde{H}}}-(\mu -\tau ) A\right) \ge 0\) and \(r\left( \widehat{{\widetilde{H}}}-(\mu +\tau ) A\right) \le 0\), then \(\mu ^*\in [\mu -\tau -\epsilon ,\mu +\tau +\epsilon )\), thereby \(\mu \) is a good approximation of \(\mu ^*\).
-
If \(r\left( \widehat{{\widetilde{H}}}-(\mu -\tau ) A\right) < 0\) and \(r\left( \widehat{{\widetilde{H}}}-(\mu +\tau ) A\right) \le 0\), then \(\mu ^*\in (-\infty ,\mu -\tau +\epsilon ]\).
-
If \(r\left( \widehat{{\widetilde{H}}}-(\mu -\tau ) A\right) \ge 0\) and \(r\left( \widehat{{\widetilde{H}}}-(\mu +\tau ) A\right) >0\), then \(\mu ^*\in [\mu +\tau -\epsilon ,+\infty )\).
-
If \(r\left( \widehat{{\widetilde{H}}}-(\mu -\tau ) A\right) <0\) and \(r\left( \widehat{{\widetilde{H}}}-(\mu +\tau ) A\right) >0\), then \(\mu ^*\in (-\infty ,\mu -\tau +\epsilon ]\cap [\mu +\tau -\epsilon ,+\infty )=\emptyset \), conflicts.
-
-
3.
If \(\mu \le -2\tau \), do symmetrically as the case when \(\mu \ge 2\tau \).
Unless \(\mu \) is already a good approximation of \(\mu ^*\), one should
-
1.
remove eigenvalues whose real parts equal to \(\mu \), or do not belong to the possible range of \(\mu ^*\) obtained from the above steps;
-
2.
find one eigenvalue with the smallest absolute imaginary part among the remaining eigenvalues, and let \(\mu \) be the real part of it;
-
3.
check whether the new \(\mu \) is a good approximation of \(\mu ^*\), and if not so, further narrow the possible range of \(\mu ^*\).
Repeat the above procedure until one of the following cases is encountered.
-
1.
If \(\mu \) is a good approximation of \(\mu ^*\), then return \(\mu \).
-
2.
Otherwise, if the possible range of \(\mu ^*\) is small enough, then return the mid-point of the range.
-
3.
If all eigenvalues are removed, and the possible range of \(\mu ^*\) is still not small enough, then apply a modified dichotomic search in the possible range.
In summary, we have Algorithm 3.1, where \(\mathfrak {R}\) and \(\mathfrak {I}\) mean the real and imaginary parts, respectively, and [a, b] is the possible range of \(\mu ^*\). The modified dichotomic search converges because \(b-a\le (b'-a')/2+\tau \), where \([a',b']\) is the possible range of \(\mu ^*\) in the previous step. \(\mu \) at \(x_i^*\) for \(i\in [2,G-1]\) can be obtained by continuity.

4 Local fluctuations and transition rates between phenotypic states in a tristable example
We apply Algorithm 3.1 and Eq. (16) to a tristable example. The LDP rate functions in the intermediate and rapid regions of gene-state switchings are calculated, and the local fluctuations obtained from the rate function are compared. Here, local fluctuations are just the second derivative of the LDP rate function at each local minimum, which corresponds to each steady fixed point of the DMFD. To be more precise, we can expand the rate function \(\phi (x)\) near any local minimum \(x^*\), i.e.
Then the asymptotic Gaussian variance of \(p^{ss}\left( x\right) \) close to \(x^*\) is \(({\mathcal {H}}\phi ''(x^*))^{-1}\).
In this section, we also show that the transition rates between phenotypic states in the intermediate region are correctly predicted by rate formula proposed in the FR model based on the Freidlin–Wentzell LDP (Freidlin and Wentzell 2014), and a direct comparison with the stationary distribution of the full CME model further appreciates the validity of the numerically obtained rate function of the FR model.
4.1 Tristable example
Let \(G=3\). By the definitions of \(g\left( x\right) \) and \(A\left( x\right) \) in Eqs. (4) and (10), the fixed points (phenotypic states) of the DMFD in Eq. (6) are the roots of
where \(\widehat{{\widetilde{H}}}^{{\bar{j}}}\left( x\right) \) is the submatrix of \(\widehat{{\widetilde{H}}}\left( x\right) \) deleting the jth row and column. Let
Then,
Eq. (24) together with \(k_1>k_2>k_3\) describes a positive feedback regulation with proteins attaching to the gene as dimers.
Substitute Eqs. (24) and (25) into Eq. (23).
Define \(u_1:=\widehat{{\widetilde{h}}}_{1,2}^c/\widehat{{\widetilde{h}}}_{2,1}^c\), \(u_2:=\widehat{{\widetilde{h}}}_{2,3}^c/\widehat{{\widetilde{h}}}_{3,2}^c\), and divide Eq. (26) by \(\widehat{{\widetilde{h}}}_{2,1}^c\widehat{{\widetilde{h}}}_{3,2}^c\).
We plot a curve
in the phase plane of \(u_1\) and \(1/u_2\) in Fig. 2a for \(k_2^0/k_1^0=0.1283\) and \(k_3^0/k_1^0=0.0078\) (remind that \(k_1^0\equiv \gamma \)).
a Curve in the phase plane of \(u_1\) and \(1/u_2\) by Eq. (27) with \(G=3\), \(k_2^0/k_1^0=0.1283\), \(k_3^0/k_1^0=0.0078\). As a function of x, the number of roots of \(f\left( x,u_1,u_2\right) =0\) changes with \(u_1\) and \(u_2\). In each region divided by the curve, we label the number of roots of \(f\left( x,u_1,u_2\right) =0\) (i.e. the fixed points of the DMFD). b Phase diagram with the same parameters as in a and \(u_1=0.3329\). It shows the fixed points of the DMFD vary with \(u_2\). Dash lines are the unstable fixed points indicated by \(x^{\left( 1.5\right) }\) and \(x^{\left( 2.5\right) }\), while real lines are those stable ones indicated by \(x^{\left( 1\right) }\), \(x^{\left( 2\right) }\) and \(x^{\left( 3\right) }\). c The rate function \(\widehat{{\widetilde{\varPhi }}}^R\left( x\right) \) of the reduced CME model, and the Gaussian approximations around \(x^{\left( i\right) }\) with \(\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 1\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 2\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 3\right) }\right) =1:0.0056:0.0027\). d The rate function \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) of the FR model, and the Gaussian approximations around \(x^{\left( i\right) }\) with \(\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 1\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 2\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 3\right) }\right) =1:0.0163:0.0170\). In c, d, \(G=3\), \(k_2^0/k_1^0=0.1283\), \(k_3^0/k_1^0=0.0078\), \(u_1=0.3329\), \(u_2=0.0054\). In d, \(\widehat{{\widetilde{h}}}^c_{3,2}/\widehat{{\widetilde{h}}}^c_{2,1}=1\) and \(k_1^0\equiv \gamma =0.01\)
The root number in each region divided by the curve is labeled in Fig. 2a. In one region, \(f\left( x,u_1,u_2\right) =0\) has five roots. Fixing \(u_1=0.3329\), we plot \(1/u_2\) as a function of x by \(f\left( x,u_1,u_2\right) =0\) in Fig. 2b. As \(u_2=0.0054\) (vertical dotted line), \(f\left( x,u_1,u_2\right) =0\) has five roots \(x^{\left( 1\right) }\), \(x^{\left( 1.5\right) }\), \(x^{\left( 2\right) }\), \(x^{\left( 2.5\right) }\), \(x^{\left( 3\right) }\). These roots are the fixed points of the DMFD (an ODE). Three stable fixed points \(x^{\left( 1\right) }\), \(x^{\left( 2\right) }\) and \(x^{\left( 3\right) }\) are interlaced by two unstable ones \(x^{\left( 1.5\right) }\) and \(x^{\left( 2.5\right) }\), which indicates that the FR model is tristable.
4.2 FR model exhibits different local fluctuations from those of the reduced CME model
In our tristable example, \(\partial _x\widehat{{\widetilde{\varPhi }}}\left( x^{\left( i\right) }\right) =\partial _x\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( i\right) }\right) =0\) at the stable fixed point \(x^{\left( i\right) }\) of the DMFD by Theorem 2 and Eq. (16). Therefore, around \(x^{\left( i\right) }\), we have the Gaussian approximations
The local fluctuations around the fixed point \(x^{\left( i\right) }\) for the FR model and the reduced CME model are indicated by \(\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( i\right) }\right) \) and \(\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( i\right) }\right) \), respectively. The larger the second derivative is, the weaker the local fluctuation will be, illustrated by the ratio \(\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 1\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 2\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}\left( x^{\left( 3\right) }\right) \) (or \(\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 1\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 2\right) }\right) :\partial _x^2\widehat{{\widetilde{\varPhi }}}^R\left( x^{\left( 3\right) }\right) \)) of the local fluctuations over all fixed points.
In Fig. 2c, we plot \(\widehat{{\widetilde{\varPhi }}}^R\left( x\right) \) of the tristable example in Sect. 4.1. Further fixing \(\widehat{{\widetilde{h}}}^c_{3,2}/\widehat{{\widetilde{h}}}^c_{2,1}=1\) and \(k_1^0\equiv \gamma =0.01\), we plot \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) of the same example in Fig. 2d. The Gaussian approximations Eq. (28) around \(x^{\left( i\right) }\) are plotted in dash line in Fig. 2c, d. The local fluctuations of \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) are different from those of \(\widehat{{\widetilde{\varPhi }}}^R\left( x\right) \) even if they share the same DMFD. Actually, we have
Thus, the local fluctuations at \(x^{\left( 2\right) }\) and \(x^{\left( 3\right) }\) are stronger in the rapid regime (the reduced CME model) of gene-state switchings than those in the intermediate one (the FR model).
4.3 Transition rates between phenotypic states
The mean first-passage time \(T_{ij}^c\) from \(x^{\left( i\right) }\) to \(x^{\left( j\right) }\) of the full CME model is numerically calculated by the conventional first-passage theory (Redner 2007). In Fig. 3, \(T_{ij}^c\) is plotted logarithmically as functions of \({\mathcal {H}}\) in \(\circ \), \(\times \), \(+\) for different \(n_{\max }\).
\(T_{ij}^c\) is insensitive to \(n_{\max }\) in the intermediate region \(n_{\max }k_i\gg {\mathcal {H}}\widehat{{\widetilde{h}}}_{i,j}\left( x\right) \gg \gamma \) of gene-state switchings. The FR model is simulated by the Doob–Gillespie method to obtain the mean first-passage time \(T_{ij}^f\) (Gillespie 1976, 1977). In Fig. 3, \(T_{ij}^f\) (real line) agrees well with \(T_{ij}^c\) for different \(n_{\max }\) (\(\circ \), \(\times \), \(+\)).
Refer to the discussion on the links between large deviations and WKB for the PDMP (Bressloff and Faugeras 2017), the transition rate \(k_{ij}\) from \(x^{\left( i\right) }\) to \(x^{\left( j\right) }\) is approximated by the general Arrhenius form
where \(\varDelta \widehat{{\widetilde{\varPhi }}}_{ij}:=\widehat{{\widetilde{\varPhi }}}\left[ x^{\left( \frac{i+j}{2}\right) }\right] -\widehat{{\widetilde{\varPhi }}}\left[ x^{\left( i\right) }\right] \) is the barrier height from \(x^{\left( i\right) }\) to \(x^{\left( j\right) }\), and \(k_{ij}^0\) is a prefactor such that \(\lim _{{\mathcal {H}}\rightarrow \infty }-\frac{1}{{\mathcal {H}}}\log k_{ij}^0=0\). Substitute \(k_{ij}=1/T_{ij}^c\) into Eq. (28).
The linear relationship between \(\log \left( T_{ij}^c\right) \) and \({\mathcal {H}}\) in Eq. (29) is observed in Fig. 3, by which we fit and plot the slope \(\varDelta \widehat{{\widetilde{\varPhi }}}_{ij}\) for different \(n_{\max }\) in the insets of Fig. 3 by dash lines. The real lines, which are calculated directly by \(\varDelta \widehat{{\widetilde{\varPhi }}}_{ij}:=\widehat{{\widetilde{\varPhi }}}\left[ x^{\left( \frac{i+j}{2}\right) }\right] -\widehat{{\widetilde{\varPhi }}}\left[ x^{\left( i\right) }\right] \), match the dash lines. In conclusion, the LDP of the FR model correctly predicts the transition rates between phenotypic states in the intermediate region of gene-state switchings.
Logarithmic mean first-passage time of the full CME model as functions of \({\mathcal {H}}\) for different \(n_{\max }\) (\(\circ \), \(\times \), \(+\)), and that of the FR model (real lines). \(k_1^0\equiv \gamma =0.01\), \(u_1=0.3329\), \(u_2=0.0054\), \(\widehat{{\widetilde{h}}}^c_{3,2}/\widehat{{\widetilde{h}}}^c_{2,1}=1\), \(k_1^0:k_2^0:k_3^0=1:0.1283:0.0078\). The insets show the barrier heights \(\varDelta \widehat{{\widetilde{\varPhi }}}_{ij}\) calculated by fitting Eq. (29) for different \(n_{\max }\) (dash lines), and those directly calculated by \(\varDelta \widehat{{\widetilde{\varPhi }}}_{ij} \,{=}\, \widehat{{\widetilde{\varPhi }}}\left[ x^{\left( \frac{i+j}{2}\right) }\right] -\widehat{{\widetilde{\varPhi }}}\left[ x^{\left( i\right) }\right] \) (real lines)
4.4 Comparison with the stationary distribution of the full CME model
By the same parameters as in Fig. 2d, we plot the stationary distribution \(p^{ss}_c\left( x\right) \) (\(x=n/n_{\max }\)) of the full CME model for \(n_{\max }=1000000\) and \({\mathcal {H}}=50\) in Fig. 4a. As expected, \(-\frac{1}{{\mathcal {H}}}\log p^{ss}_c\left( x\right) \) (real line) and \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) (dash line) are quite close (Fig. 4b).
Parameters are the same as in Fig. 2d. a Stationary distribution \(p^{ss}_c\left( x\right) \) (\(x=n/n_{\max }\)) of the full CME model for \(n_{\max }=1{,}000{,}000\) and \({\mathcal {H}}=50\). b \(-\frac{1}{{\mathcal {H}}}\log p^{ss}_c\left( x\right) \) (real line) and \(\widehat{{\widetilde{\varPhi }}}\left( x\right) \) obtained by our numerical algorithm (dash line)
5 Conclusions and remarks
A living cell usually has multiple phenotypic states to face fluctuating environments (Kussell and Leibler 2005; Acar et al. 2008). The FR model proposed in Ge et al. (2015) for single-cell dynamics with two gene states quantitatively describes the stabilities of the phenotypic states and the transition rates among them in the intermediate regime, which is the case in E. coli, but rarely studied before. Considering that genes often have more than two states due to the combinatorial nature of transcriptional regulations (Zhu et al. 2005), we generalise the FR model together with the LDP rate function of steady state to the scenario with more than two gene states. We found that the derivative of the rate function is just the unique nontrival DGE of the negative gene-state transition-rate matrix on the protein birth–death matrix.
Given that several GEs exist, a method that determines the rate function from GEs is necessary. Under appropriate assumptions, we prove that the nontrivial DGE is continuous, the derivative of the rate function equals the nontrivial DGE, and the rate function satisfies the Lyapunov property of the DMFD. The numerical results of a tristable example support the equivalence of the nontrivial DGE and the LDP rate function.
The result in Corollary 1 is interesting because under Assumption 3 the matrix \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) is discontinuous at each \(x_i^*\) but always has a continuous nontrival real eigenvalue \(\mu (x)\) in the whole interval \((x_G^*,x_1^*)\). On the contrary, Example 1 will show that even if \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) is continuous in \((x_G^*,x_1^*)\), it may have no continuous nontrivial real eigenvalue on the whole interval.
Example 1
Let \(x_i^*=\left( 5-i\right) /4\) for \(i\in \left[ 1,4\right] \), \(\gamma =1\),
It is not hard to see that the limit of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) at each \(x_i^*\) is finite, hence the matrix \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) is continuous in the whole \(\left( x_4^*,x_1^*\right) \). The eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) given in Fig. 5a–c are also continuous in \(\left( x_4^*,x_1^*\right) \) as expected, but none of them is always real.
a–c GEs of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \), or equivalently, eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) in Example 1 for \(x\in \left( x_4^*,x_1^*\right) \). a, b Real and imaginary parts of GEs. c Corresponding 3D plot
The realness of the nontrivial DGE is automatically promised by Definition A1, whereas nondominant GEs may be complex. The nontrivial DGE \(\mu \left( x\right) \) of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \) indicated by red lines in Fig. 5b, c, which should be continuous if Assumption 3 holds, is now discontinuous at \(x_3^*\) and \(x_2^*\) because \(\widehat{{\widetilde{H}}}\left( x\right) \) becomes reducible.
In this paper, we have assumed a linear A(x) in Eq. (10). However, this is not necessary. Generally, denote the ith diagonal element of A(x) by \(A_i(x)\). If no two diagonal elements \(A_i(x)\) vanish at a same x, then simply replacing \(\gamma (x_i^*-x)\) by \(A_i(x)\), the proof of Theorem 1 is still valid. After slight modification, other main results in this paper are kept.
For now, we only considered the one-dimensional case (single self-regulatory gene). For high dimensional cases, Eq. (12) becomes \(\det \left[ \widehat{{\widetilde{H}}}\left( {{\varvec{x}}}\right) -{\mathbb {F}}\left( {{\varvec{x}}}\right) \right] =0\), where
and \({\mathcal {F}}_i\left( {{\varvec{x}}}\right) \) is the vector field at gene state i. To apply the methods in this paper to high dimensional cases, many concepts must be generalised since the derivative \(\nabla \widehat{{\widetilde{\varPhi }}}\left( {{\varvec{x}}}\right) \) is now a vector but not a scalar. The most crucial step should be to properly regard the difference between \(\nabla \widehat{{\widetilde{\varPhi }}}\left( {{\varvec{x}}}\right) \) and other solutions of the underdetermined system \(\det \left[ \widehat{{\widetilde{H}}}\left( {{\varvec{x}}}\right) -{\mathbb {F}}\left( {{\varvec{x}}}\right) \right] =0\). In one-dimensional cases, it is the unique nontrivial DGE.
Abbreviations
- CME:
-
Chemical master equation
- FR:
-
Fluctuating-rate
- DMFD:
-
Deterministic mean-field dynamics
- LDP:
-
Large deviation principle
- NLF:
-
Nonequilibrium landscape function
- GE:
-
Generalised eigenvalue
- DGE:
-
Dominant generalised eigenvalue
References
Acar M, Mettetal JT, van Oudenaarden A (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40:471–475
Artyomov MN, Das J, Kardar M, Chakraborty AK (2007) Purely stochastic binary decisions in cell signaling models without underlying deterministic bistabilities. Proc Natl Acad Sci USA 104(48):18958–18963
Babloyantz A, Sanglier M (1972) Chemical instabilities of “all-or-none” type in beta—galactosidase induction and active transport. FEBS Lett 23(3):364–366
Berg OG (1978) A model for the statistical fluctuations of protein numbers in a microbial population. J Theor Biol 71:587–603
Bressloff PC (2015) Path-integral methods for analyzing the effects of fluctuations in stochastic hybrid neural networks. J Math Neurosci 5(4):1–33
Bressloff PC, Faugeras O (2017) On the Hamiltonian structure of large deviations in stochastic hybrid systems. J Stat Mech Theory Exp 2017:033206
Bressloff PC, Newby JM (2014a) Path integrals and large deviations in stochastic hybrid systems. Phys Rev E 89(042):701
Bressloff PC, Newby JM (2014b) Stochastic hybrid model of spontaneous dendritic NMDA spikes. Phys Biol 11(1):016006
Choi PJ, Cai L, Frieda K, Xie XS (2008) A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322(5900):442–446
Chu KWE (1987) Exclusion theorems and the perturbation analysis of the generalized eigenvalue problem. SIAM J Numer Anal 24(5):1114–1125
Crudu A, Debussche A, Muller A, Radulescu O (2012) Convergence of stochastic gene networks to hybrid piecewise deterministic processes. Ann Appl Probab 22(5):1822–1859
Davis MHA (1984) Piecewise-deterministic Markov processes: a general class of non-diffusion stochastic models. J R Stat Soc Ser B (Methodol) 46(3):353–388
Davis MHA (1993) Markov models and optimization, monographs on statistics and applied probability, vol 49. Chapman and Hall, London
Delbrück M (1940) Statistical fluctuations in autocatalytic reactions. J Chem Phys 8(1):120
Deutsch E, Neumann M (1984) Derivatives of the Perron root at an essentially nonnegative matrix and the group inverse of an \(M\)-matrix. J Math Anal Appl 102(1):1–29
Dürrenberger P, Gupta A, Khammash M (2019) A finite state projection method for steady-state sensitivity analysis of stochastic reaction networks. J Chem Phys 150(134):101
Dykman MI, Mori E, Ross J, Hunt PM (1994) Large fluctuations and optimal paths in chemical kinetics. J Chem Phys 100:5735
Eldar A, Elowitz MB (2010) Functional roles for noise in genetic circuits. Nature 467:167–173
Faggionato A, Gabrielli D, Crivellari MR (2009) Non-equilibrium thermodynamics of piecewise deterministic Markov processes. J Stat Phys 137:259–304
Faggionato A, Gabrielli D, Crivellari MR (2010) Averaging and large deviation principles for fully-coupled piecewise deterministic Markov processes and applications to molecular motors. Markov Process Relat Fields 16(3):497–548
Feng H, Han B, Wang J (2011) Adiabatic and non-adiabatic non-equilibrium stochastic dynamics of single regulating genes. J Phys Chem 115(5):1254–1261
Feng J, Kurtz TG (2015) Large deviations for stochastic processes, mathematical surveys and monographs, vol 131. American Mathematical Society, Providence
Frauenfelder H, Sligar SG, Wolynes PG (1991) The energy landscapes and motions of proteins. Science 254(5038):1598–1603
Freidlin MI, Wentzell AD (2014) Random perturbations of dynamical systems, Grundlehren der mathematischen Wissenschaften, vol 260, 3rd edn. Spinger, Berlin
Frobenius G (1912) Ueber matrizen aus nicht negativen elementen. Sitzungsberichte der Königlich Preussischen Akademie der Wissenschaften, pp 456–477
Ge H, Qian H (2009) Thermodynamic limit of a nonequilibrium steady state: Maxwell-type construction for a bistable biochemical system. Phys Rev Lett 103(148):103
Ge H, Qian H, Xie XS (2015) Stochastic phenotype transition of a single cell in an intermediate region of gene state switching. Phys Rev Lett 114(078):101
Ge H, Wu P, Qian H, Xie SX (2018) Relatively slow stochastic gene-state switching in the presence of positive feedback significantly broadens the region of bimodality through stabilizing the uninduced phenotypic state. PLoS Comput Biol 14(3):e1006051
Gershgorin SA (1931) über die abgrenzung der eigenwerte einer matrix. Bull l’Acad Sci l’URSS Classe Sci Math 6:749–754
Gillespie DT (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys 22(4):403–434
Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81(25):2340–2361
Grima R, Schmidt DR, Newman TJ (2012) Steady-state fluctuations of a genetic feedback loop: An exact solution. J Chem Phys 137(3):035104
Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES (2011) Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146(4):633–644
Gupta A, Mikelson J, Khammash M (2017) A finite state projection algorithm for the stationary solution of the chemical master equation. J Chem Phys 147(154):101
Hanggi P, Grabert H, Talkner P, Thomas H (1984) Bistable systems: master equation versus Fokker–Planck modeling. Phys Rev A 29:371–378
Hasenauer J, Wolf V, Kazeroonian A, Theis FJ (2014) Method of conditional moments (MCM) for the chemical master equation. J Math Biol 69:687–735
Hegland M, Hellander A, Lötstedt P (2008) Sparse grids and hybrid methods for the chemical master equation. BIT Numer Math 48:265–283
Hornos JEM, Schultz D, Innocentini GCP, Wang J, Walczak AM, Onuchic JN, Wolynes PG (2005) Self-regulating gene: an exact solution. Phys Rev E 72(051):907
Hufton PG, Lin YT, Galla T, McKane AJ (2016) Intrinsic noise in systems with switching environments. Phys Rev E 93(052):119
Hufton PG, Lin YT, Galla T (2018) Phenotypic switching of populations of cells in a stochastic environment. J Stat Mech Theory Exp 023:501
Hufton PG, Lin YT, Galla T (2019a) Classical stochastic systems with fast-switching environments: reduced master equations, their interpretation, and limits of validity. Phys Rev E 99(032):121
Hufton PG, Lin YT, Galla T (2019b) Model reduction methods for population dynamics with fast-switching environments: reduced master equations, stochastic differential equations, and applications. Phys Rev E 99(032):122
Ikramov KD (1993) Matrix pencils: theory, applications, and numerical methods. J Sov Math 64:783–853
Jia C, Qian H, Chen M, Zhang MQ (2018) Relaxation rates of gene expression kinetics reveal the feedback signs of autoregulatory gene networks. J Chem Phys 148(9):095102
Kang HW, Kurtz TG (2013) Separation of time-scales and model reduction for stochastic reaction networks. Ann Appl Probab 23(2):529–583
Karmakar R, Bose I (2004) Graded and binary responses in stochastic gene expression. Phys Biol 1(4):197
Kazeev V, adn Michael Nip MK, Schwab C (2014) Direct solution of the chemical master equation using quantized tensor trains. PLOS Comput Biol 10(3):e1003359
Keener JP, Newby JM (2011) Perturbation analysis of spontaneous action potential initiation by stochastic ion channels. Phys Rev E 84(011):918
Kepler TB, Elston TC (2001) Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys J 81(6):3116–3136
Kifer Y (2009) Large deviations and adiabatic transitions for dynamical systems and Markov processes in fully coupled averaging. Mem Am Math Soc 201(944)
Knessl C, Matkowsky BJ, Schuss Z, Tier C (1985) An asymptotic theory of large deviations for Markov jump processes. SIAM J Appl Math 45(6):1006–1028
Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309(5743):2075–2078
Li GW, Xie XS (2011) Central dogma at the single-molecule level in living cells. Nature 475:308–315
Lin YT, Doering CR (2016) Gene expression dynamics with stochastic bursts: construction and exact results for a coarse-grained model. Phys Rev E 93(022):409
Lu M, Onuchic J, Ben-Jacob E (2014) Construction of an effective landscape for multistate genetic switches. Phys Rev Lett 113(078):102
MacNamara S, Burrage K, Sidje RB (2008) Multiscale modeling of chemical kinetics via the master equation. Multiscale Model Simul 6(4):1146–1168
Mateescu M, Wolf V, Didier F, Henzinger TA (2010) Fast adaptive uniformisation of the chemical master equation. IET Syst Biol 4(6):441–452
Munskya B, Khammashb M (2006) The finite state projection algorithm for the solution of the chemical master equation. J Chem Phys 124(4):044104
Newby JM (2012) Isolating intrinsic noise sources in a stochastic genetic switch. Phys Biol 9(026):002
Newby J (2015) Bistable switching asymptotics for the self regulating gene. J Phys A Math Theor 48(18):185001
Newby J, Chapman J (2014) Metastable behavior in Markov processes with internal states. J Math Biol 69:941–976
Newby JM, Keener JP (2011) An asymptotic analysis of the spatially inhomogeneous velocity-jump process. Multiscale Model Simul 9(2):735–765
Newby JM, Bressloff PC, Keener JP (2013) Breakdown of fast–slow analysis in an excitable system with channel noise. Phys Rev Lett 111(128):101
Ochab-Marcinek A, Tabaka M (2010) Bimodal gene expression in noncooperative regulatory systems. Proc Natl Acad Sci USA 107(51):22096–22101
Olivieri E, Vares ME (2005) Large deviations and metastability, encyclopedia of mathematics and its applications, vol 100. Cambridge University Press, Cambridge
Onuchic JN, Luthey-Schulten Z, Wolynes PG (1997) Theory of protein folding: the energy landscape perspective. Ann Rev Phys Chem 48:545–600
Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A (2004) Multistability in the lactose utilization network of Escherichia coli. Nature 427:737–740
Paulsson J (2005) Models of stochastic gene expression. Phys Life Rev 2(2):157–175
Peleš S, Munsky B, Khammash M (2006) Reduction and solution of the chemical master equation using time scale separation and finite state projection. J Chem Phys 125(20):204104
Qian H (2014) Fitness and entropy production in a cell population dynamics with epigenetic phenotype switching. Quant Biol 2(1):47–53
Qian H, Shia PZ, Xing J (2009) Stochastic bifurcation, slow fluctuations, and bistability as an origin of biochemical complexity. Phys Chem Chem Phys 24(11):4861–4870
Ramos AF, Innocentini GCP, Hornos JEM (2011) Exact time-dependent solutions for a self-regulating gene. Phys Rev E 83(062):902
Redner S (2007) A guide to first-passage processes. Cambridge University Press, Cambridge
Samad HE, Khammash M, Petzold L, Gillespie D (2005) Stochastic modelling of gene regulatory networks. Int J Robust Nonlinear Control 15(15):691–711
Santillán M (2008) Bistable behavior in a model of the lac operon in Escherichia coli with variable growth rate. Biophys J 94(6):2065–2081
Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS (2010) Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329(5991):533–538
Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA 98(15):8614–8619
To TL, Maheshri N (2010) Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science 327(5969):1142–1145
Touchette H (2009) The large deviation approach to statistical mechanics. Phys Rep 478:1–69
Vellela M, Qian H (2008) Stochastic dynamics and non-equilibrium thermodynamics of a bistable chemical system: the Schlogl model revisited. J R Soc Interface 6(39):925–940
Wang J, Xu L, Wang E, Huang S (2010) The potential landscape of genetic circuits imposes the arrow of time in stem cell differentiation. Biophys J 99(1):29–39
Zhou JX, Aliyu MDS, Aurell E, Huang S (2012) Quasi-potential landscape in complex multi-stable systems. J R Soc Interface 9(77):3539–3553
Zhu Z, Shendure J, Church GM (2005) Discovering functional transcription-factor combinations in the human cell cycle. Genome Res 15(6):848–855
Acknowledgements
This work was supported by Natural Science Foundation of China, Grant No. 11971037 and 11622101 (Hao Ge), and Natural Science Foundation of China, Grant No. 11271083 (Yunxin Zhang).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Results related to dominant generalised eigenvalue problem
-
\(G>0\): matrix dimension/size.
-
\({\mathcal {G}}:=\left\{ 1,2,3,\cdots ,G\right\} \).
-
\(B=\left( b_{i,j}\right) _{G\times G}\): general \(G\times G\) matrix.
-
\(B^{{\mathcal {S}}_1,{\mathcal {S}}_2}\) for \({\mathcal {S}}_1,{\mathcal {S}}_2\subset {\mathcal {G}}\): principal submatrix of B formed by rows in \({\mathcal {S}}_1\) and columns in \({\mathcal {S}}_2\). Particularly, \(B^{{\mathcal {S}}}:=B^{{\mathcal {S}},{\mathcal {S}}}\) for \({\mathcal {S}}\subset {\mathcal {G}}\).
-
\({\bar{i}}:={\mathcal {G}}{\setminus }\left\{ i\right\} \).
-
\(|b_{\cdot ,j}|:=\sum _{i\ne j}|b_{i,j}|\).
-
Z: \(G\times G\) Z-matrix (real matrix such that off-diagonal elements are nonpositive) in the normal form (block upper-triangular form with each diagonal block irreducible).
-
\({\mathcal {I}}\): indices of irreducible diagonal blocks of Z.
-
\(Z_i\) for \(i\in {\mathcal {I}}\): irreducible diagonal blocks of Z.
-
M: \(G\times G\) M-matrix (Z-matrix such that all eigenvalues have nonnegative real parts) in the normal form. Note that we allow singular M-matrix.
-
D: \(G\times G\) real diagonal matrix.
-
\(D_i\) for \(i\in {\mathcal {I}}\): diagonal blocks of D corresponding to \(Z_i\).
-
\(d_i\) for \(i\in {\mathcal {G}}\): the ith diagonal element of D.
-
\({\mathcal {G}}_0:=\left\{ i\in {\mathcal {G}}|d_i=0\right\} \). \({\mathcal {G}}_+:=\left\{ i\in {\mathcal {G}}|d_i>0\right\} \). \({\mathcal {G}}_-:=\left\{ i\in {\mathcal {G}}|d_i<0\right\} \).
-
\(r\left( Z\right) \): dominant eigenvalue (the eigenvalue with smallest real part, which must be real by the Perron–Frobenius theorem) of Z.
-
\(m\left( D\right) \) and \(n\left( D\right) \): the number of positive and negative diagonal elements of D. \(D\ne 0\) means \(m\left( D\right) +n\left( D\right) >0\).
-
\(O\left( o,\rho \right) :=\left\{ z\in {\mathbb {C}}||z-o|\le \rho \right\} \), where \(o\in {\mathbb {C}}\) and \(\rho \ge 0\).
In this “Appendix”, we state results without proofs. Their proofs are given in “Appendix B”. We always assume \(D\ne 0\).
Definition A1
Denote the real part of \(\lambda \in {\mathbb {C}}\) by \(\mathfrak {R}(\lambda )\). Define
\(\lambda \in \varLambda \left( B|D\right) \) is called the generalised eigenvalue (GE) of B on D (Chu 1987; Ikramov 1993). For \(\lambda _0\in \varLambda \left( B|D\right) \), let \(c\left( \lambda _0|B|D\right) \) be the multiplicity of \(\lambda _0\) as a root of \(\det \left( B-\lambda D\right) =0\). We define \(R\left( Z|D\right) :=\left\{ \lambda \in {\mathbb {R}}|r\left( Z-\lambda D\right) =0\right\} \), and call \(\lambda \in R\left( Z|D\right) \) the dominant generalised eigenvalue (DGE) of Z on D. Define \(\varXi \left( Z-\lambda D\right) :=\det \left( Z-\lambda D\right) /r\left( Z-\lambda D\right) \), which is analytic even for \(\lambda \in R\left( Z|D\right) \) because they are removable singular points.
Lemma A1
Assume that M is irreducible.
-
1.
If \(n\left( D\right) =0\), then \(R\left( M|D\right) =\left\{ \mu _+\right\} \) with \(\mu _+\ge 0\) and \(c\left( \mu _+|M|D\right) =1\). \(r\left( M-\lambda D\right) >0\) for \(\lambda <\mu _+\) and \(r\left( M-\lambda D\right) <0\) for \(\lambda >\mu _+\).
-
2.
If \(m\left( D\right) =0\), then \(R\left( M|D\right) =\left\{ \mu _-\right\} \) with \(\mu _-\le 0\) and \(c\left( \mu _-|M|D\right) =1\). \(r\left( M-\lambda D\right) >0\) for \(\lambda >\mu _-\) and \(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _-\).
-
3.
Otherwise, \(R\left( M|D\right) =\left\{ \mu _-,\mu _+\right\} \) with \(\mu _+\ge 0\ge \mu _-\). \(c\left( \mu _{\pm }|M|D\right) =1\) unless \(\mu _{\pm }=0\), at which \(c\left( \mu _{\pm }|M|D\right) =c\left( 0|M|D\right) =2\). \(r\left( M-\lambda D\right) >0\) for \(\mu _-<\lambda <\mu _+\) and \(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _-\) or \(\lambda >\mu _+\).
Lemma A1 justifies Definition A2.
Definition A2
For irreducible M, define \(\mu _+\left( M|D\right) :=\max _{\lambda \in R\left( M|D\right) }\lambda \) for \(m\left( D\right) >0\), and \(\mu _-\left( M|D\right) :=\min _{\lambda \in R\left( M|D\right) }\lambda \) for \(n\left( D\right) >0\).
Definition A2 can be explained as follows. By Lemma A1, for \(m\left( D\right) >0\) and \(n\left( D\right) =0\), there is only a nonnegative DGE for M on D, which is denoted by \(\mu _+\left( M|D\right) \). Similarly, for \(m\left( D\right) =0\) and \(n\left( D\right) >0\), there is only a nonpositive DGE, which is denoted by \(\mu _-\left( M|D\right) \). If \(m\left( D\right) >0\) and \(n\left( D\right) >0\), then there are both a nonnegative DGE and a nonpositive DGE, which are respectively denoted by \(\mu _+\left( M|D\right) \) and \(\mu _-\left( M|D\right) \).
Since \(r\left( M-\lambda D\right) =\min _{i\in {\mathcal {G}}}r\left( M_i-\lambda D_i\right) \), the results in Lemma A1 can be generalised to reducible M (see Corollary A1) if for any irreducible diagonal block \(i\in {\mathcal {I}}\) such that \(D_i=0\), \(M_i\) is nonsingular. Lemma A2 provides a sufficient condition for this.
Lemma A2
\({\mathcal {K}}_0^{\mathrm{sin}}:=\left\{ i\in {\mathcal {I}}|m\left( D_i\right) =n\left( D_i\right) =0,\det \left( M_i\right) =0\right\} =\emptyset \) if and only if \(\det \left( M^{{\mathcal {G}}_0}\right) \ne 0\).
Corollary A1
Assume that \(\det \left( M^{{\mathcal {G}}_0}\right) \ne 0\).
-
1.
If \(n\left( D\right) =0\), then \(R\left( M|D\right) =\left\{ \mu _+\right\} \) with
$$\begin{aligned} \mu _+=\mu _+\left( M|D\right) :=\min _{i:m\left( D_i\right) >0}\mu _+\left( M_i|D_i\right) , \end{aligned}$$\(r\left( M-\lambda D\right) <0\) for \(\lambda >\mu _+\), and \(r\left( M-\lambda D\right) >0\) for \(\lambda <\mu _+\).
-
2.
If \(m\left( D\right) =0\), then \(R\left( M|D\right) =\left\{ \mu _-\right\} \) with
$$\begin{aligned} \mu _-=\mu _-\left( M|D\right) :=\max _{i:n\left( D_i\right) >0}\mu _-\left( M_i|D_i\right) , \end{aligned}$$\(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _-\), and \(r\left( M-\lambda D\right) >0\) for \(\lambda >\mu _-\).
-
3.
Otherwise, \(R\left( M|D\right) =\left\{ \mu _-,\mu _+\right\} \) with
$$\begin{aligned}&\mu _+=\mu _+\left( M|D\right) :=\min _{i:m\left( D_i\right)>0}\mu _+\left( M_i|D_i\right) ,\\&\mu _-=\mu _-\left( M|D\right) :=\max _{i:n\left( D_i\right) >0}\mu _-\left( M_i|D_i\right) , \end{aligned}$$\(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _-\) or \(\lambda >\mu _+\), and \(r\left( M-\lambda D\right) >0\) for \(\mu _-<\lambda <\mu _+\).
Corollary A1 extends Definition A2 to reducible M, and gives Lemma A3.
Lemma A3
Assume that \(M\left( \theta \right) \) is a continuous \(G\times G\) M-matrix function of \(\theta \), \(D\left( \theta \right) \) is a continuous \(G\times G\) diagonal matrix function of \(\theta \) with constant \({\mathcal {G}}_{\pm }\) and \({\mathcal {G}}_0\) \(\forall \theta \), and \(\det \left[ M^{{\mathcal {G}}_0}\left( \theta \right) \right] \ne 0\) \(\forall \theta \).
-
1.
If \(m\left[ D\left( \theta \right) \right] >0\), then \(\mu _+\left( \theta \right) :=\mu _+\left( M|D\right) \left( \theta \right) \) is continuous.
-
2.
If \(n\left[ D\left( \theta \right) \right] >0\), then \(\mu _-\left( \theta \right) :=\mu _-\left( M|D\right) \left( \theta \right) \) is continuous.
Lemma A4
-
1.
Assume that \(|b_{j,j}|\ge |b_{\cdot ,j}|\) \(\forall j\in {\mathcal {G}}_0\), \(B^{{\mathcal {G}}_0}\) is irreducible, and \(\exists j_0\in {\mathcal {G}}_0\) such that \(|b_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|b_{i,j_0}|\). Then
$$\begin{aligned} \bigcup _{j\in {\mathcal {G}}{\setminus }{\mathcal {G}}_0}O\left( b_{j,j}/d_j,|b_{\cdot ,j}|/|d_j|\right) =:{\mathcal {O}}\left( B|D\right) \supset \varLambda \left( B|D\right) . \end{aligned}$$ -
2.
Under the assumptions in statement 1, further assume that \(b_{j,j}\ge 0\) for \(j\notin {\mathcal {G}}_0\) and B is diagonally dominant for all columns. Then
$$\begin{aligned} \sum _{\lambda \in \left[ \varLambda \cap {\mathcal {O}}_+\right] \left( B|D\right) }c\left( \lambda |B|D\right) \ge m\left( D\right) ,\quad \sum _{\lambda \in \left[ \varLambda \cap {\mathcal {O}}_-\right] \left( B|D\right) }c\left( \lambda |B|D\right) \ge n\left( D\right) , \end{aligned}$$where \({\mathcal {O}}_{\pm }\left( B|D\right) :=\bigcup _{j\in {\mathcal {G}}_{\pm }}O\left( b_{j,j}/d_j,|b_{\cdot ,j}|/d_j\right) \).
Lemma A4 generalises the Gershgorin circle theorem to GEs.
Definition A3
Let \(T=\left( t_{i,j}\right) _{G\times G}\) be a Z-matrix with \(\sum _{i\in {\mathcal {G}}} t_{i,j}=0\) \(\forall j\in {\mathcal {G}}\). T is called a negative transition rate matrix. Let \(m\left( T|D\right) :=\sum _{\lambda \in \varLambda _+\left( T|D\right) }c\left( \lambda |T|D\right) \) and \(n\left( T|D\right) :=\sum _{\lambda \in \varLambda _-\left( T|D\right) }c\left( \lambda |T|D\right) \).
Lemmas A5 and A6, and Theorem A1 connect the signs of the real parts of GEs with the signatures of D for negative transition rate matrix T step by step. Since T is diagonally dominant, it is not hard to prove by the Gershgorin circle theorem that T is a singular M-matrix. Applying Lemma A4 to T, we have Lemma A5.
Lemma A5
Assume that \(T^{{\mathcal {G}}_0}\) is irreducible, and \(\exists j_0\in {\mathcal {G}}_0\) such that \(|t_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|t_{i,j_0}|\). Then \(\varLambda _0\left( T|D\right) \subset \left\{ 0\right\} \), and exact one of the following happens.
-
1.
\(m\left( T|D\right) =m\left( D\right) \).
-
2.
\(n\left( T|D\right) =n\left( D\right) \).
-
3.
\(m\left( T|D\right) <m\left( D\right) \), \(n\left( T|D\right) <n\left( D\right) \).
If T is irreducible, Lemma A5 becomes Lemma A6 by Lemma A1.
Lemma A6
Assume that T is irreducible, \(T^{{\mathcal {G}}_0}\) is irreducible, and \(\exists j_0\in {\mathcal {G}}_0\) such that \(|t_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|t_{i,j_0}|\). Then exact one of the following happens.
-
1.
\(m\left( T|D\right) =m\left( D\right) \), \(n\left( T|D\right) =n\left( D\right) -1\), \(\mu _-\left( T|D\right) =0\), \(c\left( \mu _-|T|D\right) =1\). If \(m\left( D\right) >0\), \(\mu _+\left( T|D\right) >0\) and \(c\left( \mu _+|T|D\right) =1\).
-
2.
\(m\left( T|D\right) =m\left( D\right) -1\), \(n\left( T|D\right) =n\left( D\right) \), \(\mu _+\left( T|D\right) =0\), \(c\left( \mu _+|T|D\right) =1\). If \(n\left( D\right) >0\), \(\mu _-\left( T|D\right) <0\) and \(c\left( \mu _-|T|D\right) =1\).
-
3.
\(m\left( T|D\right) =m\left( D\right) -1\), \(n\left( T|D\right) =n\left( D\right) -1\), \(\mu _{\pm }\left( T|D\right) =0\), \(c\left( \mu _{\pm }|T|D\right) =1\).
Theorem A1 generalises Lemma A6 to reducible T, and concretizes Lemma A5.
Theorem A1
Assume that \(T^{{\mathcal {G}}_0}\) is irreducible, and \(\exists j_0\in {\mathcal {G}}_0\) such that \(|t_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|t_{i,j_0}|\). Then exact one of the following happens.
-
1.
\(m\left( T|D\right) =m\left( D\right) \), \(\mu _-\left( T|D\right) =0\). If \(m\left( D\right) >0\), \(\mu _+\left( T|D\right) >0\).
-
2.
\(n\left( T|D\right) =n\left( D\right) \), \(\mu _+\left( T|D\right) =0\). If \(n\left( D\right) >0\), \(\mu _-\left( T|D\right) <0\).
-
3.
\(m\left( T|D\right) <m\left( D\right) \), \(n\left( T|D\right) <n\left( D\right) \), \(\mu _{\pm }\left( T|D\right) =0\).
Definition A4
\(T^{{\mathcal {G}}_0}\) is irreducible. \(\exists j_0\in {\mathcal {G}}_0\) such that \(|t_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|t_{i,j_0}|\). For \(m\left( D\right) ,n\left( D\right) >0\), define
as the nontrivial DGE of T on D.
By Theorem A1, at least one of \(\mu _{\pm }\left( T|D\right) \) is zero. Therefore, the nontrivial DGE in Definition A4 is obtained by always choosing the nonzero DGE if possible.
Theorem A2
Assume that \(T=T\left( \theta \right) \) is a continuous \(G\times G\) negative transition rate matrix function of \(\theta \), \(D\left( \theta \right) \) is a continuous \(G\times G\) diagonal matrix function of \(\theta \) with constant \({\mathcal {G}}_{\pm }\) and \({\mathcal {G}}_0\) \(\forall \theta \), and \(\det \left[ T^{{\mathcal {G}}_0}\left( \theta \right) \right] \ne 0\) \(\forall \theta \). Then \(\mu \left( \theta \right) :=\mu \left( T|D\right) \left( \theta \right) \) is continuous.
Theorem A2 proves the continuity of \(\mu \left( T|D\right) \) by Lemma A3.
Lemma A7
For \(\epsilon >0\) and \(j_0\in {\mathcal {G}}\), let \(Z^\epsilon :=\left( z^\epsilon _{i,j}\right) _{G\times G}\) with \(z^\epsilon _{i,j_0}:=\epsilon z_{i,j_0}\) and \(z^\epsilon _{i,j}:=z_{i,j}\) for \(j\ne j_0\). Then \({{\,\mathrm{sgn}\,}}\left[ r\left( Z^\epsilon \right) \right] \equiv {{\,\mathrm{sgn}\,}}\left[ r\left( Z\right) \right] \), where \({{\,\mathrm{sgn}\,}}\) is the sign function.
Lemma A8
Assume that T is irreducible. Then \(v=\left( v_j\right) _{G\times 1}\ne 0\) satisfies \(Tv=0\) iff
Appendix B: Proofs of results in “Appendix A”
Proof of Lemma A1.
Proof
Case 1: Since \(r\left( M-\lambda D\right) \) has the smallest real part among all eigenvalues of \(M-\lambda D\), it is not hard to show by the Gershgorin circle theorem that \(\lim _{\lambda \rightarrow +\infty }r\left( M-\lambda D\right) =-\infty \). By \(r\left( M\right) \ge 0\), \(\exists \mu _+\ge 0\) such that \(r\left( M-\mu _+ D\right) =0\). Because \(D>0\), \(\lambda <\mu _+ \Rightarrow M-\lambda D>M-\mu _+ D \Rightarrow r\left( M-\lambda D\right) >r\left( M-\mu _+ D\right) =0\). \(\frac{dr\left( M-\lambda D\right) }{d\lambda }|_{\lambda =\mu _+}<0\) since otherwise, by \(\frac{d^2r\left( M-\lambda D\right) }{d\lambda ^2}<0\) (Deutsch and Neumann 1984), \(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _+\), conflicts. By \(\frac{d^2r\left( M-\lambda D\right) }{d\lambda ^2}<0\), \(r\left( M-\lambda D\right) <0\) for \(\lambda >\mu _+\). \(\varXi \left( M-\mu _+ D\right) >0\) because it is the product of nondominant eigenvalues of irreducible M-matrix \(M-\mu _+ D\). Therefore,
i.e. \(c\left( \mu _+|M|D\right) =1\).
Case 2: Similar as case 1.
Case 3: Resembling case 1, \(\lim _{\lambda \rightarrow \pm \infty }r\left( M-\lambda D\right) =-\infty \). If \(r\left( M\right) >0\), \(\exists \mu _+>0>\mu _-\) such that \(r\left( M-\mu _{\pm } D\right) =0\). Because \(\frac{d^2r\left( M-\lambda D\right) }{d\lambda ^2}<0\), we have
\(r\left( M-\lambda D\right) >0\) for \(\mu _-<\lambda <\mu _+\), and \(r\left( M-\lambda D\right) <0\) for \(\lambda <\mu _-\) or \(\lambda >\mu _+\). By Eq. (A1), \(\frac{d\det \left( M-\lambda D\right) }{d\lambda }|_{\lambda =\mu _+}<0<\frac{d\det \left( M-\lambda D\right) }{d\lambda }|_{\lambda =\mu _-}\). Thus, \(c\left( \mu _{\pm }|M|D\right) =1\).
If \(r\left( M\right) =0\) and \(\frac{dr\left( M-\lambda D\right) }{d\lambda }\big |_{\lambda =0}>0\) (\(\frac{dr\left( M-\lambda D\right) }{d\lambda }\big |_{\lambda =0}<0\)), \(\exists \mu _-=0<\mu _+\) (\(\mu _-<0=\mu _+\)) such that \(r\left( M-\mu _{\pm } D\right) =0\) since \(\lim _{\lambda \rightarrow \pm \infty }r\left( M-\lambda D\right) =-\infty \). The subsequent proofs are the same as \(r\left( M\right) >0\). If \(\frac{dr\left( M-\lambda D\right) }{d\lambda }\big |_{\lambda =0}=0\), since \(\frac{d^2r\left( M-\lambda D\right) }{d\lambda ^2}<0\), \(r\left( M-\lambda D\right) <0\) for \(\lambda \ne 0\), and \(c\left( 0|M|D\right) =2\). \(\square \)
Proof of Lemma A2.
Proof
Sufficiency: Define \({\mathcal {K}}_0^{\mathrm{non}}:=\left\{ i\in {\mathcal {I}}|m\left( D_i\right) =n\left( D_i\right) =0,\det \left( M_i\right) \ne 0\right\} \). \(\forall i\in {\mathcal {K}}_0^{\mathrm{sin}}\cup {\mathcal {K}}_0^{\mathrm{non}}\), \(M_i\) is a principal submatrix of the nonsingular M-matrix \(M^{{\mathcal {G}}_0}\), thereby nonsingular. Thus, \(i\in {\mathcal {K}}_0^{\mathrm{non}}\), which means \({\mathcal {K}}_0^{\mathrm{sin}}=\emptyset \).
Necessity: Assume without loss of generality that \(M^{{\mathcal {G}}_0}\) is in the normal form. Any irreducible diagonal block \(M_j^{{\mathcal {G}}_0}\) of \(M^{{\mathcal {G}}_0}\) is either an irreducible diagonal block \(M_i\) of M, or a principal submatrix of that. For the former, \(M_j^{{\mathcal {G}}_0}\) is nonsingular since \({\mathcal {K}}_0^{\mathrm{sin}}=\emptyset \). For the latter, \(M_j^{{\mathcal {G}}_0}\) is nonsingular since \(M_i\) is an irreducible M-matrix. In summary, \(M^{{\mathcal {G}}_0}\) is nonsingular. \(\square \)
Proof of Lemma A3.
Proof
Case 1: Prove by contradiction that \(\lim _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) =\mu _+\left( \theta _0\right) \). Otherwise, since \(\mu _+\left( \theta \right) \) is bounded near \(\theta _0\), \(+\infty>\varlimsup _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right)>\varliminf _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) >-\infty \). Let \(\lim _{i\rightarrow +\infty }\theta _i=\theta _0\) be a sequence such that
By continuity of \(r\left( M-\lambda D\right) \),
So \(\varliminf _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) \in R\left( M|D\right) \left( \theta _0\right) \). Similarly, \(\varlimsup _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) \in R\left( M|D\right) \left( \theta _0\right) \). Since \(\varliminf _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) \ne \varlimsup _{\theta \rightarrow \theta _0}\mu _+\left( \theta \right) \), the only possibility is that
For i large enough, \(\mu _+\left( \theta _i\right) <\left[ \mu _+\left( \theta _0\right) +\mu _-\left( \theta _0\right) \right] /2=:{\overline{\mu }}\left( \theta _0\right) \). By Corollary A1, \(r\left[ M\left( \theta _0\right) -{\overline{\mu }}\left( \theta _0\right) D\left( \theta _0\right) \right] >0\) since \(\mu _-\left( \theta _0\right)<{\overline{\mu }}\left( \theta _0\right) <\mu _+\left( \theta _0\right) \), and for i large enough, \(r\left[ M\left( \theta _i\right) -{\overline{\mu }}\left( \theta _0\right) D\left( \theta _i\right) \right] <0\) since \({\overline{\mu }}\left( \theta _0\right) >\mu _+\left( \theta _i\right) \). By the continuity of \(r\left( M-\lambda D\right) \),
conflicts. Thus, \(\mu _+\left( \theta \right) \) is continuous, and similarly, \(\mu _-\left( \theta \right) \) is continuous.
Case 2: Similar as case 1. \(\square \)
Proof of Lemma A4.
Proof
\(\forall \lambda \in \varLambda \left( B|D\right) \), \(\exists v=\left( v_1,v_2,\cdots ,v_G\right) \) such that \(v\left( B-\lambda D\right) =0\). Prove by contradiction that \(\exists i_1\notin {\mathcal {G}}_0\) such that \(|v_{i_1}|=\max _{i'\in {\mathcal {G}}}|v_{i'}|\). Otherwise, \(|v_{i_2}|=\max _{i'\in {\mathcal {G}}}|v_{i'}|\) for some \(i_2\in {\mathcal {G}}_0\). Because \(B^{{\mathcal {G}}_0}\) is irreducible, there exists a path \(i_2={\widehat{i}}_1,{\widehat{i}}_2,\cdots ,{\widehat{i}}_a=j_0\in {\mathcal {G}}_0\) such that \(b_{{\widehat{i}}_{a'+1},{\widehat{i}}_{a'}}>0\) \(\forall a'\in \left[ 1,a-1\right] \). Because \(d_j=0\) and \(|b_{j,j}|\ge |b_{\cdot ,j}|\) \(\forall j\in {\mathcal {G}}_0\), by consecutively vanishing components \({\widehat{i}}_1,{\widehat{i}}_2,\cdots ,{\widehat{i}}_{a}\) of \(v\left( B-\lambda D\right) \), \(|b_{{\widehat{i}}_{a'},{\widehat{i}}_{a'}}|=|b_{\cdot ,{\widehat{i}}_{a'}}|\) and \(|v_{{\widehat{i}}_{a'}}|=\max _{i'\in {\mathcal {G}}}|v_{i'}|\) \(\forall a'\in \left[ 1,a\right] \). Since \(|b_{j_0,j_0}|>\sum _{i\in {\mathcal {G}}_0{\setminus }\left\{ j_0\right\} }|b_{i,j_0}|\), \(\exists i_1\notin {\mathcal {G}}_0\) such that \(b_{i_1,j_0}\ne 0\). So \(|v_{i_1}|=\max _{i'\in {\mathcal {G}}}|v_{i'}|\), conflicts.
Since component \(i_1\) of \(v\left( B-\lambda D\right) \) vanishes, \(v_{i_1}\left( \lambda d_{i_1}-b_{i_1,i_1}\right) =\sum _{i\ne i_1}v_ib_{i,i_1}\). Thus,
Therefore, \(\lambda \in O\left( b_{i_1,i_1}/d_{i_1},|b_{\cdot ,i_1}|/|d_{i_1}|\right) \subset {\mathcal {O}}\left( B|D\right) \). This is statement 1.
Let \(B^{\epsilon }=\left( b^{\epsilon }_{i,j}\right) _{G\times G}\) for \(\epsilon \in \left[ 0,1\right] \). \(b^{\epsilon }_{i,j}=\epsilon b_{i,j}\) for \(j\notin {\mathcal {G}}_0\) and \(i\ne j\); otherwise, \(b^{\epsilon }_{i,j}=b_{i,j}\). \(B^{\epsilon }\) satisfies conditions in statement 1. Define \({\mathcal {G}}_+^0:=\left\{ j|j\in {\mathcal {G}},d_j>0,b_{j,j}=0\right\} \) and \({\mathcal {G}}_-^0:=\left\{ j|j\in {\mathcal {G}},d_j<0,b_{j,j}=0\right\} \). For \(\epsilon =0\), \(\varLambda \left( B^\epsilon |D\right) =\left\{ b_{j,j}/d_j|j\notin {\mathcal {G}}_0\right\} \). For \(\epsilon <1\),
By statement 1 and the continuity of \(\varLambda \left( B^{\epsilon }|D\right) \) on \(\epsilon \),
This is statement 2. \(\square \)
Proof of Lemma A5.
Proof
By Lemma A4, \(\sum _{\lambda \in \left[ \varLambda \cap {\mathcal {O}}_+\right] \left( T|D\right) }c\left( \lambda |T|D\right) \ge m\left( D\right) \). \(\left[ \varLambda _-\cap {\mathcal {O}}_+\right] \left( T|D\right) =\emptyset \) and \(\sum _{\lambda \in \varLambda \left( T|D\right) }c\left( \lambda |T|D\right) =m\left( D\right) +n\left( D\right) \), so \(n\left( T|D\right) \le n\left( D\right) \). Similarly, \(m\left( T|D\right) \le m\left( D\right) \). Thus, if both cases 1 and 2 fail, then \(m\left( T|D\right) < m\left( D\right) \) and \(n\left( T|D\right) < n\left( D\right) \). So at least one of the three cases happens. Obviously, case 3 cannot happen simultaneously with any of cases 1 and 2. Because \(0\in \varLambda \left( T|D\right) \), cases 1 and 2 cannot happen simultaneously. In summary, exact one of the three cases happens. By Lemma A4, \(\varLambda _0\left( T|D\right) \subset {\mathcal {O}}\left( T|D\right) \). So \(\varLambda _0\left( T|D\right) \subset {\mathcal {O}}\left( T|D\right) \cap \left\{ z\in {\mathbb {C}}|\mathfrak {R}\left( z\right) =0\right\} =\left\{ 0\right\} \). \(\square \)
Proof of Lemma A6.
Proof
Case 1: Starting from case 1 in Lemma A5, \(m\left( T|D\right) =m\left( D\right) \). \(T^{{\mathcal {G}}_0}\) is a nonsingular M-matrix. So \(\det \left( T^{{\mathcal {G}}_0}\right) >0\). For \(0<\lambda <\min _{\lambda '\in \varLambda _+\left( T|D\right) }\mathfrak {R}\left( \lambda '\right) \),
where \(\lambda _i\) are the roots of \(\det \left( T-\lambda D\right) =0\). Moreover, since T is an irreducible M-matrix, \(\varXi \left( T\right) >0\) because it is the product of nondominant eigenvalues of M-matrix T. So for \(\lambda >0\) small enough, \(\varXi \left( T-\lambda D\right) >0\). In conclusion, \(r\left( T-\lambda D\right) =\det \left( T-\lambda D\right) /\varXi \left( T-\lambda D\right) >0\). By Lemma A1, if \(m\left( D\right) >0\), \(0<\lambda <\mu _+\left( T|D\right) \). Since \(r\left( T\right) =0\), \(\mu _-\left( T|D\right) =0\). Finally, \(c\left( \mu _{\pm }|T|D\right) =1\) by Lemma A1. Together with \(\varLambda _0\subset \left\{ 0\right\} \) by Lemma A5, there is \(n\left( Z|D\right) =n\left( D\right) -1\),
Case 2: Similar as case 1.
Case 3: Starting from case 3 in Lemma A5, \(m\left( T|D\right) <m\left( D\right) \) and \(n\left( T|D\right) <n\left( D\right) \). Because \(c\left( 0|T|D\right) \le 2\) by Lemma A1, the only possibility is \(m\left( T|D\right) =m\left( D\right) -1\), \(n\left( T|D\right) =n\left( D\right) -1\), and \(\mu _{\pm }\left( T|D\right) =0\). \(\square \)
Proof of Theorem A1.
Proof
Case 1: Starting from case 1 in Lemma A5, \(m\left( T|D\right) =m\left( D\right) \). \(T_i\) is irreducible and diagonally dominant in columns, so \(T_i\) is singular iff it has vanishing column sums. Also, since \(T^{{\mathcal {G}}_0}\) is irreducible, it is either an irreducible block of T, or a principal submatrix of that. As a result, singular \(T_i\) satisfies all conditions in Lemma A6. So there are three types of singular \(T_i\).
-
1.
\(m\left( T_i|D_i\right) =m\left( D_i\right) \), \(n\left( T_i|D_i\right) =n\left( D_i\right) -1\), \(\mu _-\left( T_i|D_i\right) =0\). If \(m\left( D_i\right) >0\), \(\mu _+\left( T_i|D_i\right) >0\).
-
2.
\(m\left( T_i|D_i\right) =m\left( D_i\right) -1\), \(n\left( T_i|D_i\right) =n\left( D_i\right) \), \(\mu _+\left( T_i|D_i\right) =0\). If \(n\left( D_i\right) >0\), \(\mu _-\left( T_i|D_i\right) <0\).
-
3.
\(m\left( T_i|D_i\right) =m\left( D_i\right) -1\), \(n\left( T_i|D_i\right) =n\left( D_i\right) -1\), \(\mu _{\pm }\left( T_i|D_i\right) =0\).
Since \(m\left( T|D\right) =m\left( D\right) \), only type 1 can happen. By Lemma A1, \(r\left( T_i-\lambda D_i\right) >0\) for \(0=\mu _-\left( T_i|D_i\right)<\lambda <\mu _+\left( T_i|D_i\right) \) if \(m\left( D_i\right) >0\); otherwise, \(r\left( T_i-\lambda D_i\right) >0\) for \(0=\mu _-\left( T_i|D_i\right) <\lambda \). On the other hand, \(r\left( T_i-\lambda D_i\right) >0\) for nonsingular \(T_i\) and \(\lambda >0\) small enough. In summary, \(r\left( T-\lambda D\right) >0\) for \(\lambda >0\) small enough. Because \(T^{{\mathcal {G}}_0}\) is nonsingular, by Corollary A1, \(\mu _+\left( T|D\right)>\lambda >0\) if \(m\left( D\right) >0\). Finally, \(\mu _-\left( T|D\right) =0\) since \(r\left( T\right) =0\).
Case 2: Similar as case 1.
Case 3: Starting from case 3 in Lemma A5, \(m\left( T|D\right) <m\left( D\right) \) and \(n\left( T|D\right) <n\left( D\right) \). If there exists singular \(T_i\) of type 3, then by Lemma A1, \(r\left( T_i-\lambda D_i\right) <0\) for \(\lambda \ne 0\), thereby \(r\left( T-\lambda D\right) <0\) for \(\lambda \ne 0\). Thus, \(\mu _{\pm }\left( T|D\right) =0\) by Corollary A1. Otherwise, by \(m\left( T|D\right) <m\left( D\right) \), there exists singular \(T_i\) of type 2. By Lemma A1, \(r\left( T_i-\lambda D_i\right) <0\) for \(\lambda >\mu _+\left( T_i|D_i\right) =0\), thereby \(r\left( T-\lambda D\right) <0\) for \(\lambda >0\). Similarly, by \(n\left( T|D\right) <n\left( D\right) \), there exists singular \(T_i\) of type 1, thereby \(r\left( T-\lambda D\right) <0\) for \(\lambda <0\). In summary, \(r\left( T-\lambda D\right) <0\) for \(\lambda \ne 0\). Thus, \(\mu _{\pm }\left( T|D\right) =0\) by Corollary A1. \(\square \)
Proof of Theorem A2.
Proof
By Lemma A3, \(\mu _{\pm }\left( \theta \right) \) are continuous. If \(\mu _+\left( \theta _0\right) >\mu _-\left( \theta _0\right) \), by continuity, \(\exists \delta >0\) such that \(\mu _+\left( \theta _1\right) >\mu _-\left( \theta _2\right) \) \(\forall \theta _1,\theta _2\in \left( \theta _0-\delta ,\theta _0+\delta \right) \). Therefore, either \(\mu _+\left( \theta \right) \equiv 0\) or \(\mu _-\left( \theta \right) \equiv 0\) in \(\left( \theta _0-\delta ,\theta _0+\delta \right) \). Thus, either \(\mu \left( \theta \right) \equiv \mu _-\left( \theta \right) \) or \(\mu \left( \theta \right) \equiv \mu _+\left( \theta \right) \) in \(\left( \theta _0-\delta ,\theta _0+\delta \right) \). Then \(\mu \left( \theta \right) \) is continuous at \(\theta _0\).
If \(\mu _{\pm }\left( \theta _0\right) =0\), then since \(\mu \in \{\mu _+,\mu _-\}\),
\(\square \)
Proof of Lemma A7.
Proof
First prove the case of irreducible Z.
-
1.
Assume \(r\left( Z\right) =0\). Since Z is irreducible, \(\exists v>0\) such that \(vZ=0\). Thus, \(vZ^{\epsilon }=0\). Because \(Z^{\epsilon }\) is irreducible and \(v>0\), \(r(Z^{\epsilon })=0\).
-
2.
Assume \(r\left( Z\right) \ne 0\). Prove by contradiction that \({{\,\mathrm{sgn}\,}}\left[ r\left( Z\right) \right] ={{\,\mathrm{sgn}\,}}\left[ r\left( Z^\epsilon \right) \right] \). Otherwise, by continuity, \(\exists \epsilon '>0\) such that \(r\left( Z^{\epsilon '}\right) =0\). By case 1, \(r\left( Z\right) =0\), conflicts.
If Z is reducible,
\(\square \)
Proof of Lemma A8.
Proof
Sufficiency: Because column sums of T vanish, the following three steps transform \(T^{{\bar{i}},{\bar{j}}}\) to \(T^{{\bar{j}},{\bar{j}}}\) for \(i>j\).
-
1.
Sum all rows of \(T^{{\bar{i}},{\bar{j}}}\) except row j to row j.
-
2.
Multiply row j by \(-1\).
-
3.
Exchange row j with rows \(j+1,j+2,\cdots ,i-1\) one by one.
During which there are totally \(i-j\) changes of sign. So \(T^{{\bar{j}},{\bar{j}}}=\left( -1\right) ^{i-j}T^{{\bar{i}},{\bar{j}}}=\left( -1\right) ^{i+j}T^{{\bar{i}},{\bar{j}}}\) for \(i>j\). There are similar statements for \(i<j\) as well. Then component i of Tv satisfies
Necessity: Because T is irreducible, its dominant eigenvalue 0 is simple, and the corresponding eigenvector space is of dimension 1. \(\square \)
Appendix C: Extension of Theorem 1
Theorem A3
For \(i\in \left[ 2,G-1\right] \), if \(\widehat{{\widetilde{H}}}\left( x\right) \) is continuous at \(x_i^*\), \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }{|x-x_i^*|}=+\infty \), and \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,j}\left( x\right) }{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }\) exists \(\forall j\ne i\), then:
-
1.
There exists a single root \(\lambda _{\infty }\left( x\right) \) of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) near \(x_i^*\) such that \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) =\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) \lambda _{\infty }\left( x\right) \).
-
2.
\({\mathbb {H}}_i:=\lim _{x\rightarrow x_i^*}\widehat{{\widetilde{H}}}\left( x\right) I_i\left[ 1/\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \right] \) satisfies that \(\forall \lambda \),
$$\begin{aligned} \lim _{x\rightarrow x_i^*}p\left( x\right) \left( \lambda \right) =\det \left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] \Bigg /\left[ \prod _{i'\ne i}\gamma \left( x_i^*-x_{i'}^*\right) \right] , \end{aligned}$$where \(I_i\left[ 1/\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \right] \) is obtained by replacing the ith diagonal element of the identity matrix by \(1/\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \), and for \(x\in \bigcup _{i'=1}^{G-1}\left( x_{i'+1}^*,x_{i'}^*\right) \),
$$\begin{aligned} p\left( x\right) \left( \lambda \right) :=\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] \Bigg /\left\{ \left[ \lambda -\lambda _{\infty }\left( x\right) \right] \prod _{i'=1}^G\gamma \left( x-x_{i'}^*\right) \right\} . \end{aligned}$$(A2) -
3.
\(\lim _{x\rightarrow x_i^*}\mu \left( x\right) =\mu _i:=\mu \left[ {\mathbb {H}}_i|A\left( x_i^*\right) \right] \).
Under the assumption of Theorem A3, it is possible that \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) =0\). Then the characteristic polynomial of GEs degenerates, i.e. \(\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] \equiv 0\) \(\forall \lambda \in {\mathbb {C}}\). This explains the importance of irreducibility of \(\widehat{{\widetilde{H}}}\left( x\right) \), which promises \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) >0\) in Theorem 1. By Theorem A1, \(\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \) is still well-defined for \(x\in \bigcup _{i=1}^{G-1}\left( x_{i+1}^*,x_{i}^*\right) \), and we define \(\mu \left( x\right) :=\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \). Nevertheless, \(\mu \left( x_i^*\right) \) no longer equals to \(\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x_i^*\right) \).
Proof
For \(x\in \bigcup _{i'=1}^{G-1}\left( x_{i'+1}^*,x_{i'}^*\right) \), \(A\left( x\right) \) is nonsingular, thereby
and the GEs of \(\widehat{{\widetilde{H}}}\left( x\right) \) on \(A\left( x\right) \) are the eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \). \(\forall \lambda \),
so as \(x\rightarrow x_i^*\), \(G-1\) eigenvalues (count multiplicity) of \(\gamma \left( x_i^*-x\right) \widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) tend to 0, and one tends to \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) \). The eigenvalues of \(\gamma \left( x_i^*-x\right) \widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) are just those of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) multiplying \(\gamma \left( x_i^*-x\right) \), so we have statement 1.
Multiply both sides of Eq. (A2) by \(\left[ \lambda -\lambda _{\infty }\left( x\right) \right] \prod _{i'=1}^G\gamma \left( x-x_{i'}^*\right) /\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \).
Because \({\mathbb {H}}_i:=\lim _{x\rightarrow x_i^*}\widehat{{\widetilde{H}}}\left( x\right) I_i\left[ 1/\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \right] \), \(\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) /\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) =0\), and \(\lim _{x\rightarrow x_i^*}\gamma \left( x_i^*-x\right) \lambda _{\infty }\left( x\right) =\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) \), for any fixed \(\lambda \),
This is statement 2.
The trace of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) tends to \(+\infty \) as \(x\rightarrow x_i^*-\) because
Since the GEs are the eigenvalues of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \) for \(x\in \bigcup _{i'=1}^{G-1}\left( x_{i'+1}^*,x_{i'}^*\right) \), the sum of all GEs is the trace of \(\widehat{{\widetilde{H}}}\left( x\right) A^{-1}\left( x\right) \), thereby tending to \(+\infty \) as \(x\rightarrow x_i^*-\). By statement 2, GEs other than \(\lambda _{\infty }\left( x\right) \) have finite limits as \(x\rightarrow x_i^*\). So \(\lim _{x\rightarrow x_i^*-}\lambda _{\infty }\left( x\right) =+\infty \). Similarly, \(\lim _{x\rightarrow x_i^*+}\lambda _{\infty }\left( x\right) =-\infty \). Because \(m[A(x_i^*)],n[A(x_i^*)]>0\), some diagonal element of \(\widehat{{\widetilde{H}}}\left( x\right) -\lambda _{\infty }\left( x\right) A\left( x\right) \) tends to \(-\infty \) as \(x\rightarrow x_i^*\). By the Gershgorin circle theorem (Gershgorin 1931), \(\lim _{x\rightarrow x_i^*}r\left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda _{\infty }\left( x\right) A\left( x\right) \right] =-\infty \). Thus, \(\mu \left( x\right) \ne \lambda _{\infty }\left( x\right) \) for x near \(x_i^*\). By statement 2, the roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) except \(\lambda _\infty \left( x\right) \) tend to the \(G-1\) roots of \(\det \left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] \) as \(x\rightarrow x_i^*\), thereby continuous at \(x_i^*\) and bounded near \(x_i^*\); thus, \(-\infty<\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \le \varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) <+\infty \). Let \(\lim _{j\rightarrow +\infty }y_j=x_i^*\) be a sequence such that
For j large enough, \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( y_j\right) >0\) because \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }{|x-x_i^*|}=+\infty \). By Lemma A7,
As a result,
Thus, \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \) is a DGE of \({\mathbb {H}}_i\) on \(A\left( x_i^*\right) \), and so is \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \). Prove by contradiction that \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) =\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \). Otherwise, \(\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \) and \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \) are the only two DGEs of \({\mathbb {H}}_i\) on \(A\left( x_i^*\right) \) by Corollary A1, and one must be 0 by Theorem A1. Assume without loss of generality that \(0=\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) >\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) \). Let \({\bar{\mu }}:=\varliminf _{x\rightarrow x_i^*}\mu \left( x\right) /2<0\). By Corollary A1, \(r\left[ {\mathbb {H}}_i-{\bar{\mu }} A\left( x_i^*\right) \right] >0\) since \(\varliminf _{x\rightarrow x_i^*}\mu \left( x\right)<{\bar{\mu }}<\varlimsup _{x\rightarrow x_i^*}\mu \left( x\right) \). For j large enough, \({\bar{\mu }}<\mu \left( y_j\right) \). Then
by Corollary A1 since \({\bar{\mu }}<0\) and \({\bar{\mu }}<\mu (y_j)\). As \(j\rightarrow +\infty \), \(r\left[ {\mathbb {H}}_i-{\bar{\mu }} A\left( x_i^*\right) \right] \le 0\), conflicts. Therefore,
Define \(\mu \left( x_i^*\right) :=\lim _{x\rightarrow x_i^*}\mu \left( x\right) \). Then, \(\mu \left( x\right) \) is continuous at \(x_i^*\).
Now we show that \(\mu \left( x_i^*\right) =\mu _i\). If \(\mu _i=0\), then by Definition A4, 0 is the only DGE of \({\mathbb {H}}_i\) on \(A\left( x_i^*\right) \). Then \(\mu \left( x_i^*\right) =0\) because \(\mu \left( x_i^*\right) \) is a DGE. Otherwise, assume without loss of generality that \(\mu _i>0\). By Theorem A1, \(m\left[ {\mathbb {H}}_i|A\left( x_i^*\right) \right] =m\left[ A\left( x_i^*\right) \right] =i-1\). By continuity, \(\exists \delta >0\) such that for \(x\in \left( x_i^*-\delta ,x_i^*\right) \), the \(i-1\) roots of \(\det \left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] =0\) with positive real parts at \(x_i^*\) still have positive real parts at x. By \(\lim _{x\rightarrow x_i^*-}\lambda _\infty \left( x\right) =+\infty \), it is possible to decrease \(\delta \) such that \(\lambda _\infty \left( x\right) >0\), thereby \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \ge i\) for \(x\in \left( x_i^*-\delta ,x_i^*\right) \). Conversely, for \(x\in \left( x_{i+1}^*,x_i^*\right) \), \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \le m\left[ A\left( x\right) \right] =i\) by Theorem A1. Therefore, \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) =m\left[ A\left( x\right) \right] =i\). By Theorem A1, \(\mu \left( x\right) >0\) for \(x\in \left( x_i^*-\delta ,x_i^*\right) \). Prove by contradiction that \(\mu \left( x_i^*\right) =\mu _i\). Otherwise, \(\mu \left( x_i^*\right) =0\). By continuity, the i roots of \(\det \left[ \widehat{{\widetilde{H}}}\left( x\right) -\lambda A\left( x\right) \right] =0\) with positive real parts in \(\left( x_i^*-\delta ,x_i^*\right) \), i.e. the \(i-1\) roots of \(\det \left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] =0\) with positive real parts at \(x_i^*\) and \(\lambda _{\infty }\left( x\right) \), are different from \(\mu \left( x\right) \) for \(x\in \left( x_i^*-\delta ',x_i^*\right) \) with \(0<\delta '<\delta \) small enough. Thus, \(m\left( \widehat{{\widetilde{H}}}|A\right) \left( x\right) \ge i+1>i=m\left[ A\left( x\right) \right] \), conflicts. Thus, we have statement 3. \(\square \)
Similar to Corollary 1, we have Corollary A2.
Corollary A2
Assume \(\widehat{{\widetilde{H}}}\left( x\right) \) is continuous in \(\left( x_G^*,x_1^*\right) \), \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }{|x-x_i^*|}=+\infty \) \(\forall i\in \left[ 2,G-1\right] \), and \(\lim _{x\rightarrow x_i^*}\frac{\widehat{{\widetilde{h}}}_{i,j}\left( x\right) }{\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) }\) exists \(\forall j\ne i\). Let \(\mu \left( x_i^*\right) :=\mu _i\) for \(i\in \left[ 2,G-1\right] \). Then, \(\mu \left( x\right) \) is continuous in \(\left( x_G^*,x_1^*\right) \).
Remark A1
Assume \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) >0\). Then the characteristic polynomial of GEs is nondegenerate. Since \(\left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] I_i\left[ 1/\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \right] ={\mathbb {H}}_i-\lambda A\left( x_i^*\right) \), we have \(\det \left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] =\det \left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] /\widehat{{\widetilde{h}}}_{i,\cdot }\left( x\right) \). By Lemma A7, \(\forall \lambda \), \(r\left[ \widehat{{\widetilde{H}}}\left( x_i^*\right) -\lambda A\left( x_i^*\right) \right] =0\) iff \(r\left[ {\mathbb {H}}_i-\lambda A\left( x_i^*\right) \right] =0\). Thus, GEs and DGEs of \(\widehat{{\widetilde{H}}}\left( x_i^*\right) \) on \(A\left( x_i^*\right) \) are the same as those of \({\mathbb {H}}_i\) on \(A\left( x_i^*\right) \). So \(\mu \left( \widehat{{\widetilde{H}}}|A\right) \left( x_i^*\right) =\mu \left[ {\mathbb {H}}_i|A\left( x_i^*\right) \right] =\mu _i\). In summary, the statements in Theorem A3 are equivalent to those in Theorem 1 for \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) >0\). Since \(\widehat{{\widetilde{h}}}_{i,\cdot }\left( x_i^*\right) >0\) for irreducible \(\widehat{{\widetilde{H}}}\left( x_i^*\right) \), Theorem A3 implies Theorem 1.
Rights and permissions
About this article
Cite this article
Li, J., Ge, H. & Zhang, Y. Fluctuating-rate model with multiple gene states. J. Math. Biol. 81, 1099–1141 (2020). https://doi.org/10.1007/s00285-020-01538-2
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-020-01538-2
Keywords
- Fluctuating-rate model
- Nonequilibrium landscape function
- Transition rates
- Dominant generalised eigenvalue