Abstract
Doctor utilizes various kinds of clinical technologies like MRI, endoscopy, CT scan, etc., to identify patient’s deformity during the review time. Among set of clinical technologies, wireless capsule endoscopy (WCE) is an advanced procedures used for digestive track malformation. During this complete process, more than 57,000 frames are captured and doctors need to examine a complete video frame by frame which is a tedious task even for an experienced gastrologist. In this article, a novel computerized automated method is proposed for the classification of abdominal infections of gastrointestinal track from WCE images. Three core steps of the suggested system belong to the category of segmentation, deep features extraction and fusion followed by robust features selection. The ulcer abnormalities from WCE videos are initially extracted through a proposed color features based low level and high-level saliency (CFbLHS) estimation method. Later, DenseNet CNN model is utilized and through transfer learning (TL) features are computed prior to feature optimization using Kapur’s entropy. A parallel fusion methodology is opted for the selection of maximum feature value (PMFV). For feature selection, Tsallis entropy is calculated later sorted into descending order. Finally, top 50% high ranked features are selected for classification using multilayered feedforward neural network classifier for recognition. Simulation is performed on collected WCE dataset and achieved maximum accuracy of 99.5% in 21.15 s.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Amongst various types, colorectal cancer occurs more frequently both in men and women. In 2015 only, approximately 132, 000 new cases of colorectal cancer are registered in USA [1]. Since 2017, on average 135,430 new gastrointestinal (GIT) infections occurred only in USA, which mostly include ulcer, bleeding, and polyps - as a most common neoplasm. According to statistics, 1.6 M people face hurting bowel infection and approximately 200,000 new cases appear each year. These GIT infections can be controlled and even cured if diagnosed at an early stage [2]. Recently, doctors utilize WCE technology [3] but this complete process contains several challenges, existence of irrelevant and redundant information, which makes this detection process more complex, expensive equipment, and time-consuming. These infections are life threatening, therefore it is obligatory to identify and diagnose GIT diseases at an early stage [4].
Recent articles in the area of computer vision (CV) introduced various computerized techniques for the diagnosis of GIT infections using WCE images [5]. In the existing methods, preprocessing step is given much importance due to its significant role in the achieving a high segmentation accuracy. A few of famous segmentation techniques are uniform segmentation [6], normal distribution based segmentation [7], optimized weighted segmentation [8], improved binomial thresholding [9], saliency-based techniques [10], and few more [11]. These segmented images are further used for features extraction for classification of relevant class. The well-known features are point, shape, texture, and color. The color features show significant performance as compared to other types of features in the endoscopy data due to RGB format. But the actual performance of system is depends on the number of selected features. Various features selection techniques are introduced by researchers such as wavelet fractal and automatic correlation [12], Genetic Algorithm based selection (GAS) [13], multi-versus-optimizer (MVO) [14], fisher criterion [15], and name a few more. Recently, deep learning based techniques outperforms in the area of CV and also successfully entered in medical imaging. Through deep learning, features are extracted in a hierarchy of layers in which higher-level features are attained by comprising lower-level convolutional layers. Various pre-trained deep convolutional neural network (DCNN) models are introduced by several researchers in the CV community among which famous ones are AlexNet [16], ResNet [17], VGG [18], GoogleNet [19], and YOLO [20].
Related work
A lot of techniques are introduced in the area of CV and machine learning for the diagnosis of medical diseases such as breast tumor [21], lungs cancer [22, 23], skin cancer [24,25,26,27], blood infections [28], brain tumor from MRI [29,30,31], stomach abnormalities from WCE images [32, 33], to name but a few [34,35,36]. From these diseases, stomach is more important human organ. The prominent stomach infections are ulcer, polyps, and bleeding among which ulcer and bleeding are more vulnerable types. Sivakumar et al. [37] identified bleeding regions in WCE video frames through superpixels segmentation. A CMYK color format is used to detect bleeding area clearly. Later Naive Bayes and superpixel segmentation methods are utilized to create an automatic concealed bleeding perception process. The presented method works efficiently by applying it on several extracted frames of few endoscopic videos.
Yuan et al. [38] presented two-phase fully automated system to identify ulcer through WCE. In the first phase, for segmentation of ulcer aspirants, multilevel superpixel based saliency map extraction method is presented. In the second phase, the acquired saliency vector is merged with image features for doing ulcer image identification process. Saliency max-pooling approach is merged with Locality-constrained Linear Coding (LLC) approach to categorize the images into relevant category. The acquired results show the effectiveness of presented approach with 92.65% accuracy and 94.12% sensitivity rates. Charfi et al. [39] suggested a two-phase technique for ulcer detection in WCE videos. For better depiction of WCE images, the method contains incorporation of texture feature extraction phase that involves Complete Local Binary Pattern (CLBP) and a color feature extraction phase that involves Global Local Oriented Edge Magnitude Pattern (GLOEMP). The presented method is experimented on WCE videos which includes both normal and irregular frames. The obtained results show the effectiveness of suggested technique with 94.07% accuracy.
Suman et al. [40] proposed an automated color feature-based method of bleeding identification from WCE frames. The presented system uses statistical color feature examination and for classification, SVM classifier is utilized. The test results illustrate the effectiveness of proposed approach by giving higher accuracy as compared to existing methods. Sainju et al. [41] introduced a supervised learning technique for computerized identification of bleeding areas from WCE video frames. The presented approach describes the image areas through statistical measures taken through first-order histogram probabilities of three channels of RGB color space. Then a semi-automatic area illustration technique is presented to train data proficiently. Extracted features are examined thoroughly to identify the best feature set and during this process, the segmentation technique is applied to extract areas from images. In the end, neural network is employed for final detection. The suggested method gives reasonable better recognition results by classifying bleeding and non-bleeding areas in WCE images. Zhang et al. [42] showed an infinite curriculum learning approach for categorizing WCE frames with respect to the occurrence of gastric ulcers. A method is designed to efficiently evaluate the intricacy of each sample through its sample size of the patch. The training time is scheduled although the patch size is increasing gradually till it becomes equivalent to actual image size and the method achieves promising better accuracy rates.
Fan et al. [43] introduced an automated CNN based method which is able to accurately identify minor ulcers and erosions from WCE frames. The AlexNet was trained over a dataset of WCE images to classify lesion and normal tissues. The experimental result shows notable accuracy as compared to recent techniques. Hajabdollahi et al. [44] proposed an automated segmentation technique for bleeding areas in WCE images. In this approach, color channels are picked and classified through ANN. During the training process, NN containing specified weights are used beyond any preprocessing and post-preprocessing. The test results show the efficacy of presented method by giving promising accuracy results.
Xing et al. [45] suggested automatic bleeding frame identification and area segmentation with the superpixel color histogram feature and a subspace K-nearest neighbor classifier. To minimize the execution cost, superpixel segmentation and key frame extraction methods are presented. The introduced 9-D color feature method merges the extracted data taken from HSV and RGB color channels. The test result shows the effectiveness of proposed system in contrast to existing methods with 99% of accuracy. Maghsoudi et al. [46] introduced an approach to detect tumor and bleeding regions in frames and categorize the normal and affected regions. GLCM, statistical, LBP, Law’s features and Gabor filters are applied to extract texture and geometric features. Later, normal and affected areas are discriminated in images through extracted features. The presented method is compared with CNN and it shows the importance of applying wide of features to identify numerous affected regions in WCE images in contrast to CNN which can only be used to train on specific databases. Moreover, CNN’s internal features cannot be persistent to enhance the applications (Table 1).
Problem statement & contributions
The irregularity in WCE images is a challenging task, and even experienced doctors spend on average 2 h for analyzing 55,000 frames of one patient [47]. Additionally, GIT infections are of different shapes, size, color, and texture, which increase the features similarities, and in turn chances of improved classification accuracy decreases. An improved classification system is completely relying upon the good features. Therefore, in this work, major challenge is to extract most principle features because, in medical imaging, the performance of a system relies upon the classification of abnormalities.
In this paper, a novel framework is proposed for stomach abnormalities classification using WCE images through DCNN. Major contributions in this work are:
- 1)
An ulcer segmentation technique is introduced named color features based low-level and high-level saliency (CFbLHS) estimation. The HSV and LAB color transformations are applied in the very first step and SVD features are extracted. Then low level saliency is computed by utilizing these SVD features. Further, high level saliency map is also constructed and combined along low level saliency map to produce an initial map for ulcer detection. Finally, an existing CRF model is employed for refinement of segmented image.
- 2)
The cropped and original images are feed to modified Densenet CNN architecture and perform activation on the average pooling and fully connected layers for deep features extraction.
- 3)
The extracted multi layered DCNN features are optimized through Kapur entropy approach - fused through parallel maximum feature value (PMFV). After fusion, Tsallis entropy features are calculated and sorted into descending order. Finally, top 50% features are selected and feed to MLP for final classification.
- 4)
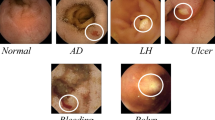
For experimental results, a new WCE dataset of 12,000 images is generated for three stomachs abnormalities such as ulcer, bleeding, and health where each class contains 4000 images.
Proposed work
The proposed automated stomach irregularities detection and recognition system is presented in this section which includes three primary steps such as ulcer detection from ulcer frames, CNN features extraction and features fusion, and best features selection and recognition. The prime structure is shown in Fig. 1 which explains that initially frames are extracted from WCE videos and then separated into ulcer, bleeding, and healthy classes through manual process with the help of an endoscopic expert. Then ulcer is detected through a saliency method. Later, CNN features are extracted through mapped ulcer and original bleeding and healthy frames followed by PMFV based features fusion and KEcTF based best features selection which are classified through MLP. The comprehensive explanation of each step is given below.
Saliency based ulcer detection
As shown in Fig. 1, color features are extracted in the first step through HSV and LAB color transformations using ulcer images. The extracted color features are later concatenated for information fusion. After that, low-level and high-level saliency is estimated by utilizing information of fused image. Finally, a threshold operation is performed to obtain a binary image which is later refined through existing CRF model.
Let φ(i) and φ(j) denote extracted color features through HSV and LAB color transformations, respectively. Two metrics such as variance and SVD are calculated from both transformations for color features which are defined through Eq. (1).
Where, i, j denote rows and columns values of each image, μ is mean value of image, and N denotes total number of pixel values of an input image. Then SVD is calculated through following Eq. (2).
Where, Um × n is m × n orthogonal column matrix over eigenvectors \( \overset{\check{} }{\mathrm{S}}{\left(\overset{\check{} }{\mathrm{S}}\right)}^{\mathrm{T}} \) and Vn × n is n × n matrix over eigenvectors \( {\left(\overset{\check{} }{\mathrm{S}}\right)}^{\mathrm{T}}\overset{\check{} }{\mathrm{S}} \) which are defined as follows in Eqs. (3) and (4).
Where, d is positive real value matrix recognized as singular values matrix and formulated as in Eq. (5).
Where, i ∈ 1, 2, 3, …n and \( \overrightarrow{\upalpha_{\mathrm{i}}} \) is a column vector. Hence, the final SVD is computed as below in Eq. (6).
The dimension of resultant SVD feature vector \( \overset{\check{} }{\mathrm{S}\ } \) is N × M, where M denotes the number of columns. In this work, a fixed dimension of 256 × 256 is used, therefore the resultant vector output must be1 × 256 for each channel and as HSV and LAB transformations consist of total 6 channels, therefore the resulted fused vector is 1 × (1536 + 6) = 1542 and defined through Eq. (7).
Where, Fv(Sij) represents the concatenated information image and Sim, Snj ∈ φ(i), φ(j), respectively. The visual output after concatenation of these features is shown in Fig. 2.
Later, low level saliency is computed by employing \( {\overset{\check{} }{\mathrm{S}}}_{\mathrm{i}} \) as follows in Eq. (8).
Where, W(i, j) shows the weighted factor of abnormal regions in the concatenated image which is defined through infection size and spatial distance of infected region. G and B denote the abnormal regions and distance of abnormal region and border regions, respectively. W(i, j) is computed by Eq. (9).
Where, Sci and Scjrepresent the spatial center position of FV(Sij), α is a static parameter of value 0.2, and L is a diagonal length of concatenated image. The effects of low level saliency are shown in Fig. 3.
After that, a hig-hlevel saliency (HLS) is estimated from two essential information as (a) complex background, and (b) presence of abnormal regions at center point of the image. HLS is most essential for complex background and low and small abnormal regions. Based on these two points, location based object prior (OP) for each region of Sij are computed as given in Eq. (10).
Where, Sd denotes the spatial Euclidean distance, ΔBij explains the number of border pixels, ΔBmaxshows the maximum number of image pixels on border regions, and β is a static parameter which scales the location of abnormal regions in the image. Finally, λOP(Sij) is utilized into Eq. (10) and final HLS is estimated as follows in Eq. (11).
Where, Nχ2max represents the maximum of Chi-square distance (χ2(i, j)) between abnormal and healthy regions. Finally, HLS map FS(λOP) and lowlevel map are simply combined and Otsu thresholding operation is performed whose effects are shown in Fig. 4.
Later, the resultant segmented images are refined through an existing conditional random field (CRF) [48] approach. The CRF approach removes the boundary and small unwanted pixels through following energy function given in Eq. (12).
Where, xi, xjshow the label assignment of all abnormal pixels and P(FS(λi)) is a probability value of ith pixels along θij(xi, xj) which is formulated as in Eq. (13).
The final ulcer segmentation results after CRF refinement are shown in Fig. 5.
Deep CNN features
CNN has become imperative machine learning approach from last few years and showing successful achievement for object classification. Through CNN, various new research problems have risen such as passing an input image to a network and after crossing all layers, it can be dissolved. Recently, several researchers try to resolve this kind of problems and introduced various CNN models among which few are AlexNet, VGG, ResNet, Inception, and Yolo. Through these models, they achieved significant accuracy. In this work, an existing CNN model named as DenseNet [49] is utilized which has simple connectivity patterns and gives the best learning among hidden layers. The connection between each layer is done through a feedforward approach. In this model, as compared to existing ones, less number of parameters is required and redundant features are avoided to learn. Major advantage is that each layer has straight access to gradients from the loss function.
DenseNet architecture is trained on 4 large publicly available datasets including CIFAR-10, 100, SVHN, and ImageNet. A total of 709 layers are involved in this network containing convolutional layers, depth concatenation layers, ReLu layers, batch normalization layers, average pooling layers, fully connected layers (FC), and output function called softmax. The basic flow of DenseNet architecture with 3Dense blocks is shown in Fig. 6.
Six steps are performed for implementation of DenseNet architecture including ResNets implementation, dense connectivity, composite function, pooling layers, growth rate, and bottleneck layers. AP layer and FC layer for deep CNN features extraction are applied in this work. As shown in Fig. 1, mapped ulcer segmented and RGB WCE images are utilized for features extraction through transfer learning [50]. The parameters involved in this process are given below in Table 2 which includes input size, pooling size, stride, padding mode, weights, learning factor, and bias learning factor. The softmax is used as activation function and feature vectors of dimensions N × 1920 and N × 1000are extracted for AP layer and FC layer, respectively. Later, best features are selected and fused through PMFV approach for recognition accuracy and minimum computational time.
Features selection and fusion
In machine learning, various types of features are utilized for recognition of objects into their relevant class [51]. However, the all extracted features are not important and many of them are irrelevant for current classification problem. These unimportant features deteriorate the classification performance and also increases the overall computation time. To motivate with these challenges, a new deep CNN features selection approach named as Kapur Entropy controlled Tsallis Entropy along higher Probability (KEcTaHP) is introduced in this work. The proposed KEcTaHP approach is formulated as follows:
There are two extracted CNN feature vectors such as AP layer vector and FC layer vector denoted by ξi(F1) and ξj(F2) of dimensions N × 1920 and N × 1000, respectively, where N denotes the total number for samples utilized for features extraction in the training and testing process. Let ξk(F3) and ξk1(F4) represent the selected feature vectors of dimensionsN × K1 and N × K2, respectively, where K1, K2 ∈ R. The Kapur entropy [52] is computed in the first step through following Eqs. (14) and (15).
Where, ξE(F1) and ξE(F2) are Kapur’s entropy vectors for ξi(F1) and ξj(F2). N1 and N2 are occurrence levels of features in F1 and F2 vectors. Hi(F1) and Hi(F2) are computed through following Eqs. (16) and (17).
Where, P(.) shows the probability distribution of each feature, WN1c and WN2c denote the probability occurrences for N1and N2 levels, respectively. Later, both entropy vectors ξE(F1) and ξE(F2) are fused through parallel maximum feature value (PMFV) approach. Through PMFV approach, initially features are combined through Eqs. (18) to (20) as given below.
Where, ξfus(ij) denote fused entropy feature vector of dimension N × 1920. Later, discrete probability distribution (DPD) of fused vector is computed and defined as Pi = P0, P1, …PN. Then, by utilizing probabilities values, Tallis entropy is computed through following Eqs. (21) to (23).
Where, SEN(fu(k) representss Tallis entropy vector, q > 0 AND < 1&0 > q < 1 and \( {\mathrm{P}}_{\mathrm{k}}=\sum \limits_{\mathrm{k}=1}^{\mathrm{N}}{\mathrm{P}}_{\mathrm{k}} \) are subject to the following constraints given in Eq. (24).
Finally, Tallis entropy vector (SEN(fu(k)) is sorted into descending order and top 50% features are selected for final recognition process which are fed to multi-layer perceptron (MLP) [53, 54]. The proposed labeled results are shown in Fig. 7. These results are computed after selection of best 50% features.
Results and discussion
The proposed system is validated on a newly generated dataset of WCE images which consists of total 12,000 video frames of three stomach abnormalities. Three stomach abnormalities are ulcer, bleeding, healthy regions and each class includes 4000 frames. The resolution of generated video frames is 760 × 1240. Through proposed features optimization approach, the performance of top 50% features is also compared with 70% and all DCNN features. MLP method of neural network is utilized and its performance is compared with few popular supervised learning classification methods like decision trees (DT) [55], Cubic SVM (CSVM) [56], weighted KNN (WKNN) [57], and ensemble trees (ET) [58]. The performance of selected features is analyzed through prominent parameters such as recall rate, specificity, precision, AUC, false positive rate, accuracy, and classification computational cost. All simulations are conducted on the personal desktop computer of CoreI7, 16 GM of RAM and 8GB graphics card.
Experiment 1: All optimized features
In the first experiment, all optimized features are considered for computation of classification performance. A 70:30 approach is opted which explains that 2800 WCE frames (70%) of each class are utilized for training the system and remaining 12,00 WCE frames (30%) of each class are employed for testing of proposed system. All testing results are held through 10-fold cross validation (10CV). The classification results of all optimal features extracted through multilayers features selection (MLFS) are presented in Table 2. The maximum accuracy of proposed system through MLFS is 97.9%, whereas other methods such as DT, CSVM, WKNN, and ET gained accuracy of 77.2%, 96.4%, 96.3%, and 88.1%, respectively. From results, it is noticed that DT performed worst as compared to other methods and reported an error rate of 22.8%. Additionally, the classification performance is also computed through single FC layer optimal features and shows best accuracy of 95.0% for MLP and worst accuracy of 73.8 using DT. The performance of MLP for both FC and MLFS is also endorsed by confusion matrices (CM’s) in Fig. 8 to represent the sensitivity rate, precision, FPR, specificity, and AUC. In addition, the classification execution time is also computed for all classifiers as presented in Table 3 and also plotted in Fig. 9 which clarifies that proposed system outperforms using MLP.
Experiment 2: 70% optimized features
In this experiment, top 70%, optimal features are selected for analysis of classification performance. 10CV is performed on same 70:30 approach for validation of the proposed system. Major intention in this work is to achieve maximum accuracy in less computation time. To do this, it is essential to provide a low dimensional feature vector which holds only relevant features. The classification results of 70% optimal features are presented in Table 4 for various classifiers which shows best accuracy of 99.3%, recall rate of 99.07%, specificity of 99%, precision rate 99.05%, AUC is 0.993, and FP rate of 0.003, respectively. The accuracy of other classification methods through proposed optimal MLFS such as DT, CSVM, WKNN, and ET is 88%, 98.6%, 98%, and 94.1%, respectively. Additionally, the classification accuracy of the proposed optimal MLFS features is compared with FC layer features and obtained a maximum accuracy of 99.2% through MLP. The accuracy of MLP through FC and MLFS are also verified through confusion matrices, presented in Fig. 10 and clearly demonstrate that minimum features produced good accuracy. Moreover, the analysis of the top 70% features and all features is conducted in terms of classification time as plotted in Fig. 11 which verifies that the reduction in features minimizes the system execution time for all classifiers.
Experiment 3: 50% optimal features
In the third experiment, top 50% features are selected for classification results. 70:30 approach is utilized and 10CV is performed. The results are shown in Table 5 which shows the best accuracy of 99.5% for MLP classifier. The other classifiers such as DT, CSVM, WKNN, and ET also perform well and give sufficient accuracy of 86.3%, 99.3%, 99.1%, and 99.5%, respectively for proposed MLFS approach. The performance analysis of MLP classifier using MLFS is also conducted through a confusion matrix given in Fig. 12 which depicts that each class provides approximately 99% of accuracy.
Additionally, the classification accuracy of FC layer features is also calculated and achieves the highest accuracy of 99.3% for MLP classifier. The other classifiers also give significant classification performance in terms of accuracy which is 84.1%, 99.1%, 98.3%, and 97.9%, respectively. The accuracy of MLP classifier using FC layer features is verified through CM, plotted in Fig. 12. The results presented in Table 5 explain that the selection of 50% optimal features provides better classification accuracy and good execution time as compared to all and 70% selected MLFS approach. The time comparison of each classifier for all MLFS, 70% MLFS, and 50% MLFS features is depicted in Fig. 13 to show that the optimal number of features improves the overall system performance.
Analysis
The analysis of proposed system is directed in terms of overall recognition performance (visual and numerical), change in recognition accuracy after 100 iterations of the proposed algorithm, and finally comparison with few latest techniques. In Fig. 1, overall flow of the proposed system is presented including saliency estimation for ulcer detection, multilayers CNN features extraction, best CNN features selection and fusion, and finally classification through MLP. The initial ulcer detection results are presented in Figs. 2, 3, 4, and 5. Then, CNN features are extracted through DenseNet model whose architecture is shown in Fig. 6. The multilayer features are extracted and best features are selected through the proposed selection approach and results are validated on newly designed WCE dataset. The results are analyzed in three different rounds: (a) all multilayers features selection (MLFS) and comparison with FC selected features; (b) 70% MLFS and comparison with FC 70% selected features, and (c) 50% MLFS and comparison with 50 FC selected features.
The results of first round are presented in Table 3 and confirmed by CM’s in Fig. 8. The bestachieved recognition accuracy of all MLFS approach is 97.9% whereas the FC selected features reached 95.0%. In Table 4, the results of second round are presented and achieved the best accuracy of 99.3% on MLP classifier through proposed MLFS approach. The results are also confirmed by CM’s given in Fig. 10. In the final round, 50% features are selected through MLFS approach as presented in Table 5 and reached the best accuracy of 99.5% which is also affirmed by CM’s in Fig. 12. In addition, the recognition time for all classifiers is also computed and plotted in Figs. 9, 11, and 13 which explains that the less and useful number of features gives the best accuracy and minimizes the overall system execution time.
Moreover, the iterations based comparison of proposed system is conducted with FC selected features as shown in Fig. 14 in which system is iterated up to 100 times and very minium change is noted which confirms the authenticity of proposed system.
In the last, the comparison of proposed system is also conducted with latest existing techniques as presented in Table 6 where the latest existing accuracy is 98.49% achieved by Amna et al. [5] in 2018. They classify three of stomach infection classes including ulcer, bleeding, and healthy but with very low computational time of 17.193 s on only 448 WCE frames (255 healthy, 119 bleeding and 68 ulcer), whereas in the presented work, same type of 12,000 WCE images are used and reached maximum accuracy of 99.50% and computational time of 21.15 s which is clearly good as compared to existing ones.
Conclusion
Ulcer and bleeding are the most frequently occurring deformities of the human digestive tract. An ulcer is more common GI tract infection and approximately 10% of people in the entire world are suffering from it. In this article, a new automated system is proposed using the best CNN feature selection. The ulcer regions are segmented through CFbLHS approach and then compute CNN features. From them, the best features are selected and provides to MLP for classification. The experiments are performed on selected Private Dataset and achieve maximum accuracy of 99.5%, recall rate 99.40%, specificity 99.20%, and computation time 21.15 s. From overall system results, we conclude that the segmentation process helps in to extract the useful features of the important region. The proposed method also shows that the fusion process increase in the classification performance but on the other end, this step increase the computational time due to more number of features. This kind of problem is resolved through the selection process which minimized the computational time along with consistent accuracy. In the future, the segmentation of ulcer regions is performed through mask RCNN and then extracts features. Moreover, the addition of a few latest performance measures is also computed for more precise analysis.
References
Siegel, R. L., Miller, K. D., and Jemal, A., Cancer statistics, 2017. CA Cancer J. Clin. 67:7–30, 2017.
Fu, Y., Zhang, W., Mandal, M., and Meng, M. Q.-H., Computer-aided bleeding detection in WCE video. IEEE journal of biomedical and health informatics 18:636–642, 2014.
Iddan, G., Meron, G., Glukhovsky, A., and Swain, P., Wireless capsule endoscopy. Nature 405:417, 2000.
Mergener, K., Update on the use of capsule endoscopy. Gastroenterol. Hepatol. 4:107, 2008.
Liaqat, A., Khan, M. A., Shah, J. H., Sharif, M., Yasmin, M., and Fernandes, A. S. L., Automated ulcer and bleeding classification from Wce images using multiple features fusion and selection. Journal of Mechanics in Medicine and Biology 18:1850038, 2018.
Nasir, M., Attique Khan, M., Sharif, M., Lali, I. U., Saba, T., and Iqbal, T., An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microsc. Res. Tech. 81:528–543, 2018.
Khan, M. A., Akram, T., Sharif, M., Shahzad, A., Aurangzeb, K., Alhussein, M. et al., An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification. BMC Cancer 18:638, 2018.
Sharif, M., Khan, M. A., Iqbal, Z., Azam, M. F., Lali, M. I. U., and Javed, M. Y., Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Comput. Electron. Agric. 150:220–234, 2018.
Sharif, M., Tanvir, U., Munir, E. U., Khan, M. A., and Yasmin, M., Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J. Ambient. Intell. Humaniz. Comput.:1–20, 2018.
Akram, T., Khan, M. A., Sharif, M., and Yasmin, M., Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J. Ambient. Intell. Humaniz. Comput.:1–20, 2018.
Nur, N., and Tjandrasa, H., Exudate segmentation in retinal images of diabetic retinopathy using saliency method based on region. In: Journal of Physics: Conference Series, 2018, 012110.
Chatterjee, S., Dey, D., and Munshi, S., Optimal selection of features using wavelet fractal descriptors and automatic correlation bias reduction for classifying skin lesions. Biomedical signal processing and control 40:252–262, 2018.
Sharif, M., Khan, M. A., Faisal, M., Yasmin, M., and Fernandes, S. L., A framework for offline signature verification system: Best features selection approach. Pattern Recogn. Lett., 2018.
Faris, H., Hassonah, M. A., Ala’M, A.-Z., Mirjalili, S., and Aljarah, I., A multi-verse optimizer approach for feature selection and optimizing SVM parameters based on a robust system architecture. Neural Comput. & Applic. 30:2355–2369, 2018.
Kaur, T., Saini, B. S., and Gupta, S., A novel feature selection method for brain tumor MR image classification based on the fisher criterion and parameter-free bat optimization. Neural Comput. & Applic. 29:193–206, 2018.
Iandola, F. N., Han, S., Moskewicz, M. W., Ashraf, K., Dally, W. J., and Keutzer, K., Squeezenet: Alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv preprint arXiv:1602.07360, 2016.
He, K., Zhang, X., Ren, S., and Sun, J., Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, 770–778.
Simonyan, K., and Zisserman, A., Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D. et al., Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, 1–9.
Redmon, J., Divvala, S., Girshick, R., and Farhadi, A., You only look once: Unified, real-time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, 779–788.
Fernandes, S. L., Rajinikanth, V., and Kadry, S., A hybrid framework to evaluate breast abnormality using infrared thermal images. IEEE Consumer Electronics Magazine 8:31–36, 2019.
Fernandes, S. L., Gurupur, V. P., Lin, H., and Martis, R. J., A novel fusion approach for early lung cancer detection using computer aided diagnosis techniques. Journal of Medical Imaging and Health Informatics 7:1841–1850, 2017.
Khan, S. A., Nazir, M., Khan, M. A., Saba, T., Javed, K., Rehman, A. et al., Lungs nodule detection framework from computed tomography images using support vector machine. Microsc. Res. Tech. 82:1256–1266, 2019.
Saba, T., Khan, M. A., Rehman, A., and Marie-Sainte, S. L., Region extraction and classification of skin cancer: A heterogeneous framework of deep CNN features fusion and reduction. J. Med. Syst. 43:289, 2019.
Khan, M. A., Akram, T., Sharif, M., Saba, T., Javed, K., Lali, I. U. et al., Construction of saliency map and hybrid set of features for efficient segmentation and classification of skin lesion. Microsc. Res. Tech. 82:741–763, 2019.
Afza, F., Khan, M. A., Sharif, M., and Rehman, A., Microscopic skin laceration segmentation and classification: A framework of statistical normal distribution and optimal feature selection. Microsc. Res. Tech. 82:1471–1488, 2019.
Rajinikanth, V., Madhavaraja, N., Satapathy, S. C., and Fernandes, S. L., Otsu's multi-thresholding and active contour snake model to segment dermoscopy images. Journal of Medical Imaging and Health Informatics 7:1837–1840, 2017.
Naz, I., Muhammad, N., Yasmin, M., Sharif, M., Shah, J. H., and Fernandes, S. L., Robust discrimination of leukocytes protuberant types for early diagnosis of leukemia. Journal of Mechanics in Medicine and Biology 19:1950055, 2019.
Fernandes, S. L., Tanik, U. J., Rajinikanth, V., and Karthik, K. A., A reliable framework for accurate brain image examination and treatment planning based on early diagnosis support for clinicians. Neural Comput. & Applic.:1–12, 2019.
Amin, J., Sharif, M., Yasmin, M., and Fernandes, S. L., Big data analysis for brain tumor detection: Deep convolutional neural networks. Futur. Gener. Comput. Syst. 87:290–297, 2018.
Khan, M. A., Lali, I. U., Rehman, A., Ishaq, M., Sharif, M., Saba, T. et al., Brain tumor detection and classification: A framework of marker-based watershed algorithm and multilevel priority features selection. Microsc. Res. Tech. 82:909–922, 2019.
Khan, M. A., Rashid, M., Sharif, M., Javed, K., and Akram, T., Classification of gastrointestinal diseases of stomach from WCE using improved saliency-based method and discriminant features selection. Multimed. Tools Appl.:1–28, 2019.
Sharif, M., Attique Khan, M., Rashid, M., Yasmin, M., Afza, F., and Tanik, U. J., Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images. Journal of Experimental & Theoretical Artificial Intelligence:1–23, 2019.
Acharya, U. R., Fernandes, S. L., WeiKoh, J. E., Ciaccio, E. J., Fabell, M. K. M., Tanik, U. J. et al., Automated detection of Alzheimer’s disease using brain MRI images–a study with various feature extraction techniques. J. Med. Syst. 43:302, 2019.
Rajinikanth, V., Thanaraj, K. P., Satapathy, S. C., Fernandes, S. L., and Dey, N., Shannon’s entropy and watershed algorithm based technique to inspect ischemic stroke wound. In: Smart Intelligent Computing and Applications. Springer, 2019, 23–31.
Khan, M. A., Javed, M. Y., Sharif, M., Saba, T., and Rehman, A., Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In: 2019 International Conference on Computer and Information Sciences (ICCIS), 2019, 1–7.
Sivakumar, P., and Kumar, B. M., A novel method to detect bleeding frame and region in wireless capsule endoscopy video. Clust. Comput.:1–7, 2018.
Yuan, Y., Wang, J., Li, B., and Meng, M. Q.-H., Saliency based ulcer detection for wireless capsule endoscopy diagnosis. IEEE Trans. Med. Imaging 34:2046–2057, 2015.
Charfi, S., and El Ansari, M., Computer-aided diagnosis system for ulcer detection in wireless capsule endoscopy videos. In: 2017 International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), 2017, 1–5.
Suman, S., Malik, A. S., Pogorelov, K., Riegler, M., Ho, S. H., Hilmi, I. et al., Detection and classification of bleeding region in WCE images using color feature. In: Proceedings of the 15th International Workshop on Content-Based Multimedia Indexing, 2017, 17.
Sainju, S., Bui, F. M., and Wahid, K. A., Automated bleeding detection in capsule endoscopy videos using statistical features and region growing. J. Med. Syst. 38:25, 2014.
Zhang, X., Zhao, S., and Xie, L., Infinite curriculum learning for efficiently detecting gastric ulcers in WCE images. arXiv preprint arXiv:1809.02371, 2018.
Fan, S., Xu, L., Fan, Y., Wei, K., and Li, L., Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys. Med. Biol. 63:165001, 2018.
Hajabdollahi, M., Esfandiarpoor, R., Soroushmehr, S., Karimi, N., Samavi, S., and Najarian, K., Segmentation of bleeding regions in wireless capsule endoscopy images an approach for inside capsule video summarization. arXiv preprint arXiv:1802.07788, 2018.
Xing, X., Jia, X., and Meng, M.-H., Bleeding detection in wireless capsule endoscopy image video using Superpixel-color histogram and a subspace KNN classifier. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2018, 1–4.
Maghsoudi, O. H., and Alizadeh, M., Feature based framework to detect diseases, tumor, and bleeding in wireless capsule endoscopy. arXiv preprint arXiv:1802.02232, 2018.
Koulaouzidis, A., Iakovidis, D. K., Karargyris, A., and Plevris, J. N., Optimizing lesion detection in small-bowel capsule endoscopy: From present problems to future solutions. Expert review of gastroenterology & hepatology 9:217–235, 2015.
Fulkerson, B., Vedaldi, A., and Soatto, S., Class segmentation and object localization with superpixel neighborhoods. In: Computer Vision, 2009 IEEE 12th International Conference on, 2009, 670–677.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger, K. Q., Densely connected convolutional networks. In: CVPR, 2017, 3.
Yin, X., Yu, X., Sohn, K., Liu, X., and Chandraker, M., Feature transfer learning for deep face recognition with long-tail data. arXiv preprint arXiv:1803.09014, 2018.
Rashid, M., Khan, M. A., Sharif, M., Raza, M., Sarfraz, M. M., and Afza, F., Object detection and classification: A joint selection and fusion strategy of deep convolutional neural network and SIFT point features. Multimed. Tools Appl. 78:15751–15777, 2019.
Raja, N., Rajinikanth, V., Fernandes, S. L., and Satapathy, S. C., Segmentation of breast thermal images using Kapur's entropy and hidden Markov random field. Journal of Medical Imaging and Health Informatics 7:1825–1829, 2017.
Heidari, A. A., Faris, H., Aljarah, I., and Mirjalili, S., An efficient hybrid multilayer perceptron neural network with grasshopper optimization. Soft. Comput.:1–18, 2018.
Khan, M. A., Akram, T., Sharif, M., Javed, M. Y., Muhammad, N., and Yasmin, M., An implementation of optimized framework for action classification using multilayers neural network on selected fused features. Pattern. Anal. Applic.:1–21, 2018.
Lavanya, D., and Rani, K. U., Performance evaluation of decision tree classifiers on medical datasets. Int. J. Comput. Appl. 26:1–4, 2011.
Khan, M. A., Sharif, M., Javed, M. Y., Akram, T., Yasmin, M., and Saba, T., License number plate recognition system using entropy-based features selection approach with SVM. IET Image Process. 12:200–209, 2017.
Sharmila, R., and Velaga N. R., A weighted k-NN based approach for corridor level travel-time prediction, 2019.
Adeel, A., Khan, M. A., Sharif, M., Azam, F., Umer, T., and Wan, S., Diagnosis and recognition of grape leaf diseases: An automated system based on a novel saliency approach and canonical correlation analysis based multiple features fusion. Sustainable Computing: Informatics and Systems, 2019. https://doi.org/10.1016/j.suscom.2019.08.002.
Suman, S., Hussin, F. A., Malik, A. S., Ho, S. H., Hilmi, I., Leow, A. H.-R. et al., Feature selection and classification of ulcerated lesions using statistical analysis for WCE images. Appl. Sci. 7:1097, 2017.
Kundu, A., Bhattacharjee, A., Fattah, S., and Shahnaz, C., An automatic ulcer detection scheme using histogram in YIQ domain from wireless capsule endoscopy images. In: Region 10 Conference, TENCON 2017-2017 IEEE, 2017, 1300–1303.
Yuan, Y., Li, B., and Meng, M. Q.-H., WCE abnormality detection based on saliency and adaptive locality-constrained linear coding. IEEE Trans. Autom. Sci. Eng. 14:149–159, 2017.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest and contribute equally in this work for results compilation and other technical support.
Ethical approval (for animals)
Not Applicable.
Ethical approval (for human)
The datasets which are used in this work are publically available such as PH2, ISBI 2016 and ISBI 2017.
Informed consent
Not Applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Khan, M.A., Sharif, M., Akram, T. et al. Stomach Deformities Recognition Using Rank-Based Deep Features Selection. J Med Syst 43, 329 (2019). https://doi.org/10.1007/s10916-019-1466-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-019-1466-3