Abstract
In this paper, a computer-aided method is proposed for abnormality detection in Wireless Capsule Endoscopy (WCE) video frames. Common abnormalities in WCE images include ulcers, bleeding, Angiodysplasia, Lymphoid Hyperplasia, and polyp. In this paper, deep features and Hand-crafted features are combined to detect these abnormalities in WCE images. There are not sufficient images available to train deep structures, therefore the ResNet50 pre-trained model is used to extract deep features. Hand-crafted features are associated with color, shape, and texture. We used a novel idea to reveal unexpected color changes in the background due to existing lesions as a color feature set. Histogram of gradient (HOG) and local binary pattern (LBP) were used respectively for shape and texture features. They are extracted from the region of interest (ROI), i.e. suspicious region. The expectation Maximization (EM) algorithm is used to extract more distinct areas in the background as ROI. The expectation Maximization (EM) algorithm is configured in a way that can extract areas with a distinct texture and color as ROI. The EM algorithm is also initialized with a new fast method which leads to an increase in the accuracy of the method. A large number of features are created by the method, so the minimum redundancy maximum relevance approach is used to select a subset of more effective features. These selected features are then fed to a Support Vector Machine for classification. The results show that the proposed approach can detect mentioned abnormalities in WCE frames with the accuracy of 97.82%
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Wireless capsule endoscopy (WCE) is a capsule shape and non-invasive tool that is equipped with a camera to record video from the entire digestive tract [34]. This device has many advantages over other conventional methods. It can access organs like small bowel that are inaccessible with other common methods [27, 45]. It also can take realistic images from body organs compared to other non-invasive devices such as CT-scan.
The recorded video of this technology usually contains more than 50,000 frames [26]. Some frames may contain abnormality, but the abnormalities are present in a few video frames, and also, the size of these abnormalities is usually small compared to the background. Inspecting recorded video to find the symptoms of a disease (abnormality) is a tedious task for physicians because it usually takes more than 1.5 h [18, 39]. These can lead to a notable miss detection rate by the specialist. Therefore, a computer-aided method is essential to detect suspicious frames containing abnormality automatically and suggest them to a physician for further investigation.
Different abnormalities may be present in the WCE images, but the most important abnormalities are bleeding, ulcers, angiodysplasia (AD), polyps, and lymphoid hyperplasia (LH). These lesions sometimes occur in the small intestine. This organ is investigated by WCE very easily, but it is so difficult with other common methods [45]. In this study, we focused on providing a computer-aided method to identify these lesions.
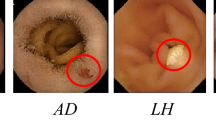
The most common abnormality is bleeding, which is seen as red and brown spots in the image [23]. AD lesions are small vascular malformations of the gut with a cherry-red appearance, and they are one important reason for bleeding in the gastrointestinal tract [25]. Ten percent of the world's population suffers from ulcers. Ulcers can be a symptom of very serious diseases [49]. It usually appears in light-gray or white color and with a spot pattern [37]. LH occurs in the rapidly increasing lymphocytic cells. It has a spherical shape with color of light yellow or light gray [21]. Polyps may be precancerous lesions, which can be seen as protruding from the mucosal wall [6]. In Fig. 1, different WCE images with abnormalities considered in this study are shown.
There are two approaches to designing a computer-aided diagnosis system. Traditional approaches are based on Hand-crafted feature extraction, and recent approaches are based on deep learning methods. There are many traditional approaches for WCE abnormality detection. In our recent works, we also focused on the traditional methods for abnormality detection [4, 6, 7]. Recently, deep learning methods based on the Convolutional Neural Networks (CNNs) have surpassed traditional methods on the medical image processing and diagnosis system. However, lack of large and public dataset with annotation for WCE images is a great challenge in training CNNs for abnormality detection. Indeed, training a deep structure needs a large and well-balanced dataset to optimize a tremendous number of parameters.
Due to this challenge, using pre-trained models and transfer learning is an effective technique to take advantage of deep learning methods. In transfer learning, a CNNs model, that is pre-trained with a dataset with sufficient data from other domains, is used to extract features from real data. In this paper, ResNet50 pre-trained model is used to extract deep features, and also hand-crafted features are extracted. We will show that we can surpass the current WCE classification systems, which focus on one traditional or deep approach.
In the proposed method, to extract deep features, we used ResNet50 pre-trained structure as the feature extractors, and the features are extracted from the AveragePooling2D layer. For extracting hand-crafted features, firstly, the region of interest (ROI), i.e., the suspicious region is extracted by using Expectation–Maximization (EM) algorithm. The proposed ROI extraction method is able to extract distinct areas in terms of color and texture from the background. Then, suitable features associated with the color, texture, and shape are extracted from the ROI.
To extract the color features, first the more worthy color channels in which the lesions are seen more distinctly are identified, and eight statistical features such as the mean of the pixels from these channels are extracted. Also, the histogram of these channels is used to extract one other color feature type. Uniform Local Binary Pattern (uniform-LBP) [46] has been used to extract texture features. Finally, Histogram of Gradient (HOG) [11] has been used to describe shape of ROI.
To reduce the number of features produced from feature extraction step, a subset of features is selected using the maximum relevance and minimum redundancy (MRMR) algorithm. Finally, to classify images into six classes (normal, image with bleeding, AD, polyp, LH, or ulcer lesion) the selected features are fed to a Support Vector Machine (SVM). The main contributions and novelty of this study are listed below:
-
Introducing a novel method to extract more distinct area in image background as ROI considering the color and texture characteristics.
-
Proposing a new fast segmentation method for WCE image that is used for EM initialization.
-
Combining deep and Hand-crafted features to simultaneously classify several types of WCE images, including bleeding, AD, polyp, LH, ulcer, and normal for the first time.
This paper is organized as follows. The hypothesis and limitations are mentioned in Sect. 2, In Sect. 3, the related works are reviewed. Then, Sect. 4 is devoted to explaining the proposed method. Several experiments are done, and the method is evaluated in Sect. 5. Finally, in Sect. 6, the conclusions of the research and future works are mentioned.
2 Hypothesis and Limitations
WCE abnormality detection system faces several limitations and challenges. The produced images by this technology have less quality with respect to traditional endoscopy or colonoscopy. The high similarity between some WCE frames with different abnormalities complicates the identification process (see Fig. 2). WCE images may also suffer from low brightness, noise, blurriness, and low resolution [29].
The assumption considered to solve the problem is that there is only one type of abnormality in each frame. The abnormalities considered include bleeding, AD, polyp, LH and ulcer.
3 Related Works
Many researchers introduced computer-aided systems for abnormality detection in WCE frames [5, 22, 41, 43]. Various types of abnormalities, including ulcers, LH, polyps, tumors, bleeding, AD, and Crohn's disease, can exist in WCE images. The most of existing researches considered bleeding or ulcer lesions in their study [5, 29, 41]. Few studies existed in the AD or polyp detection in WCE images [45, 46, 49]. According to our research, LH was considered just in our recent work [6]. Whereas, due to the similarity of this lesion to malignant lymphoma, it is very important for doctors to diagnose it.
Bleeding detection was investigated in various studies. In Yuan’s method [50], a bleeding detection method based on color histogram analysis was proposed. In Yuan’s method, colors such as blue, which were rarely seen in WCE images, were not present in the color histogram. For this purpose, the colors in the images were divided into k clusters using the k-means algorithm, and the centers of these clusters were considered as the words in the histogram. Then, each pixel was mapped to the nearest word and the words with the number of pixels assigned to them, formed a histogram. The produced histogram was considered as the feature vector, and SVM algorithm classified WCE images into bleeding or normal ones. This method could only be used to identify lesions with a distinct color from the background.
Caroppo et al. [8] introduced a deep transfer learning method for bleeding detection. In the introduced method the features were extracted from three CNN models, including ResNet50, InceptionV3, and VGG19. Then, a supervised machine learning method classified features into normal and bleeding classes. Another bleeding detection method was introduced by Hajabdollahi et al. in Ref. [20]. In this method, at first, the informative components in different color spaces were recognized. Then, a simpified structure of CNN was used for the detection process. The simplification was done by using simultaneous quantization and pruning methods. In the result section, we compare our method with these three bleeding detection methods. In [33] also a deep transfer learning method was used to detect bleeding frames in WCE images. In this method in Xception pre-trained model the fully connected layer was removed and it was replaced with layers that is compatible with the number of existed classes. Then network is learned with a faster learning rate in the new layers and very slow learning rate in the remaining layers.
Different computer-aided methods investigated AD lesions. Deeba et al. [15] introduced a saliency map-based method to detect AD lesions. The saliency map combine a color distinctness map and a pattern distinctness map. The color map was the logarithmic ratio of the red component with respect to the green component in RGB color mode. The pattern map also was acquired by computing the distance of all overlapping patches with the average patches in the image. The accuracy of this method was considerable on a dataset with 3602 images, but this method just could be used for lesions like AD that has red color. While our method is capable to detect lesions with different colors.
In Fonseca et al. [17] method also transfer learning method was used to classify WCE into normal or abnormal classes containing three different abnormalities (Angiectasia, Blood-Fresh, and Polyp). In this method, the features vector was extracted from the deepest layer of the pre-trained CNN model and then the focal loss function was used for binary classification. In our recent work [7], we introduced a method for bleeding and AD lesion detection. In the method, firstly, the ROI was extracted by EM algorithm, then, color features based on histogram and statistical properties were extracted from ROI, finally, multilayer perceptron was used to classify WCE images. In this work, we were limited to detecting red lesions. The present study has been developed with respect to our recent work [7] in several aspects in order to detect lesions with different colors and textures. EM algorithm is developed to extract lesions with distinct colors and textures and is not limited to red lesions. Also, we proposed a fast method for EM initialization. Furthermore, we extend the color histogram-based features that reveal unexpected red color changes in background to revels the color change of the lesions that is used in present study.
4 Proposed Method
In this research, a novel method is proposed to detect the most common abnormalities in WCE images, including ulcer, bleeding, AD, LH, and polyp. The main steps of the proposed method are shown in Fig. 3. Each of these steps will be described below.
4.1 Preprocessing
Some information is reported in the boundary area of WCE images. In the preprocessing step, a circular mask is applied to the image for eliminating these texts that can affect the detection process. In Fig. 4, the process of eliminating these texts in a frame margin is shown. In the considered mask, each pixel inside the circle has a value of one, and outside the circle has a value of zero. This mask is multiplied in the original image. As a result, the values inside the circle will be the values of the original image, and the values outside the circle will be zero.
4.2 Extract Hand-crafted features
Hand-crafted feature extraction consists of three steps including image enhancement, ROI extraction, and feature extraction.
4.2.1 Image Enhancement
WCE frames usually suffer from low lightening. Therefore, to have a more accurate classification, we use fast efficient algorithm introduced in [47] for the enhancement of low-brightness images. This method, at first, inverts the image as Eq. (1) where I is the low-lighting image and \(P\) is the inverted image. \({I}^{c}(x)\) is also the intensity of the image pixels in one color channel. Then the de-haze algorithm is applied to the inverted image using Eqs. (2) and (3). In Eq. (2) the hazy image is modeled. In this equation,\(O\left(x\right)\) and \(t\left(x\right)\) are respectively the intensity of original object and the amount of light reaches the camera from object. A is also the global atmospheric light.
In the de-hazing algorithm, \(O\left(x\right)\) can be recovered from \(I\left(x\right)\) by estimating \(A\) and \(t(x)\). To estimate A, firstly 100 pixels whose minimum intensities among all three channels of RGB color space are highest in the image are selected. Then among the selected pixels, the pixels that the sum of RGB values is highest is chosen. \(t(x)\) is also estimated by Eq. (3) where \(\theta\) is set to 0.8 and \(\beta \left(x\right)\) is a square block around x with the size of 9. This setting is borrowed from the literature [47]. Finally, the image is inverted again. In Fig. 5, the results of applying this method on two frames are shown.
4.2.2 ROI Extraction
ROI selection is an important step in Hand-crafted feature extraction. The lesion areas are very small relative to the background in most WCE frames. So, extracting features from the full image can lead to low accuracy in detecting lesions. Therefore, one solution is dividing images into patches and extracting features from each patch, which leads to the extraction of a large number of features. Another solution is selecting the ROI and then extracting features from this area. In the proposed method, ROIs are extracted based on the combination of the distinctive characteristics of the texture and color of lesions. In the following, before introducing the ROI extraction method, the texture map used in the ROI extraction method is explained.
4.2.3 Texture map
The texture map is extracted based on the Margolin et al. method introduced in [28]. In the method, the WCE frame is divided into overlapping patches with the size of 3 × 3 pixels, and the average of all patches (average patch) is calculated. Then, the L1 norm (Manhattan distance) of each patch with the average patch in the PCA coordinate is calculated, and the length of this path is considered as the distance of the patch. A patch is more distinctive if its distance is longer than other patches. In the first column of Fig. 6, four WCE images with different lesions are shown, and the related texture maps extracted by this method can be seen in the second column. Finally, a Gaussian filter with a standard deviation of 3 is applied to the extracted texture map to make the lesion area smoother, which helps the segmentation in the next step. In the experimental result section, we will show that the best value for the standard deviation is 3. This value was obtained by trial and error. In the third column of Fig. 6, the blurred texture maps after applying this filter are shown.
It is notable that the heterogeneous patches in an image are just used to calculate PCA more quickly. For selecting heterogeneous patches, at first, the simple linear iterative clustering (SLIC) algorithm is used to divide the image into 200 patches, and then 50% of patches with higher variance are kept.
4.2.4 EM algorithm for image segmentation
Investigation of the distribution of the pixels in the lesion and non-lesion areas shows that they have different distributions in terms of color and texture. This finding is illustrated in Fig. 7. It can be deducted from the figure, the distributions of pixels in all channels of RGB color space and texture map in both areas are near normal distribution but with different parameters (mean and variance). Therefore, to create discrimination between areas with and without lesion, WCE image pixels in the different channels of RGB color space and texture map can be considered in a combination of several multivariable normal distributions.
Therefore, WCE images can be segmented using this finding that the pixel’s distribution of lesion areas is different from other areas. Indeed, the parameters of the different normal distributions in the image must be calculated, and then each pixel can be assigned to the distribution that pixels have the most probability to occur in that distribution. The EM algorithm can solve this problem. Indeed, when we have a training set D=\(\left\{{x}^{1},{x}^{2},\dots ,{x}^{n}\right\}\) without any labels for data, but just we know they are generated by k distinct distributions, the EM algorithm can estimate the parameters of the distributions.
In this algorithm, pixels are data points, and the label of pixel \({x}^{i}\) is defined as \({l}^{i}\). The probability of belonging \({x}^{i}\) to class j \(\left(p\left({l}^{i} =j\right)\right)\) is considered as \({\theta }_{j}\), which \(\theta\) has a multinomial distribution, where \(\theta_j\geq0\;\mathrm{and}\;\sum\nolimits_{j=1}^k\theta_j=1\). We also know that a multivariable normal distribution, with \({\mu }_{j}\) and \({\Sigma }_{j}\) as mean and covariance, is the distribution of data in a class j \(\left(x^i\left|l^i=j\right.\right)\). EM algorithm models data by using Eq. (4) which is the log-likelihood of data.
This equation is estimated with two iterative steps, including E-step and M-step. In the E-step, Eq. (5) is evaluated using the current estimate for the parameters. Equation (5) is a function for the expectation of the log-likelihood, and shows the probability of \({x}^{i}\) belonging to class j. In M-Step in Eqs. (6)-(8), the parameters are updated based on guesses in E-step. After convergence of the algorithm, each pixel belongs to the most probable distribution. Therefore, the segmented image is extracted.
4.2.5 EM algorithm initialization
An important step in the EM algorithm for more accurate and faster convergence is choosing an appropriate initial starting point for the EM algorithm. For this purpose, the image pixels are divided into k segments by using a very fast method and this segmentation is considered as the starting point of the EM algorithm.
For EM initialization, we propose a fast method for ROI extraction. In this method, at first, a joint normal distribution is fitted on the image pixels, which is noted by \(X\sim \mathcal{N}\left(\mu ,\Sigma \right).\) Where X is a three-dimensional vector of image pixels in RGB color space (\(X={\left[R,G,B\right]}^{T}\)). Indeed, the size of X is a \(3\times (m.n)\) matrix, where the image size is \((m\times n)\). The parameters in the joint normal distribution are mean vector (\(\mu\)) and \(3\times 3\) covariance matrix (\(\Sigma\)).
where \(E[.]\) is expected value and covariance is \(Cov[.,.]\). \({\Sigma }_{i,j}\) is the covariance between values of the pixels in component i and j in color space. Then, the probability density function (PDF) of each image’s pixel is calculated by the Eq. (11). In this equation, \(|.|\) is the matrix determinant.
To fast segmentation, at first, the complement of \(f\left(x\right)\) is calculated by \(\overline{f(x)}=1-f(x)\) for all pixels and then image is then divided into k levels using the Otsu multi-level thresholding method [32]. For an image with L gray levels \(\{0, 1, . . . , L-1\}\) the image histogram can be defined as \(\left\{{f}_{0}, {f}_{1}, \dots , {f}_{L-1}\right\}\) where \({f}_{i}\) is the occurrence frequency of level\(i\). In Otsu method image is segmented into k clusters by selecting optimal threshold values from the set\(T = \left\{\left({t}_{1}, {t}_{2}, . . . , {t}_{K}\right)\right|0 < {t}_{1} < {t}_{2} < . . . < {t}_{K}< L\}\). The optimal thresholds are those that maximize the variance between clusters. The best value for k is 5, which is specified in the second experiment in the result section. So, this segmentation is considered as an initial state for EM algorithms. In Fig. 8, initial segmentation for one WCE image with AD lesion is shown.
4.2.6 ROI selection
A segmented image is the output of the EM algorithm. Now, one segment must be selected as the ROI. As mentioned before, in the texture map, the lesion area is more distinctive versus other areas, and this area has a higher value than the background. Therefore, to select the ROI segment, for each segment, the mean value of pixels in the texture map is calculated, and the ROI is the segment with the highest mean value. Finally, in the selected ROI may some small dots or lines exist; hence, blobs with eccentricity > 0.9 (ellipse with eccentricity zero is a circle, while ellipse with eccentricity one is a line segment) and those smaller than 100 pixels are removed to extract final ROI segment. The result of applying the proposed ROI extraction method on WCE images with different lesions is illustrated in Fig. 9.
ROI extraction for WCE images with different lesions. The columns, from left to right, are respectively, (a) the original image, (b) the lesion selected area from the database, (c) the initial segmentation for EM algorithm, (d) the selected segment using the EM algorithm, and (e) the final result of the proposed segmentation method. The rows are image with LH lesion, AD lesion, Ulcer, Bleeding and Polyp
4.2.7 Feature Extraction
In this step, we extract three types of descriptors, including color, texture, and shape to detect different lesions.
The extract color descriptors, at first the more worthy color channels in different color spaces for representing each abnormality is identified by the method that we introduced in [7]. In this method, for each abnormality and for each channel of RGB, LAB, HSV and YCbCr color spaces, we calculate one worthiness measure. To calculate this measure, for one abnormality in a specific channel, 30 percent of existing images with this abnormality in the dataset are selected randomly. For each image the Normalized Cumulative Histogram (NCH) is calculated, and then the lesion area is removed from this image, and the NCH is calculated for this image again. In the next step, the mean of absolute difference (MAD) of the two NCHs related to this image and the corresponding abnormal region deleted image is calculated. Finally, the average of MAD for all selected images is considered as the measure for that channel.
In Table 1, the first two worthy color channels for each abnormality are reported. From the table, the more worthy channels for identifying abnormalities are ‘a’ and ‘b’ in CIELab، ‘R’ in RGB,‘Cr’ and ‘Cb’ in YCbCr and ‘V’ in HSV. In continue, two sets of color features are extracted from these channels.
In the first set, eight parameters, including contrast, mode, mean, median, entropy, variance, minimum and maximum are extracted from the ROI pixels in the introduced six-worthy channels. These parameters have been used in several WCE abnormality detection studies [31]. The number of features that are extracted in this step is 48.
In the second set, we extend our work introduced in [7] to reveal unexpected color changes in the background due to existing lesions. The main idea of this work was that in an image with AD lesion there was a sudden change in the NCH of the ROI+ area (ROI and surrounding areas) in the ‘a’ of CIELab color space. Our previous work can be used for red lesions like AD or bleeding because these lesions are highlighted in the ‘a’ channel,- and in the NCH a sudden change can be seen around the lesion in this color space. In the present work, we extract sudden change from the NCH of ROI + for the introduced six-worthy channels.
To extract sudden change from a specific channel, at first, ROI + is obtained by applying a morphological dilation operator with a 20 × 20 square structuring element on the ROI. Then, the NCH for the ROI + is computed. Finally, the NCH values are quantized in 10 levels and the number of NCH values that exist in each level is the features of the considered channel. Indeed, for each channel 10 features are extracted and totally 60 features are extracted from these channels.
Uniform-LBP is used to extract texture features from ROI (non-ROI areas are set to zero). LBP is a famous method [2, 3] for feature extraction that is used in many WCE abnormality detection methods [10, 30]. In the LBP algorithm, eight-pixels with a radius of one around the pixel are considered as the neighbors. (see Fig. 10). Each pixel is compared with its neighbor along a circle in a clockwise direction and it produces an 8-digit binary number. An 8-bit binary number can have 256 different values. The feature vector in the LBP algorithm is the histogram of these values. In the uniform, LBP method just uniform patterns have a separate bin in the histogram and a single bin is devoted to non-uniform patterns. The uniform patterns have at most two 0–1 or 1–0 transitions in the binary number. Therefore, in uniform-LBP, there are 59 different bins in the histogram that is the length of the feature vector.
To describe the shape of ROIs HOG is used. To extract an equal number of HOG features from the ROI, this region is fitted into a specific square with a size of \(48\times 48\) pixels. The parameters that are used in the HOG algorithm are reported in Table 2. 180 shape features are extracted from this step.
4.3 Extract Feature from Pre-trained Model
In this section, we used pre-trained ResNet50 structure as the feature extractors for abnormality detection in WCE images. The basic idea of Residual Networks (ResNets) [40] is to skip some blocks of convolutional layers by using shortcut connections that form blocks called residual blocks. This structure mitigates the degradation problem existing in deep networks and improves training efficiency. The basic blocks in ResNet50 follow two design rules: 1) The number of filters is same in the layers for the same size of output feature map 2) The number of filters is doubled if the feature map size is halved.
ResNet50 is pre-trained on ImageNet dataset and can be used to extract discriminative features from WCE frames based on the transfer learning theory. Due to the small number of WCE datasets and the limited number of images in them, fine-tuning step is not considered to avoid overfitting [8] and images are directly fed to the architectures and a feature vector of 1 × 2048 is extracted from AveragePooling2D layer of ResNet50.
4.4 Feature selection
Totally 2395 features are extracted from previous steps, including 2048 deep featured and 346 Hand-crafted features. We use maximum relevance and minimum redundancy (MRMR) [16] feature selection method to select a subset of features. This method tries to minimize the redundancy of a feature set and also maximize the relevance of a feature set to the response variable y (the classes). This method ranks all features and returns the indices and scores of them ordered by importance. An important issue is selecting the number of features from the ranked features in the output of the MRMR algorithm.
One important issue is selecting the appropriate size of the feature subset (\(\xi\)). To select this size, we draw the features score graph in descending order, and then the first derivative of a signal is plotted. This graph shows the instantaneous slope of the signal. In this graph, a moving average with a length of 15 is applied to smooth the graph. Finally, in the smoothed graph, \(\xi\) is equal to the first point in the graph where the slope is less than a threshold (T). In Sect. 4, the value of T is estimated by the trial-and-error method.
4.5 Abnormality Detection
The selected features from the previous step are then fed to SVM for classifying WCE images into different classes, including normal, AD, ulcer, polyp, LH, and bleeding. The SVM classification method was used in many medical image classification approaches [14, 38]. The kernel used in the SVM algorithm is a polynomial kernel with degree 2.
5 Experimental Results and Discussion
In this section, at first, the datasets used in this research are introduced. Then, the evaluation metrics are introduced. Then the experiments and results are discussed.
5.1 Datasets
In this research, four publicly available datasets were used to evaluate and compare the proposed method with other existing methods. The Kvasir dataset [36], The Gastrointestinal Image Analysis Challenge 2017 (GIANA 2017) [1], The Red-lesion endoscopy dataset [9], and the KID dataset [24].
Kvasir-Capsule is a publicly available dataset with different labeled frames that was created in 2020 and updated in 2021 [39]. In the Kvasir- capsule dataset, 47238 images with different classes exist. Among the existing class, 10 classes are related to pathological findings (Angiectasia, Blood, Erythematous, Hematin, Erosion, Foreign Bodies, Ulcer, Polyp) or normal classes. Images in this dataset are annotated by medical doctors (experienced endoscopists) and the lesion area is specified within a box. Also, the class of each frame is specified. The size of images in this dataset is \(336\times 336\) pixels. In Table 3, the number of different lesions existing in the dataset is specified. Some image samples from this dataset can be seen in Fig. 11.
The GIANA 2017 contains 1200 WCE frames, including 600 normal and 600 frames with AD lesions. The size of the frames is \(704\times 704\) pixels and the frames are fully annotated by experts. The Red-lesion endoscopy dataset consists of 3895 images including 2325 normal and 1570 bleeding WCE frames. The binary ground truth mask of each frame is also available in the dataset. The size of frames in the dataset is \(320\times 320\) pixels. In the proposed method, we resize all images into \(320\times 320\) pixels. Some frames examples in this dataset are shown in Fig. 12.
The public KID dataset contains two albums with different lesions (kid dataset 1 and kid dataset 2). The images with a size \(360\times 360\) of pixels in this dataset are also fully annotated by experts. The lesions in kid dataset 1 are AD, Apthae, Bleeding, Chylous Cysts, and Lymphangiectasis Nodular. The kid dataset 2 contains Inflammatory, Normal, polypoid, and vascular frames. Figures 13 and 14 show the image samples from the Red-lesion endoscopy dataset and KID dataset, respectively.
We combined these four datasets to evaluate our proposed method. In Table 4, the number of participating frames from each dataset in the final fusion dataset is illustrated.
5.2 Evaluation Criteria
Several metrics are used in this research, which were widely used in medical image segmentation and classification. These metrics are accuracy (AC), precision, recall, false-positive rate (FPR), false-negative rate (FNR) Matthews Correlation Coefficient (MCC), F-measure, intersection over union (IoU) and dice score (DS), which are introduced in Eq. (12) to (20). The metrics are based on four well-known basic metrics, including true positive (TP), false positive (FP), false negative (FN) and true negative (TN).
Note that the values of the metrics are in [0–1] range. All metrics except FPR show a better evaluation when they are closer to one. It is vice versa for the FPR.
5.3 Results
In this section, the proposed method is evaluated and compared with other existing methods in terms of segmentation and classification. Our investigations show that no method exists that detects these six classes together. Hence, the comparisons are with the researches that consider some of these lesions in their methods.
In the first experiment, the ROI extraction method was evaluated. Since on the existing datasets, the AD lesions were annotated accurately in the GIANA 2017 dataset and KID dataset and other methods also reported their result on this lesion. Therefore, we compared our method with other existing AD lesion segmentation methods. At first, we considered an experiment to choose the best value for standard deviation in the texture map extraction step. In our experiment, we considered the GIANA 2017 dataset and the images with AD lesions were segmented by the ROI extraction method and the value of standard deviation was considered 1 to 6 in the texture map extraction step. The average value of DS for all images with AD lesion was calculated for each standard deviation and the best value belonged to σ = 3 as can be seen in Fig. 15.
The best number of distributions (k) in the ROI extraction method was calculated in the next experiment. To achieve this aim, for each value of k in the range of [25, 45] a test was considered. In each test, ROI was extracted and DS was calculated for 30% of frames with accurate annotation (AD lesions and bleeding lesions in GINANA 2017, Red-lesion endoscopy and KID dataset). For each test, the average of DS was calculated. These values were shown in Fig. 16 and can be deducted from the figure, the best value for k is 5.
Vieira et al. introduced two methods (method 1 [43] and method 2 [44]) for AD lesions segmentation and detection-based EM algorithm and hidden Markov model. In Fig. 17 our methods with and without considering initialization were compared with these two methods on the KID dataset. In the boxplot of the proposed method, the value of the first, second and third quartile, minimum and maximum were higher than other methods. Therefore, it can be concluded that the proposed method with initialization could segment AD lesions more accurately than other methods. In addition, the boxplot was more compact compared with other methods, so the proposed method was more reliable. Also, the use of initialization had a positive impact on increasing the accuracy of the proposed method.
The ROI extraction method was also compared on the AD lesions in the GIANA 2017 dataset with Deeba et al. [15] method. The Authors in [15] shared their implementation code in GitHub.Footnote 1 From the results in Fig. 18, it can be concluded, that our proposed method was more accurate in extracting ROI.
Compariton of the ROI extraction method with Deeba’s methods [15]
We also compared our method with two other methods that their results were reported on the GIANA 2017 dataset in the term of DS and IoU in Table 5. In Shvets’s method [35] and Gobpradit’s method [19] the AD lesions were segmented using the deep learning methods. The table showed the superiority of the present method over these two methods.
One experiment was considered to select the appropriate size of features subset (\(\xi\)) in the feature selection step. In this experiment, at first the best value for \(\xi\) was acquired by trial and error. Then, in the smoothed slop graph of scores, the corresponding value of \(\xi\) in the graph was selected as Threshold for all other experiments. In the trial-and-error step, the accuracy of the method versus the different values of \(\xi\) was calculated. These values were plotted in Fig. 19. From the figure, the max value was related to selecting a subset with 900 features. In Fig. 20, the scores of ranked features from the MRMR algorithm for classifying the fusion dataset were plotted. The corresponding smoothed slope graph was also shown in Fig. 21. This graph has a value of 0.0005 at point 900. So, this value is chosen for threshold (T).
The abnormality detection system is tested on the fusion dataset and the results are given in Table 6 and the related confusion matrix is shown in Table 7. The results show the high performance of the proposed method in the detection of different abnormalities.
In the next experiment, we evaluated the impact of different steps of the proposed method containing Hand-crafted feature extraction, deep feature extraction, and feature selection. To investigate each step, it was removed from the proposed methods and the remained steps were tested on the fusion dataset. The results in Table 8 showed that removing each step decreased the performance of the proposed method in all evaluation criteria. Therefore, each step had a positive impact on the proposed method.
We also tested the effectiveness of different steps of Hand-crafted feature extraction similar to the previous experiment. For this purpose, we considered the proposed method without deep features, which contained Hand-crafted features. In this new structure, the impact of removing Image Enhancement and ROI extraction were tested. From the results reported in Table 9, it can be concluded that these steps also had a positive effect on the performance of the proposed method.
In the next experiment, the ResNet50 pre-trained structure is compared with other pre-trained structures. In order to perform this evaluation, the images in the hybrid database are given to the input and the deep features are extracted from the fully connected layer of each considered structure. The results of the classification evaluation of different structures are reported in Table 10. As can be seen, the Resnet50 deep network performed best in all metrics.
We also considered a simple CNN [48] and a deep CNN to compare them with our proposed method. The structure of simple CNN was shown in Table 11. This network was trained with the fusion dataset. The ResNet50 structure was also considered for deep structure. Transfer learning was used to retrain the ResNet50 model with the fusion dataset. To retrain the network, the last layers of the network were replaced to contain the same number of nodes as the number of our classes. Then, we froze the weights of earlier layers in the network by setting the learning rates in those layers to zero. The results in Table 12 convince the superiority of our proposed method.
Since the used fusion dataset was unbalanced, the effectiveness of the method was checked in the next experiment with a balanced dataset. For this purpose, 590 images were randomly selected for each class of normal, bleeding, ulcer, AD and LH. The method was evaluated on these images and the classification results were reported in \* MERGEFORMAT Table 13. The results show that the proposed method performed well for the balanced dataset.
We also evaluated the method on all lesions existing in the Kvasir dataset. The results of classifying 10 existing classes in this dataset were shown in Table 14. The reported results showed that although the method was designed to identify specific lesions, it was not limited to the considered lesions and it was able to identify more lesions. We also compared our results on this dataset with our recent work [6] in Table 15. Our method in [6] was also evaluated on the Kvasir dataset. As can be seen, the proposed method had improved significantly compared to our previous method.
In continue the proposed method were compared with other methods on different datasets. Firstly, our proposed method was compared with four other method including Deeba’s method, Yuan’s method and our two recent works on the GIANA2017 dataset. The reported results in Table 16 showed that the Yuan’s method and Deeba’s method had poor performance but our recent works and the proposed method had high performance on this dataset. Of course, the best performance in all evaluation criteria belonged to the proposed method.
In the next experiment, the proposed method was compared with Caroppo et al. [8] method on the Red-lesion endoscopy dataset. As mentioned in the related work section, they proposed a deep transfer learning method for bleeding detection. They reported the result on the Red-lesion endoscopy dataset. Therefore, we also tested our proposed method on this dataset. From the results reported in Table 17 the superiority of our proposed method in all evaluation criteria was inferred.
The proposed method also was compared with Hajabdollahi et al. method [20]. The results of this method were reported on the fusion of two publicly available datasets. 728 normal, five bleeding from KID dataset 1 and 50 bleeding frames from deeba’s dataset [13]. We also evaluated our methods on this dataset and the results were shown in Table 18. The results showed that our method had better performance compared to this method in all evaluation criteria.
We also compared our method with Fonseca et al. [17] method. In this method, the author used Kvasir capsule dataset for classifying images into normal and abnormal classes. The abnormal class was containing all existing images in polyp, AD and bleeding classes. We also used this dataset to validate and compare our method with the Fonseca et al. method. The results that were shown in Table 19 shows superiority of the proposed method.
Finally, the proposed method was compared with De Maissin et al. [12] method on the.
CrohnIPI dataset [42] contains several types of abnormality caused by Crohn's disease. This dataset includes seven types of abnormalities and three of which are related to ulcer abnormalities. We evaluate our method on all existing abnormalities in this dataset. De Maissin et al. have reported the confusion matrix resulting from their method. We have calculated the evaluation criteria from this matrix for their method. The results of these evaluations are reported in Table 20. As can be seen, the proposed method has performed better in all evaluation criteria except FPR which in abnormality detection in the medical images the most important metric is FNR. Indeed, the loss of an abnormal image is much more important than suggesting a frame without abnormality as a suspicious frame. Table 21 summarized all comparisons done in this study. Indeed, the main research achievements in WCE image processing were shown.
6 Conclusions
In this paper, a new computer aided method was proposed to detect different lesions in WCE images. The proposed method was based on the combination of deep and traditional learning methods. For this purpose, Deep features were extracted from ResNet50 pre-trained model. Due to the lack of a public and large database of endoscopic capsule images, most existing methods have used the traditional learning method to abnormality detection. The Hand-crafted features also extracted in three main step containing image enhancement, ROI extraction and feature extraction from ROI and the ROI extraction is the most important step. But the most of existing methods are limited to extract abnormalities with a specific color or texture, so they could identify few abnormalities. But the results in this research showed that the proposed method has the ability to extract different types of abnormalities with no restrictions on color and texture. The results also showed that using the combination of traditional and deep features increased the performance of the proposed method. In fact, we cloud surpass the current WCE classification systems which focuses on one traditional or deep approaches.
In the Hand-crafted feature extraction steps, we proposed an EM based method to extract ROIs. The EM method was initialized with a new novel fast segmentation method. We also proposed a novel method to extracting sudden changes due to existing lesions with different color in the background. The proposed method was evaluated on our different datasets. The results show the ability of the proposed method on abnormalities detection in WCE images. Also, the proposed method is compared with several existing methods for lesion detection, and in all comparisons, the superiority of the proposed method is concluded.
Data availability
The first dataset analyzed during the current study is available in the GIANA challenge, [https://endovissub2017-giana.grand-challenge.org/].
The second dataset analyzed during the current study is available in the public dataset KID repository, [https://mdss.uth.gr/datasets/endoscopy/kid/].
References
“Gastrointestinal Image Analysis” (2021) [Online]. Available: https://endovissub2017-giana.grand-challenge.org. [Accessed 2021 July 10].
Ajam A, Forghani M, Alyan Nezhadi M, Qazanfari H, Amiri Z (2019) “Content-based Image Retrieval using Color Difference Histogram in Image Textures”, in 5th Iranian Conference on Signal Processing and Intelligent Systems (ICSPIS), IEEE, Shahrood, Iran.
AlyanNezhadi M, Qazanfari H, Ajam A, Amiri Z (2020) Content-based Image Retrieval Considering Color Difference Histogram of Image Texture and Edge Orientation. Int J Eng 33(5):949–958
Amiri Z, Hassanpour H, Beghdadi A (2019) “A Computer-Aided Method to Detect Bleeding Frames in Capsule Endoscopy Images”, in 8th European Workshop on Visual Information Processing (EUVIP) pp. 217–221
Amiri Z, Hassanpour H, Beghdadi A (2019) “Feature Selection for Bleeding Detection in Capsule Endoscopy Images using Genetic Algorithm”, in 5th Iranian Conference on Signal Processing and Intelligent Systems (ICSPIS), IEEE, Shahrood, Iran.
Amiri Z, Hassanpour H, Beghdadi A (2021) “A Computer-Aided Method for Digestive System Abnormality Detection in WCE Images”, J Healthcare Eng, vol. 2021
Amiri Z, Hassanpour H, Beghdadi A (2022) Feature Extraction for Abnormality Detection in Capsule Endoscopy images. Biomed Signal Process Control 71:103219
Caroppo A, Leone A, Siciliano P (2021) Deep Transfer Learning Approaches for Bleeding Detection in Endoscopy Images. Comput Med Imaging Graph 88:101852
Coelho P, Pereira A, Salgado M, Cunha A (2018) “A Deep Learning Approach for Red Lesions Detection in Video Capsule Endoscopies”, in International Conference Image Analysis and Recognition, pp. 553–56
Constantinescu AFIMRI, Ciurea ME, Streba CT, Iovanescu VF, Vere CC (2015) Analysis of Wireless Capsule Endoscopy Images using Local Binary Patterns. Appl Med Inf 36(2):31–42
Dalal N, Triggs B (2005) “Histograms of Oriented Gradients for Human Detection”, in IEEE Computer Society Conference on Computer Vision and Pattern Recognition
de Maissin A, Vallée R, Flamant M, Fondain-Bossiere M, Le Berre C, C. A. and A. Bourreille (2021) Multi-expert Annotation of Crohn’s Disease Images of the Small Bowel for Automatic Detection using a Convolutional Recurrent Attention Neural Network. Endosc Int Open 09(71):1136–1144
Deeba F (n.d.) “Bleeding images and corresponding ground truth of CE images” [Online]. Available: https://sites.google.com/site/farahdeeba073/Research/resources
Deeba F, Islam M, Bui FM, Wahid KA (2018) Performance assessment of a bleeding detection algorithm for endoscopic video based on classifier fusion method and exhaustive feature selection. Biomed Signal Process Control 40(2):415
Deeba F, Mohammed SK, Bui FM, Wahid KA (2018) A Saliency-based Unsupervised Method for Angiectasia Detection in Endoscopic Video Frames. J Med Biol Eng 38(2):325–335
Ding C, P. H (2005) Minimum Redundancy Feature Selection from Microarray Gene Expression Data. J Bioinf Comput Biol 3(2):185–205
Fonseca F, Nunes B, Salgado M, Cunha A (2022) Abnormality Classification in Small Datasets of Capsule Endoscopy Images. Proced Comput Sci 196:469–476
Gobpradit S, Vateekul P (2020) “Angiodysplasia Segmentation on Capsule Endoscopy Images Using AlbuNet with Squeeze-and-Excitation Blocks”, in Intelligent Information and Database Systems (ACIIDS)
Gobpradit S, Vateekul P (2020) Angiodysplasia Segmentation on Capsule Endoscopy Images Using AlbuNet with Squeeze-and-Excitation Blocks in In Asian Conference on Intelligent Information and Database Systems (pp. 283-293). Springer, Cham
Hajabdollahi M, Esfandiarpoor R, Khadivi P, Soroushmehr SR, Karimi N, Najarian K, Samavi S (2019) Segmentation of Bleeding Regions in Wireless Capsule Endoscopy for Detection of Informative Frames. Biomed Signal Process Control 53:101565
Iwamuro M, Hiraoka S, Okada H, Kawai Y, Miyabe Y, Takata K, Yamamoto K (2016) Lymphoid Hyperplasia of the Colon and Its Association with Underlying Allergic Airway Diseases. Int J Colorectal Dis 31(2):313–317
Jia X, Meng Q-HM (2017) “A Study on Automated Segmentation of Blood Regions in Wireless Capsule Endoscopy Images using Fully Convolutional Networks”, in IEEE 14th International Symposium on Biomedical Imaging
Jia X, Cai L, Liu J, Dai W, Meng MQH (2016) “GI Bleeding Detection in Wireless Capsule Endoscopy Images based on Pattern Recognition and a MapReduce Framework”, in IEEE International Conference on Real-time Computing and Robotics, Angkor Wat, Cambodia
Koulaouzidis A, Iakovidis DK, Yung DE, Rondonotti E, Kopylov U, Plevris JN, Toth E, Eliakim A (2017) “KID Project: An Internet-based Digital Video Atlas of Capsule Endoscopy for Research Purposes”, Endosc Int Open, vol. 5, no. 6
Lecleire S, Iwanicki-Caron I, Di-Fiore A, Elie C, Alhameedi R, Ramirez S, Antonietti M (2012) Yield and Impact of Emergency Capsule Enteroscopy, in Severe Obscure-Overt Gastrointestinal Bleeding. Endosc 44(04):337–342
Li ZC, Eliakim R, Zou W, Wu H, Liao Z, Gong Z, Wang J, C. J. W. and S. Y. Song (2014) “The Current Main Types of Capsule Endoscopy”, in Handbook of Capsule Endoscopy. Springer, Dordrecht
Liao C, Wang C, Bai J, Lan L, Wu X (2021) “Deep Learning for Registration of Region of Interest in Consecutive Wireless Capsule Endoscopy Frames”, Comput Methods Programs Biomed, vol. 208
Margolin R, Tal A, Zelnik-Manor L (2013) “What Makes a Patch Distinct?”, in IEEE Conference on Computer Vision and Pattern Recognition (pp. 1139–1146)
Mohammed A, Farup I, Pedersen M, Hovde Ø, YildirimYayilgan S (2018) Stochastic Capsule Endoscopy Image Enhancement. J Imaging 4(6):75
Nawarathna R, Oh J, Muthukudage J, Tavanapong W, Wong J, De Groen PC, Tang SJ (2014) Abnormal Image Detection in Endoscopy Videos using a Filter Bank and Local Binary Patterns. Neurocomputing 144:70–91
Noya F, Alvarez-Gonzalez MA, Benitez R (2017) “Automated Angiodysplasia Detection from Wireless Capsule Endoscopy”, in 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society
Otsu N (1979) A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Saraiva MM, Ribeiro T, Afonso J, Ferreira JP, Cardoso H, Andrade P, Macedo G (2022) Artificial Intelligence and Capsule Endoscopy: Automatic Detection of Small Bowel Blood Content Using a Convolutional Neural Network. GE-Portuguese J Gastroenterol 29(5):331–338
Shahril R, Baharun S, Islam A (2016) Pre-processing Technique for Wireless Capsule Endoscopy Image Enhancement. Int J Electric Comput Eng 6:1617–1626
Shvets AA, Iglovikov VI, Rakhlin A, Kalinin AA (2018) “Angiodysplasia Detection and Localization using Deep Convolutional Neural Networks”, in 2018 17th IEEE International Conference on Machine Learning and Applications (icmla). pp. 612–617
Smedsrud PH, Thambawita V, Hicks SA, Gjestang H, Nedrejord OO, Næss E, Halvorsen P (2020) Kvasir-Capsule, a Video Capsule Endoscopy Dataset. Scientific Data 8(1):1–10
Souaidi M, Abdelouahed AA, El Ansari M (2019) Multi-scale Completed Local Binary Patterns for Ulcer Detection in Wireless Capsule Endoscopy Images. Multimed Tools Appl 78(10):13091–13108
Sundaram PS, Santhiyakumari N (2019) An Enhancement of Computer Aided Approach for Colon Cancer Detection in WCE Images Using ROI Based Color Histogram and SVM2. J Med Syst 43(2):29
Szczypiński P, Klepaczkoa A, P. P. and P. Danielb (2014) Texture and Color based Image Segmentation and Pathology Detection in Capsule Endoscopy Videos. Comput Methods Programs Biomed 113(1):396–411
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) “Inception-v4, Inception-Resnet and the Impact of Residual Connections on Learning”, in Thirty-first AAAI conference on artificial intelligence, San Francisco
Usman M, Satrya G, Usman M, Shin S (2016) “Detection of Small Colon Bleeding in Wireless Capsule Endoscopy Videos”, Computerized Medical Imaging and Graphics, vol. 09, no. 5
Vallée R, De Maissin A, Coutrot A, Mouchère H, Bourreille A, Normand N (2020) “CrohnIPI: An Endoscopic Image Database for the Evaluation of Automatic Crohn's Disease Lesions Recognition Algorithms”, Medical Imaging 2020: Biomed Appl Mole Struct Function Imaging, vol. 11317
Vieira PM, Gonçalves B, Gonçalves CR, Lima CS (2016) “Segmentation of Angiodysplasia Lesions in WCE Images using a MAP Approach with Markov Random Fields”, in IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA
Vieira P, Silva C, Costa D (2019) Automatic Segmentation and Detection of Small Bowel Angioectasias in WCE Images. Ann Biomed Eng 47:1446–1462
Voderholzer WA, Beinhoelzl J, Rogalla P, Murrer S, Schachschal G, Lochs H, Ortner MA (2005) Small Bowel Involvement in Crohn’s disease: a Prospective Comparison of Wireless Capsule Endoscopy and Computed Tomography Enteroclysis. Gut 54(3):369–373
Wang G, Fang C, Ming Y (2015) “Uniform Local Binary Pattern Based Texture-Edge Feature for 3D Human Behavior Recognition”, PLoS One 10(5), vol. 10, no. 5
Xuan D, Guan W, Yi P (2011) “Fast Efficient Algorithm for Enhancement of Low Lighting Video” in IEEE International Conference on Multimedia, Barcelona
Yu JS, Chen J, Xiang ZQ, Zou YX (2015) “A Hybrid Convolutional Neural Networks with Extreme Learning Machine for WCE Image Classification”, in IEEE International Conference on Robotics and Biomimetics (ROBIO) (pp. 1822–1827)
Yuan Y, Wang J, Li B, Meng MQH (2015) “Saliency based Ulcer Detection for Wireless Capsule Endoscopy Diagnosis” IEEE Trans Med Imaging, vol. 34, no. 10
Yuan Y, Li B, Meng Q (2015) Bleeding Frame and Region Detection in the Wireless Capsule Endoscopy Video. IEEE J Biomed Health Inform 20(2):624–630
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Zahra Amiri and Hamid Hassanpour. The first draft of the manuscript was written by Zahra Amiri. Hamid Hassanpour and Azzedine Beghdadi commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
We have no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Amiri, Z., Hassanpour, H. & Beghdadi, A. Combining deep features and hand-crafted features for abnormality detection in WCE images. Multimed Tools Appl 83, 5837–5870 (2024). https://doi.org/10.1007/s11042-023-15198-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15198-z