Abstract
The present paper proposes a novel feature selection technique for the MR brain tumor image classification that aims to choose the optimal feature subset with maximum discriminatory ability in the minimum amount of time. It is based on the fusion of the Fisher and the parameter-free Bat (PFree Bat) optimization algorithm. As the conventional Bat algorithm is bad at exploration, a modification is proposed that guides the Bat by the pulse frequency, global best and the local best position. This improved version of Bat referred to as the PFree Bat algorithm eliminates the velocity equation and directly updates the Bat position. Subsequently, this method in conjunction with the Fisher criteria has been used to select the best set of features for brain tumor classification. The chosen features are then fed to the commonly used least square (LS) support vector machine (SVM) classifier to categorize the area of interest into the high or low grade. For the evaluation of the proposed attribute selection method, tenfold cross-validation has been conducted on a set of 95 ROIs taken from the BRATS 2012 dataset. On an extensive comparison with the other hybrid approaches, the proposed approach brought about the 100% recognition rate in the smallest amount of time. Furthermore, an integrated index is proposed that uniquely identifies the best performing algorithm, taking into account the accuracy, number of features and the computational time. For the fair comparison, the performance of the proposed method has also been examined on breast cancer dataset taken from UCI repository. The obtained results validate that the designed algorithm has better average accuracy than existing state-of-the-art works.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Feature selection refers to the mechanism of choosing the optimal set of features according to some defined feature evaluation criteria. The sorted features result in the reduction of both the data dimensionality and the computation time. Feature selection methods are categorized as feature ranking and feature subset selection methods [1]. These techniques generate a feature vector free from both the less relevant and redundant information [2].
Georgiadis et al. [3] used exhaustive search methods for the feature reduction for classification of solitary dural metastasis and meningioma type of brain tumor in MR images. The method requires a computational time of 11 h for classification. This substantial increase in computational load attributes to the various feature selection and the classifier training procedures. Zhang et al. [4] proposed a method based upon Kernel class separability for the feature reduction. The features leading to the larger class separability were considered for the classification of the tumor from non-tumor regions. The limitation of the approach was that it was validated on a limited dataset of 11 patients. Ahmed et al. [5] explored four different feature selection methods, namely principal feature analysis (PFA), KL divergence (KLD) measure, boosting and entropy. The authors concluded that the best posterior fossa tumor segmentation results were obtained using KL–expectation maximization (EM) method. The entire process required 10 min for normalization, 30 min for feature extraction, 30 min for feature selection using KLD and finally 40 min for segmentation using EM. Sachdeva et al. [6] employed genetic algorithm (GA) for choosing the subset of the most informative features. The efficacy of the approach was justified by the increase in classification accuracy from 56.3 to 91.7% when used in conjunction with support vector machine (SVM). The mechanism was a wrapper-based approach in which the feature selection stage is bound with the classifier to obtain the most optimal set of the attributes. Arakeri et al. [2] employed both the feature ranking and feature subset selection techniques for the differentiation of benign and malignant tumors in MR images. Different combinations were explored, namely information gain (IG) method used in combination with independent component analysis (ICA), principal component analysis and GA. From the experiments, it was concluded that most significant features were selected using IG in conjunction with ICA. The feature selection process required a time nearly equal to 1.2 s.
In a more recent study by Jothi and Inbarani [7], the authors have concluded that hybrid feature selection methods provide better predictive accuracy than the wrapper- or filter-based methods. The authors devised a new hybrid feature selection approach named as tolerance rough set firefly-based quick reduct by combining the supervised tolerance rough set with the firefly algorithm. The method resulted in the selection of the minimal number of attributes.
The systematic analysis of the literature shows a trade-off between the accuracy, number of attributes and the computational time. Choosing the best subset of features delivering the maximum accuracy in the minimum amount of time is still an open challenge in the field of the medical image classification. To address all these issues simultaneously, the present paper proposes a novel feature selection method that incorporates Fisher criteria with a modified version of the Bat algorithm referred to as parameter-free Bat (PFree Bat) algorithm. Instead of classification accuracy, the trace obtained via the Fisher approach is used as fitness criteria to be optimized by the PFree Bat algorithm. The key novelty of the proposed work is that this framework eliminates the need to incorporate a classifier at each stage of the feature selection. Moreover, an integrated measure is proposed that uniquely identifies the best performing algorithm via taking into consideration the number of attributes, accuracy and the computational time.
Rest of the paper is structured as follows: Sect. 2 describes the formulation of the proposed feature selection method, Sect. 3 provides the block diagram of the proposed methodology along with the performance measures, Sect. 4 provides the results, Sect. 5 gives the discussion, and finally the last section concludes the paper.
2 Design of the feature selection method based on Fisher criterion and the proposed PFree Bat optimization algorithm
Most of the metaheuristic-based attribute selection techniques measure the importance of the feature subset by using the metric of classification accuracy. Solely using the classification accuracy as the fitness criteria has two drawbacks. First, it makes the attribute selection technique dependent on the chosen classifier. The generated feature subset obtained using one classifier may not be appropriate for another one. Secondly, for getting the fitness value, the classifier must be retrained with the corresponding feature subset and then used to perform classification on the subset to obtain the classification accuracy. To address these issues, an embedded approach is designed using Fisher criteria and the modified Bat algorithm. Although both the Fisher criteria and the Bat algorithm have been investigated in recent works on feature selection, this scheme of combining the Fisher with the PFree version of the Bat algorithm is first one to our knowledge.
Different from the conventional approaches of assigning a crisp value of 0 or 1 to the features, a weight in the range {0–1} is given to each attribute which is then evolved according to some selected criteria. Instead of choosing the classification accuracy as the fitness function, the trace obtained via the Fisher criteria is used as a fitness function.
2.1 Formulation of the objective function based on Fisher criterion
The Fisher criterion measures the potential of the feature in the absence of any classifier. The objective function based on the Fisher approach has been used for feature selection and then applied to the disease classification.
Let d be the attribute number, and \(w = (w_{1} ,w_{2} , \ldots \ldots \ldots w_{d} )\) be the feature weight vector, where w k refers to the eminence of the kth feature. According to the PFree Bat algorithm, w is an individual Bat position that is required to be calculated that results in the minimization of the fitness value. The sequence of steps followed to compute the fitness of the individuals Bat’s is as follows [8].
Let \(Z^{i,j} = (z_{1}^{i,\,j} ,z_{2}^{i,\,j} , \ldots \ldots \ldots z_{d}^{i,\,j} )\) be the complete feature vector of the jth example of the ith group, n i be the number of the examples of the ith group, and C be the number of groups. In the first stage, the mean of the feature vector belonging to the ith group is calculated as
where mi is the mean of group i.
And the combined average of the feature vector for all the training examples is given as:
The group mean and the combined mean are both a d-dimensional vector and are denoted as \(m^{i} = \{ m_{1}^{i} ,m_{2}^{i} , \ldots \ldots \ldots m_{d}^{i} \}\) and \(m = \{ m^{1} ,m^{2} , \ldots \ldots \ldots m^{d} )\), respectively. In the second stage, the weighted average distance, S w, between all the training examples and the group mean is calculated as
And the weighted distance between different groups, S B, is calculated as
The Fisher criterion is formulated as maximizing the weighted distance between different groups, i.e., maximizing S B and minimizing S w. Using both these distance criteria, the fitness function can be formulated as a function of w and is expressed as
The goal of the optimization algorithm would be to find the optimal value of w at which f(w)is minimum. After the minimization process, the features are selected according to some threshold. The threshold is varied from 0.1 to 1 with a step of 0.05. Each resulting feature subset is fed to the least square support vector machine (LS-SVM) classifier to perform the categorization. The feature subset resulting in maximum classification accuracy is chosen as the optimal one, and correspondingly that threshold value is selected.
2.2 Parameter-free Bat (PFree Bat) algorithm for feature selection
Bat algorithm (Bat) proposed by Yang [9] is a recent metaheuristic algorithm based on the echolocation ability of the microbats that guides them on their foraging behavior. The Bat uses a combination of the major advantages of the harmony search and the particle swarm optimization algorithm.
Bat follows certain rules to update the position and the velocity of the bats in accordance with the pulse frequency Q i. The position x i and velocity v i for the ith Bat are defined in a d-dimensional search space. The new solutions are obtained by updating x i and v i at each time step, t, and are given as [9]:
In the above Eq. (6), β is a random number drawn from uniform distribution (range {0 1}), Q i is the frequency value associated with the ith Bat, and Q max and Q min are the maximum and minimum frequency values. The velocity and the position of the ith Bat at the tth time step are denoted as \(v_{i}^{t}\) and \(x_{i}^{t}\). x* is the global best solution obtained by comparing the fitness value for all the Bat positions. For the purpose of favoring the exploitation around the best position obtained so far Yang [9] has proposed a modification via which a Bat can improve the solution near the best position. It is mathematically given as:
Here, ɛ is the randomly generated value in the interval [− 1, 1], and A t is the mean loudness value for all the bats at the tth time step. Furthermore, the rate of pulse emission, r i, and the loudness, A i, are updated with iterations as follows:
In the above expression γ and α are fixed constants and \(r_{i}^{0}\) is the initial pulse emission rate of the ith Bat.
The Bat algorithm is poor at the exploitation and exploration [10]. The algorithm’s position and velocity update equations have few similarities with the PSO. Similar to the PSO, the standard Bat also has some deficiencies. To counteract with this limitation, the following modification structure is proposed, inspired by the study in [11] [12]. The proposed mechanism does not need the velocity update equation as the standard Bat. The Bat locations are directly updated using a new position update equation as given below:
The improved equation guides the Bat under the influence of the global best solution and the previous best solution (pbest i). Further, this update process is modulated by the Bat frequency Q i. The entire process works like a mutation operator to accelerate the search toward the global optima. During the initial stages, the ratio of \(\frac{x*}{{x_{i}^{t - 1} }}\) is small leading to large step size, and during the later phases of the search, this step size is decreased because x i becomes almost equal to x*. As the modified Bat algorithm eliminates the velocity update equation, it is termed as PFree Bat algorithm.
The usage of the parameter-free frequency modulation scheme accelerates the global performance of the conventional Bat and hence avoids the premature convergence. This mechanism combines the advantages of the Bat and the parameter-free PSO and thereby has the potential to reach an optimal solution and increase the diversity of the search space.
The pseudo-code for the proposed algorithm for feature selection application is given below:

The x* denotes the optimal weight for each of the feature.
3 Methods
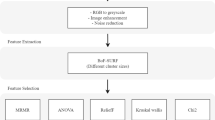
The entire workflow using the above methodology is given as a flowchart in Fig. 1. The block diagram for MR image classification using the proposed feature selection approach is shown in Fig. 2. The preprocessing steps include the acquisition of the low and the high-grade glioma tumor volumes from the BRATS 2012 dataset [13], ROI delineation and the feature extraction. The features were extracted using the standard texture models. The extracted features were normalized, and a set of most informative features were obtained using the proposed Fisher + PFree Bat optimization algorithm. The reduced feature set was then fed to the LS-SVM classifier for the final categorization.
The prediction performance of the proposed feature mechanism has been tested using four different performance evaluation criteria, the mathematical formulations of which are detailed below. These measures include sensitivity, specificity, accuracy and an integrated index, AFCN, respectively.
Sensitivity and specificity comprise of the following terms, true positive (TP), false negative (FN), true negative (TN) and false positive (FP). TP is the number of high-grade tumors classified as high grade, FP is the number of the low-grade tumors classified as high grade, TN is the number of low-grade tumors classified as low grade, and FN is the number of high-grade tumors classified as low grade. Mathematically sensitivity is computed as
Alternatively, the specificity is mathematically represented as [14]
The accuracy is given as
Furthermore, for the fair comparison of the proposed algorithm a new metric is proposed that simultaneously reflects the efficacy by taking into account the number of attributes, classification accuracy and the computational time referred to as AFC. Higher the value of AFC better is the algorithm performance.
where N refers to the total number of algorithms to be compared and \(F_{i}^{ * }\) is given below:
In the above expression, CT specifies the computational time. The typical range of the accuracy is 0–1, and F i* is 1/(Length of Feature Vector) to 1. The minimum value of F i* represents that only a single feature is selected out of the total feature entries, whereas 1 represents the inclusion of all features in the final feature vector by the attribute selection algorithm.
From this AFC, a normalized value is computed as:
AFCN i index uniquely identifies the attribute selection algorithm that performs the best while taking into consideration the factors of accuracy, the number of selected features and the computational time. In the present work, the AFCN i index is computed at different values of thresholds ranging from 0.1 to 1, and that particular optimal threshold has been chosen that results in the maximum value of this index.
4 Results
The proposed framework has three broad applications. Firstly, the proposed PFree Bat optimization algorithm has been successfully applied for the numerical function optimization. Secondly, this modified algorithm in conjunction with the Fisher criteria has been used as feature selection method for the classification of the MR brain tumors (into low- and high-grade categories), and thirdly the same has been tested for classification of the breast cancer tumors (into malignant and the benign categories). Primarily, the work is focused on the design of a feature selection technique for the MR brain tumor image classification, but for the validation of the modification proposed in the Bat algorithm the verification has been done on the standard set of the benchmark functions [10]. Moreover, for the fair comparison with the recent state-of-the-art works on feature selection, the results have been computed on breast cancer dataset from the UCI repository.
For the benchmark functions, the analysis has been carried out by taking the dimension value equal to 10. The details regarding the mathematical formulation and range restriction of the test functions are given in Table 1 [15]. All the chosen functions have the global optima at a value equal to 0.
Table 2 shows the comparison of the proposed optimization algorithm with the different versions of the Bat algorithm existing in the literature. It reports the ‘best,’ ‘worst,’ ‘mean’ and the ‘standard deviation (SD)’ achieved by the various algorithms. The entries clearly show that the proposed optimization approach has outperformed the other techniques on the four out of five benchmark functions as shown in bold. Typically, for the Griewank function the algorithm achieved the best value equal to 0 and the minimum SD equal to 0 in contrast to the other approaches.
4.1 Feature selection for classification of brain tumors
This section presents the results of the experimentations conducted on tumorous images taken from BRATS 2012 database [13]. The data comprised of the real high- and low-grade glioma volumes. From these volumes, a total of 95 2D image slices were chosen exhibiting a significant contrast between T1 and T1 contrast-enhanced (CE) images. The latest datasets like BRATS 2013 and BRATS 2015 also comprise of the cases continued from BRATS 2012.
The entire abnormal region was extracted from the fluid attenuation inversion recovery (FLAIR) set of images. The whole extracted region was mapped onto the T1 and T1-CE images. Finally, from the mapped region, a difference image was constructed using T1 and T1-CE. Figures 3 and 4 briefly outline the steps for the generation of the difference image. In both of these tables, (a), (c) and (e) represent the original FLAIR, T1 and T1-CE image while (b), (d) and (f) show the corresponding abnormal region mapped from the FLAIR onto the T1 and T1-CE set of images.
A total of 52 features were extracted from the difference image by applying the standard texture models. These include the gray-level co-occurrence matrix (GLCM) [19] [20], gray-level run-length matrix (GLRM) [21], gray tone difference matrix (GTDM) [22], Law’s texture features (LTF) [23], fractal [24], Gabor filters [25], Gabor wavelet [26], first-order statistics and the empirical mode decomposition (EMD) [27]-based features. The features with the respective count are enlisted in Table 3. The last attribute, i.e., density measure, was computed by applying the improved CEEMDAN [27] algorithm to the difference image followed by the Hilbert transform.
After extraction, the proposed feature selection method was applied resulting in the selection of the most informative attributes. Table 10 in appendix section presents the optimum PFree Bat algorithm parameters for this application. These parameters have been chosen after experimenting on the training data that results in maximum classification accuracy in the minimum amount of time with a minimal number of features.
After the selection of the appropriate feature subset, then a tenfold cross-validation approach has been applied to evaluate the classification accuracy via the use of the LS-SVM. For the present work, the MATLAB 15a implementation of LS-SVM has been done by using LS-SVMlab toolbox [28]. Moreover, the computational time has also been estimated using MATLAB 15 platform with 64-bit Windows 7 Professional operating system, 3.10 GHz Intel Core i7 processor with 8 GB RAM.
Table 4 gives the performance of the different hybrid approaches using various metrics at a threshold value equal to 0.75. The MATLAB source codes for PSO, BBO, DE, real-coded GA, firefly algorithm, were obtained from the yarpiz.com (http://yarpiz.com/). Moreover, Table 5 enlists the prominent features that were selected. From the tabular analysis, it is concluded that the proposed method achieved the best value of accuracy, sensitivity, specificity, number of features and the computational time. The obtained values were 1, 1, 1, 1 and 0.17 s for the proposed method. A value equal to 1 for the accuracy signifies that all the test cases were accurately characterized into low- and high-grade categories. Though the algorithm A5 that employs Fisher and the ABC algorithm has accomplished a competitive performance with accuracy, sensitivity and specificity equal to 1, the computational time was very large equal to 170 s. The key advantage of the proposed method is the minimal computational time, the minimum number of features and maximum accuracy.
Apart from measuring the accuracy at a fixed value of the threshold of 0.75, algorithm efficiency is also analyzed at varying levels of thresholds starting from 0.1 to 1 using the formulated AFCN index that uniquely identifies the best performing algorithm by taking into account the accuracy, number of features and the computational time simultaneously. Figure 5 gives an insight of the algorithm competence in terms of the AFCN index
Closer the value to 1 better are the classification results. Typically for a threshold value equal to 0.5 the value of AFCN index is 0.041, 0.040, 0.164, 0.032, 0.098, 0.071, 0.095, 0.443, 0.113 and 1, respectively, for the different algorithms. On simultaneously considering all the effectual performance metrics the proposed algorithm has resulted in the maximum value of AFCN index.
4.2 Feature selection for classification of breast tumors
It is emphasized that the prime focus of the present work is to design a feature selection technique for the MR brain tumor image classification into low- and high-grade categories. To the best of the author’s knowledge, no current state-of-the-art works have reported the results for BRATS 2012 dataset. So to validate the versatility of the proposed feature selection method on different types of tumors and for a fair comparison with the recent state-of-the-art works on feature selection, the algorithm has been tested on the Wisconsin Breast Cancer Dataset (WBCD) [39] and Wisconsin Diagnosis Breast Cancer Database (WDBC) [40, 41] taken from the UCI machine learning repository. The WBCD contains a total of 699 samples each having a total of 9 features. A total of 458 samples belong to the benign class, and 241 belong to the malignant class. The WDBC consists of 30 attributes and 569 instances. These examples either have a benign or a malignant class label. A total of 357 cases were lying in the benign class and 212 in the malignant class.
Table 6 recapitulates the performance of the proposed feature selection approach on the WBCD and WDBC datasets using the chosen performance metrics. The tabular findings clearly show that Fisher + PFree Bat approach achieved the largest mean accuracy and the sensitivity equal to 98.54, 99.10 and 98.60, 100 for both the datasets.
Table 7 gives the best results obtained using the designed approach. For the fair comparison the best value of the accuracy, sensitivity and specificity is reported at a different number of features (top-ranked features in accordance with the threshold value). The tabular entries clearly show that the best results are obtained using the proposed technique. The resulting metric values (in % age) are 100, 100 and 100 at the lower and the higher set of features.
As a lot many feature selection and classification approaches exist in the literature, Tables 8 and 9 provide an extensive analysis of best accuracy achieved so far for both the datasets using the proposed and the existing algorithms.
The results clearly show that the designed approach was successful in achieving 100% classification accuracy on both the datasets, thereby clearly differentiating the benign and the malignant cases.
5 Discussion
In this paper, we have successfully formulated a new algorithm for feature selection based on the fusion of the Fisher and PFree Bat optimization algorithm that aids in the classification of the brain tumors into different categories. From the comparison with the recent metaheuristic optimization algorithms given in Table 4, it is seen that the proposed feature selection method is potentially more efficient in terms of the accuracy, sensitivity, specificity and the computational time that were equal to 100, 100, 100% and 0.17 s.
Therefore, the proposed method is fully capable of detecting the glioma grade, i.e., low or high than the other presented approaches with the minimum feature count and the minimal computational cost that are highly desirable in clinical diagnosis. On simultaneously considering all the performance measures using the proposed AFCN index, the designed feature selection mechanism obtained the best result at the different value of the thresholds as indicative from Fig. 5. The key to the success of the fused approach is the utilization of the PFree Bat algorithm for increasing the exploration capabilities of the algorithm resulting in the selection of appropriate feature subset that improves the runtime performance and the accuracy of the classifier.
The performance of the proposed method is also compared with the studies in the literature which uses the same dataset, i.e., the breast tumors. In a comprehensive comparison using the WBCD and WDBC database from the UCI machine learning repository, the proposed feature selection method obtained a mean accuracy of 98.54 and 98.60%, respectively, and the best accuracy of 100% for both datasets as inferred from Tables 6 and 7.
The obtained values using the proposed method are higher than the mean value equal to 97.88%, 98.11% and the best value equal to 98.53 and 98.45% achieved via a most recent approach proposed by [30].
Future works will be concentrated on the validation of the proposed feature selection method on a larger dataset containing multiple classes of the tumor and investigating its applicability in real-time clinical diagnosis.
6 Conclusion
This paper has proposed a new attribute selection method based on Fisher criterion and PFree Bat algorithm for the disease classification. The advantage of this embedded technique is that it is independent of the classifier, and it is used only once after the trace computed by the Fisher criteria is minimized by the PFree Bat optimization algorithm. The experimental results on a diversified set of medical images have demonstrated that designed approach, when coupled with the LS-SVM, has brought out the best recognition rate at the minimum computational cost. In a tenfold cross-validation experiment on different tumorous ROIs that were manually extracted from the real glioma tumor images, the hybrid proposed technique achieved the best classification accuracy equal to 100%.
Change history
14 May 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s00521-024-09943-0
References
Zacharaki EI, Wang S, Chawla S, Soo D (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62:1609–1618
Arakeri MP, Reddy GRM (2013) Computer-aided diagnosis system for tissue characterization of brain tumor on magnetic resonance images. Signal Image Video Process 7:1–17
Georgiadis P, Cavouras D, Kalatzis I et al (2009) Enhancing the discrimination accuracy between metastases, gliomas and meningiomas on brain MRI by volumetric textural features and ensemble pattern recognition methods. Magn Reson Imaging 27:120–130
Zhang N, Ruan S, Lebonvallet S et al (2011) Kernel feature selection to fuse multi-spectral MRI images for brain tumor segmentation. Comput Vis Image Underst 115:256–269
Ahmed S, Iftekharuddin KM, Vossough A (2011) Efficacy of texture, shape, and intensity feature fusion for posterior-fossa tumor segmentation in MRI. IEEE Trans Inf Technol Biomed 15:206–213
Sachdeva J, Kumar V, Gupta I et al (2011) Multiclass brain tumor classification using GA-SVM. Dev E-Syst Eng 2011:182–187
Jothi G, Inbarani H (2015) Hybrid Tolerance Rough Set-Firefly based supervised feature selection for MRI brain tumor image classification. Appl Soft Comput J 46:639–651
Liu X, Ma L, Song L et al (2015) Recognizing common CT imaging signs of lung diseases through a new feature selection method based on Fisher criterion and genetic optimization. IEEE J Biomed Heal Inform 19:635–647
Yang X-S (2010) A new metaheuristic bat-inspired algorithm. In: Nature inspired cooperative strategies for optimization (NICSO 2010). Springer, Berlin, pp 65–74
Yilmaz S, Kucuksille EU (2015) A new modification approach on bat algorithm for solving optimization problems. Appl Soft Comput 28:259–275
Bakwad KM, Pattnaik SS, Sohi BS et al (2009) Hybrid bacterial foraging with parameter free PSO. In: Nature and biologically inspired computing 2009. NaBIC 2009. World Congress on, pp 1077–1081
Ramana Murthy G, Senthil Arumugam M, Loo CK (2009) Hybrid particle swarm optimization algorithm with fine tuning operators. Int J Bio-Inspired Comput 1:14–31
Menze BH, Jakab A, Bauer S et al (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024
Lee MC, Nelson SJ (2008) Supervised pattern recognition for the prediction of contrast-enhancement appearance in brain tumors from multivariate magnetic resonance imaging and spectroscopy. Artif Intell Med 43:61–74
Yao X, Liu Y, Lin G (1999) Evolutionary programming made faster. IEEE Trans Evol Comput 3:82–102
Yilmaz S, Kucuksille EU (2013) Improved bat algorithm (IBA) on continuous optimization problems. Lect Notes Softw Eng 1:279–283
Fister Jr I, Fister D, Yang X-S (2013) A hybrid bat algorithm. arXiv Prepr. arXiv1303.6310
Fister Jr I, Fister D, Fister I (2013) Differential evolution strategies with random forest regression in the bat algorithm. In: Proceedings of the 15th annual conference companion on Genetic and evolutionary computation, pp 1703–1706
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC 3:610–621
Vidya KS, Ng EY, Acharya UR et al (2015) Computer-aided diagnosis of myocardial infarction using ultrasound images with DWT, GLCM and HOS methods: a comparative study. Comput Biol Med 62:86–93
Tang X (1998) Texture Information in run-length matrices. IEEE Trans Image Process 7:1602–1609
Amadasun M, King R (1989) Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 19:1264–1274
Tencer L, Reznakova M, Cheriet M (2012) A new framework for online sketch-based image retrieval in web environment. In: Information science, signal processing and their applications (ISSPA), 2012 11th international conference on. IEEE, Montreal, QC, pp 1430–1431
Costa AF, Humpire-mamani G, Juci A et al (2012) An efficient algorithm for fractal analysis of textures. In: 25th SIBGRAPI conference on graphics patterns images. IEEE, Ouro Preto, Brazil, pp 39–46
Hong L, Wan Y, Jain A (1998) Fingerprint image enhancement: algorithm and performance evaluation. IEEE Trans Pattern Anal Mach Intell 20:777–789
Liu Y, Muftah M, Das T et al (2012) Classification of MR tumor images based on Gabor wavelet analysis. J Med Biol Eng 32:22–28
Colominas MA, Schlotthauer G, Torres ME (2014) Improved complete ensemble EMD: a suitable tool for biomedical signal processing. Biomed Signal Process Control 14:19–29
De Brabanter K, Karsmakers P, Ojeda F, et al (2011) LS-SVMlab toolbox user’s guide. ESAT-SISTA technical report 10
Yang X-S, Press L (2010) Nature-inspired metaheuristic algorithms, second edition
Yang X-S, Hossein Gandomi A (2012) Bat algorithm: a novel approach for global engineering optimization. Eng Comput 29:464–483
Meng X-B, Gao XZ, Liu Y, Zhang H (2015) A novel bat algorithm with habitat selection and Doppler effect in echoes for optimization. Expert Syst Appl 42:6350–6364
Karaboga D (2005) An idea based on honey bee swarm for numerical optimization
Karaboga D, Basturk B (2007) A powerful and efficient algorithm for numerical function optimization: artificial bee colony (ABC) algorithm. J Glob Optim 39:459–471
Karaboga D, Basturk B (2008) On the performance of artificial bee colony (ABC) algorithm. Appl Soft Comput 8:687–697
Karaboga D, Akay B (2009) A comparative study of artificial bee colony algorithm. Appl Math Comput 214:108–132
Yang X-S, Deb S (2009) Cuckoo search via Lévy flights. In: Nature & biologically inspired computing 2009. NaBIC 2009. World Congress on, pp 210–214
Yang X-S, Deb S (2010) Engineering optimisation by cuckoo search. Int J Math Model Numer Optim 1:330–343
Mahdavi M, Fesanghary M, Damangir E (2007) An improved harmony search algorithm for solving optimization problems. Appl Math Comput 188:1567–1579
Mangasarian OL, Setiono R, Wolberg WH (1990) Pattern recognition via linear programming: theory and application to medical diagnosis. Large-Scale Numer Optim 22–31
Mangasarian OL, Street WN, Wolberg WH (1995) Breast cancer diagnosis and prognosis via linear programming. Oper Res 43:570–577
Wolberg WH, Street WN, Heisey DM, Mangasarian OL (1995) Computer-derived nuclear features distinguish malignant from benign breast cytology. Hum Pathol 26:792–796
Ratanamahatana CA, Gunopulos D (2003) Feature selection for the naive bayesian classifier using decision trees. Appl Artif Intell 17:475–487
Palaniappan S, Pushparaj T (2013) A novel prediction on breast cancer from the basis of association rules and neural network. Int J Comput Sci Mob Comput 2:269–277
Miao D, Gao C, Zhang N, Zhang Z (2011) Diverse reduct subspaces based co-training for partially labeled data. Int J Approx Reason 52:1103–1117
Abdel-Aal RE (2005) GMDH-based feature ranking and selection for improved classification of medical data. J Biomed Inform 38:456–468
Luukka P, Leppälampi T (2006) Similarity classifier with generalized mean applied to medical data. Comput Biol Med 36:1026–1040
Ahmad F, Isa NAM, Hussain Z, Sulaiman SN (2012) A genetic algorithm-based multi-objective optimization of an artificial neural network classifier for breast cancer diagnosis. Neural Comput Appl 23:1427–1435
Sheikhpour R, Agha M, Sheikhpour R (2016) Particle swarm optimization for bandwidth determination and feature selection of kernel density estimation based classifiers in diagnosis of breast cancer. Appl Soft Comput 40:113–131
Mojtaba S, Bamakan H, Gholami P (2014) A novel feature selection method based on an integrated data envelopment analysis and entropy model. Procedia Comput Sci 31:632–638
Xue B, Zhang M, Browne WN (2012) New fitness functions in binary particle swarm optimisation for feature selection. In: IEEE congress on evolutionary computation (CEC), 2012, pp 1–8
Xue B, Zhang M, Browne WN (2014) Particle swarm optimisation for feature selection in classification: novel initialisation and updating mechanisms. Appl Soft Comput 18:261–276
Maldonado S, Weber R, Basak J (2011) Simultaneous feature selection and classification using kernel-penalized support vector machines. Inf Sci 181:115–128
Miao D, Gao C, Zhang N, Zhang Z (2011) Diverse reduct subspaces based co-training for partially labeled data. Int J Approx Reason 52:1103–1117
Luukka P, Leppalampi T (2006) Similarity classifier with generalized mean applied to medical data. Comput Biol Med 36:1026–1040
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
‘The authors declare that they have no conflict of interest.
Appendix
Appendix
See Table 10.
About this article
Cite this article
Kaur, T., Saini, B.S. & Gupta, S. RETRACTED ARTICLE: A novel feature selection method for brain tumor MR image classification based on the Fisher criterion and parameter-free Bat optimization. Neural Comput & Applic 29, 193–206 (2018). https://doi.org/10.1007/s00521-017-2869-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-017-2869-z