Abstract
Wireless capsule endoscopy (WCE) is a new imaging procedure that is used to record internal conditions of gastrointestinal tract for medical diagnosis. However, due to the presence of bulk of WCE image data, it becomes difficult for the physician to investigate it thoroughly. Therefore, considering aforementioned constraint, lately gastrointestinal diseases are identified by computer-aided methods and with better classification accuracy. In this research, a new computer-based diagnosis method is proposed for the detection and classification of gastrointestinal diseases from WCE images. The proposed approach comprises of four fundamentalsteps:1) HSI color transformation before implementing automatic active contour segmentation; 2) implementation of a novel saliency-based method in YIQ color space; 3) fusion of images using proposed maximizing a posterior probability method; 4) fusion of extracted features, calculated using SVD, LBP, and GLCM, prior to final classification step. We perform our simulations on our own collected dataset – containing total 9000 samples of ulcer, bleeding and healthy. To prove the authenticity of proposed work, list of statistical measures is considered including classification accuracy, FNR, sensitivity, AUC, and Time. Further, a fair comparison of state-of-the-art classifiers is also provided which will be giving readers a deep inside of classifier’s selection for this application. Simulation results clearly reveal that the proposed method shows improved performance in terms of segmentation and classification accuracy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer, also known as bowel cancer, is a general type of cancer which affects both men and women [29]. The number of mortalities due to colorectal cancer is 694,000 in less developed countries of the world [30]. An American cancer society estimated about 132,000 new cases of colorectal cancer, appeared in 2015 [36]. A few gastrointestinal infections are directly linked with the colorectal cancer, such as hemorrhoids, short bowel, to name but a few. These infections can be diagnosed through colonoscopy method, but this complete procedure is time consuming and also having a constraint of limited number of specialists [25]. Recently, push gastroscopy methods are utilized in the clinics for diagnosis of gastrointestinal diseases like an ulcer, polyp, and bleeding. Although, these methods are not suitable to identify small bowels due to complex structure [17]. In 2000, this sort of problem was solved by a new technology named, wireless capsule endoscopy (WCE), which has an ability to identify gastrointestinal diseases right from the small bowels [8]. WCE technology is now rapidly used in hospitals for the diagnosis of gastrointestinal diseases like ulcer and bleeding [21]. Recently reported that about 1Million patients are successfully treated with WCE [17].
One of the constraints in this complete procedure is that it takes a lot of time, which makes this procedure somewhat arduous, because ulcer shows itself only for a short duration in the entire video. It might be possible; physician misses an ulcer region during this complete process. Moreover, few irregularities are hidden to the naked eyes due to set of challenges like a change of shape, texture, and color similarities.
For an accurate diagnosis, several researchers proposed computer-aided diagnosis (CAD) methods, which consist of few fundamental steps including lesion segmentation, feature extraction & reduction, and classification. The segmentation of lesion region is important for the detection of diseased part from WCE images. For this reason, several methods are introduced in the literature. In [16], authors introduced a new color features-based approach for detection of gastrointestinal diseases like ulcer and bleeding from WCE images. In color features, they calculated chromaticity moment to distinguish the normal and abnormal regions. Finally, the performance of the color features is analyzed using neural networks (NN). In [20], authors introduced a new technique for the detection of ulcer and bleeding from WCE images, in which texture and HSV color features are extracted, which are later utilized by Fisher scoring (FS) method. Through this FS approach, features with maximum information are selected, which are later classified using multilayered NN.
For features extraction, several methods are utilized by the scholars including color features [33], discriminative joint features [38], scale invariant feature transform (SIFT) features [39], texture features [19], Log filter bank [2], to name but a few [24, 27]. Feature reduction step plays it vital role and several methods exist such as principal component analysis (PCA), linear discriminant analysis (LDA), etc. Set of classifiers are working robustly to accurately identify infectious regions such as artificial neural network [10],support vector machine (SVM) [22], K-nearest neighbor (KNN), naïve Bayes [31], to name but a few.
However, in computer-based methods, several challenges still exist which degrade the system’s accuracy. In the segmentation process, several challenges include lesion irregularity, texture, color similarity, complex background, and border – making segmentation process more complex. Moreover, in the classification phase, robust features produce accurate results, but the selection of most appropriate features out of pool of features is still a greater challenge.
To address these challenges, we propose a new technique for gastrointestinal diseases detection and classification from WCE images namely CCSFM (color-based contour saliency fusion method). The proposed approach is an integration of four primary phases. In the first phase, HSI transformation is performed on the original RGB image to select Hue, saturated and intensity channels. Then a threshold based weighted function is performed, which selects the channel with maximum information, which is later segmented using active contour model. In the second phase, YIQ transformation is performed on RGB image prior computing max and min pixel values against each channel, which produces a mask function. Finally, thresholding is applied to a mask image and later fuse their pixels with an active contour image. In the third phase, color, local binary patterns (LBP), and GLCM features are extracted from mapped RGB segmented images. The extracted features are later fused by a simple concatenation method, prior to reduction steps. Only those feature values having high probability are selected for further processing. Finally, a multi perceptron neural network is employed for the classification of reduced vector. Our major contributions are enumerated below:
-
I-
A new saliency-based segmentation technique is proposed based on YIQ color transformation, which is carried out on RGB image. A mask function is structured which utilizes maximum and minimum pixel values against each channel to support a segmentation process.
-
II-
A new maximum a posteriori probability (MAP) estimation method is implemented for the fusion of proposed saliency image and active contour segmented image.
-
III-
A feature selection methodology is proposed, which defines a threshold value based on probability of maximum feature occurring to select most discriminant features.
-
IV-
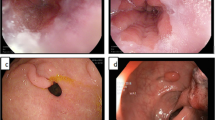
A new database is constructed which comprises 9000 RGB images of gastrointestinal diseases such as ulcer and bleeding. Moreover, for a fair comparison, 3000 healthy RGB images are also provided. Selected image samples can be seen in Fig. 1, which are; (a) ulcer, (b) bleeding, and (c) healthy.
2 Related work
A substantial amount of work has been done in the field of medical imaging to develop computerized methods – having capability to assist physicians [11,12,13]. Several computer-based methods have been proposed for the identification and classification of GI diseases from WCE images, which make the diagnostics easier for the doctors. Kundu et al. [15] utilized Y plane in YIQ color transformation for automatic detection of ulcer from WCE images. Y-plane total pixels are taken as features, which are classified using SVM by implementing Gaussian RBF kernel function. Shipra et al. [34] introduced a statistical color-based feature for bleeding detection from the WCE images. These statistical color features are extracted from RGB images and characterized into bleeding and non-bleeding images using SVM classifier. Suman et al. [33] extracted color features from several color spaces including RGB, CMYK, LAB, HSV, XYZ, and YUV to make ulcer and non-ulcer regions distinct. Then these features are combined using cross-correlation approach in order to provide a fair comparison between two patterns. Finally, SVM is used for classification to achieve a classification accuracy of 97.89%.Said et al. [5] presented texture features based approach for anomalies identification from WCE images. Initially, the texture features such as LBP variance and discrete wavelet transform (DWT) are extracted from GI disease images to tackle illuminations changes. The extracted features are finally classified using SVM and MLP to achieve promising results. Charfi et al. [4] followed a hybrid feature extraction methodology for ulcer recognition from WCE images. These features include complete local binary patterns (CLBP) and global local oriented edge magnitude patterns (GLOEMP). The CLBP features are extracted for the texture information of ulcer images, whereas GLOEMP features are calculated for the color information. Thereafter, both CLBP and GLOEMP features are integrated in the form of vector and classified using SVM and MLP. Yuan et al. [37] introduced an automated approach for ulcer identification from WCE images. The introduced method consists of two phases. In the first phase, a multi-level superpixels based saliency approach is proposed, which draw the outline of the ulcer region. Color and texture features are extracted from each level and then all levels are integrated to construct a final saliency map. In the second phase, saliency max-pooling (SMP) method is proposed and combined with the locality-constrained linear coding (LLC) method to achieve a classification accuracy of 92.65%. Fu et al. [7] presented a computer-based method for bleeding identification from WCE images. The image pixels are grouped using superpixels segmentation approach. The features are extracted from superpixel and later fed to SVM for classification. Addition to that, several algorithms are proposed in this domain to generate pool of solutions [34].
The above-mentioned techniques are mostly relying upon color and texture features. Inspire from the aforementioned methods, we propose a new approach for the detection and classification of GI diseases like ulcer and bleeding from WCE images based on improved saliency method and MLPNN.
3 Materials and methods
In this section, a novel approach is presented for GI diseases detection and classification from WCE images. Fundamental steps of proposed approach are: a) active contour-based segmentation using HSI color transformation; b) proposed saliency method based on YIQ color space; c) fusion of segmented images; d) features extraction and reduction, and e) classification using artificial neural networks. A detailed description of each step is provided and schema of proposed framework can be seen in Fig. 2.
3.1 Active contour-based segmentation
In few WCE images, there exists a smooth color variation between the diseased and healthy regions, which is one of the reasons of wrong segmentation. To tackle this problem, we implemented an active contour model without edge segmentation [3] using HSI color transformation. The entire process comprised of three sub-phases. In the first phase, HSI color transformation is performed followed by an implementation of weighted function to extract best suitable channel in the second phase. Finally, the selected channel is fed into an active contour segmentation method, with no edge function being used to stop the evolving curve at the desired boundary locations.
Let ξ(x, y) ∈ ℝ(R × C × 3) denotes an input RGB image. Their HIS conversion is defined using Eq. (1-3) as:
where, \( n=\frac{1}{2}\left(\left({\xi}_R-{\xi}_G\right)+\left({\xi}_R-{\xi}_B\right)\right) \) and \( d=\sqrt{\left({\left({\xi}_R-{\xi}_G\right)}^2+\left({\xi}_R-{\xi}_B\right)\left({\xi}_G-{\xi}_B\right)\right)} \). ξH represents hue channel, n denotes the sum of difference between RGB channels, and d is a distance of RGB pixels. Three channels red, green, and blue are denoted by ξR, ξG, and ξB - calculated as \( {\xi}_R=\frac{r}{\sum_{k=1}^m{\xi}_k} \), \( {\xi}_G=\frac{g}{\sum_{k=1}^m{\xi}_k} \),&\( {\xi}_B=\frac{b}{\sum_{k=1}^m{\xi}_k} \). The given parameters include m = 3 and k ∈ {1, 2, 3} - representing indices of each RGB channel.
where, n1 = φ(φ(ξR, ξG, ξB)), and φ is a minimum operator which selects minimum value from each index. At the first time, when φ operator is used, it returns three values, one from each extracted channel (red, green, and blue). The second φ selects the minimum value from all three computed values. Moreover, d1 is the sum of pixels of all ξR, ξG, and ξB channels as defined as d1 = ξR + ξG + ξB.
Where, ξI is an intensity channel. To select a channel with maximum information, we utilized our published work [1, 6], previously tested on natural images – now using for medical application. In this technique, a weighting criterion is implemented to identify a gray channel incorporating maximum information regarding foreground object. These weights are calculated based on object’s distance from center (wdc), number of connected components (wcl), boundary connection (wbc), and a generated distance matrix (wdm). Therefore, the cumulative weight relies on four conditions mentioned below:
Later, an active contour method is implemented on the selected channel, to identify the healthy and diseased region. The energy function of the active contour method is defined as:
Where,λ1 = 2 & λ2 = 4 are the constants to control brightness affects, μ is a mean value, v≥0, C is an evolving curve, L is the curve length, A is the region area inside C, in(C) is the inside boundary of the curve, out(C) denotes the outside boundary of the curve and s1, s2 are left and right snake contours, depends on the curve C and defined as:
The process started with the initialization of a mask (φ0). We introduce an automatic mask initialization technique described in Algorithm 1:

It is observed that the average diameter of the diseased region is about 2 mm. Therefore, we initialized the mask close to the lesion boundary with the size of 3 mm approximately. In this method, we considered maximum pixel value, as the lesion area is darker than the healthy skin. Next, compute the s1(φn) and s2(φn). The time dependent partial differential equations (PDE) are utilized to get the φn + 1. Finally, apply the exit condition to stop the process, which states, when there is no change in solution for three consecutive iterations. The output of resultant active contour image ξact(s, C) is shown in Fig. 3.
3.2 Improved saliency-based segmentation
In this section, we present a new improved saliency-based method for ulcer detection from WCE images. The proposed saliency method consists of following series of steps: a) RGB to YIQ conversion; b) extraction of YIQ channels and find max and min pixel values, and c) generation of mask function calculated from max and min values. A detailed flow of proposed saliency method is shown in Fig. 4.
Initially, YIQ transformation is performed, in which Y-channel represents luminance factor and I & Q show the chrominance factor. One of the reasons to select this color space is it property of being near to human visual system and also shows more sensitivity in I-axis compared to Q-axis. The conversion of RGB to YIQ is defined as:
where, ξY, ξl, and ξQrepresent luminance, hue, and saturation channel respectively.(α1, … , α9) denote transformed values, in the range of −1 to 1. The exact values used are, ξY ∈ {0.299, 0.587, 0.114}, ξI ∈ {0.596, −0.274, −0.321}, and ξQ ∈ {0.211, −0.523, 0.311} [9]. Next step is to find max (ϱmax) and min(ϱmax)pixel values from extracted channels:
where, index i ∈ {1, 2, 3}, shows three extracted channels Y, I, and Q, respectively. The mass function is defined as follows:
where, ξmask(BW)is a binary image, F(x, y) is a mask function, which is generated by the max and min pixel values. The effects of saliency-based ulcer detection results are shown in the Fig. 5. Moreover, as a refinement step, we utilized some morphological operations such as opening, closing to extricate extraneous pixels.
3.3 Maximizing a posterior probability (MAP) based pixels fusion
The obtained image from different algorithms may have distinct patterns, therefore, to improve its quality; we combine the characteristics of both images. Several methods have been proposed for image fusion including pixel-based, region-based, pyramid transform based fusion, and few other methods. In order to take advantage, we adopted maximizing a posterior probability based fusion method. To inspire with these methods, in this article, we implement a Maximizing a Posterior (MAP) based fusion method, which combines valuable pixels of segmented images into a single matrix.
Let β1 represents pixels from an active contour segmented image, ξact(s, C),\( \overline{\beta} \) representing pixels from proposed saliency method, and ξpro(x, y) is the fused image, having dimension R(256 × 256). According to the Bayesian estimation, the joint priori distribution (JPD) of β1 and \( \overline{\beta} \) is defined as [35].
where, d representing total points of both segmented images, defined as follows:
Further, modify the above equation to get the final fusion image as follows:
Here, \( P\left({\beta}_1,\overline{\beta}\right) \) follows joint probability distribution of β1and \( \overline{\beta} \). For the given total pixels of segmented images d, the MAP estimates \( {\overline{\beta}}_{map} \) and βmap are calculated as:
where, ξpro(x, y)is fused segmented binary image, which is mapped to original RGB image U(x, y) as:
Figure 6 is showing series of steps from active contour segmentation to final RGB image.
3.4 Features extraction
Features give pattern information of the images, which can be later utilized in the classification phase [10, 26,27,28]. In the domain of medical, classification GI diseases showed much attention from last few years. For this purpose, several feature extraction and reduction techniques are proposed by researchers but still it contain set of challenges: a) color similarity between diseased and healthy regions; b) shape of diseased regions; and c) selection of most discriminant features. These problems are somewhat addressed in this article by combining three different types of features including color, LBP, and gray level co-occurrences matrices (GLCM) features.
In the first stage, we extract singular value decomposition (SVD) based color features, which are extracted from RGB mapped image. The purpose of SVD is to reduce degree of freedom in a complex system [32, 40]. As ξprorgb(x, y)is an RGB mapped image, and their extracted channels are ξR, ξG, and ξB.Let the rank of an RGB mapped image matrix isΔA of size (N × N), whereΔA∈ ξprorgb(x, y)andN2represents total number of pixels for each extracted channel.
whereH and V are (N × N) oorthogonal matrix and T denotes the transposition of matrix V. Moreover, hi and vi denotes the H’s and V’s column vector, respectively. The diagonal elements of δ denotes by δi are called the singular values of ΔA and satisfied δ1 ≥ δ2 ≥ ⋯ ≥ δj ≥ δj + 1 = δN = 0. The size of extracted SVD color feature for each channel is (1 × 125) as shown in the Fig. 7. Thereafter, add three mean features for each channel in the SVD matrix, which is later used for classification.
Secondly, we extract LBP and GLCM features for texture analysis. Mostly, LBP features are utilized for face recognition [23] but from last few years, LBP features are used in the domain of medical imaging for disease classification [39]. The LBP operators label the pixels of given images by thresholding function. The thresholding function is performed on the neighborhood of each pixel of the given image and gives the output in binary form. The LBP features are calculated as follows:
Where, Op denotes the neighborhood pixels, Oc denotes the central pixels, s(x) is a sign function, P denotes the symmetric neighborhood pixels, and R denotes the radius of a circle. The LBP gives the output vector of dimension N × 59, where N denotes the number of images which are utilized for feature extraction.
Finally, we extract 22 GLCM features [19] including contrast, entropy, difference variance, difference entropy, information measure of correlation 1, information measure of correlation 2, and few more. The extracted features are finally fused by simple concatenation method, which gives the output fused vector of size (N × 206). The fused vector is defined by Ψfused.
Thereafter, we implement a new but simple method of feature reduction-based probability distribution. The proposed feature reduction method is based on two steps. In the first step, we calculate the probability of a fused feature vector. Then select the higher probability features, which are later utilized in reduction function. The probability of a fused vector is defined as follows:
Let Ψfused(i) denote the features index of the fused vector, Pr denotes the probability value of each extracted feature i, which is defined as:
Wherex(i) denote the number of favorable features, K denotes the total number of features, and Pr(i) is the probability of each feature index. Then select a higher probability feature and put into reduction function to remove the irrelevant features.
Where ξsel(FV) is the final selected feature vector, which is later plugin to MLPNN [10] for classification. The cost function of MLPNN is:
4 Experimental results and discussion
In this section, experimental results of the proposed method are presented in terms of both numerical and graphical plots. To show authenticity of proposed method, we collected 9000 WCE images from 6 patients, which are provided by POF Hospital Wah Cantt, Pakistan. The collected WCE images are divided into three different categories: a) ulcer, b) bleeding, and c) healthy, selected samples are shown in Fig. 1. These 9000 WCE images (3000 Ulcer, 3000 Bleeding, and 3000 Healthy), having resolution (381 × 321), are separated from 18 videos of 6 subjects – belong to any of the aforementioned category. The ground truth images are provided by a specialist doctor who assigned images a label, few sample ground truth images are shown in Fig. 8. Classification results are generated on MLPNN, but to provide a fair comparison, set of classifiers are selected including fine tree (FTree), quadratic discriminant analysis (QDA), linear SVM (LSVM), quadratic SVM (QSVM), cubic SVM (CSVM), Fine Gaussian SVM (FGSVM), Medium Gaussian SVM (MGSVM), fine KNN, medium KNN (MKNN), cosine KNN, cubic KNN, weighted KNN, Boosted Tree, and Bagged Tree. The performance of these classification methods is analyzed based on six statistical measures including sensitivity, AUC, FPR, FNR, accuracy, and computation time. The results are calculated in two different steps: a) ulcer segmentation results and b) classification results. All simulations are being done on MATLAB 2017b using personal desktop core I7 with 8 GB of RAM.
Description of classifiers in terms of selected parameters
Classifier | Description |
FTree | Preset: Fine tree, Maximum number of splits: 100 Split criterion: Gini’s diversity index, Surrogate decision splits: off |
QDA | Preset: Quadratic discriminant, Covariance structure: Full |
LSVM | Preset: Linear SVM, Kernel function: Linear, Kernel scale: Automatic Box constraint level: 1, Multi-class method: One-vs-One Standardized data: true |
QSVM | Preset: Quadratic SVM, Kernel function: Quadratic, Kernel scale: Automatic, Box constraint level: 1, Multi-class method: One-vs-One Standardized data: true |
CSVM | Preset: Cubic SVM, Kernel function: Cubic, Kernel scale: Automatic Box constraint level: 1, Multi-class method: One-vs-One Standardized data: true |
FGSVM | Preset: Fine Gaussian SVM, Kernel function: Gaussian, Kernel scale: 11 Box constraint level: 1, Multi-class method: One-vs-One Standardized data: true |
MGSVM | Preset: Medium Gaussian SVM, Kernel function: Gaussian, Kernel scale: 44, Box constraint level: 1, Multi-class method: One-vs-One Standardized data: true |
Fine KNN | Preset: Fine KNN, Number of Neighbors: 1, Distance Metric: Euclidean Distance Weight: Equal, Standardize data: true |
MKNN | Preset: Medium KNN, Number of Neighbors: 10, Distance Metric: Euclidean, Distance Weight: Equal, Standardize data: true |
Cosine KNN | Preset: Cosine KNN, Number of Neighbors: 10, Distance Metric: Cosine Distance Weight: Equal, Standardize data: true |
Cubic KNN | Preset: Cubic KNN, Number of Neighbors: 10, Distance Metric: Minkowski, Distance Weight: Equal, Standardize data: true |
WKNN | Preset: Weighted KNN, Number of Neighbors: 10, Distance Metric: Euclidean, Distance Weight: Square Inverse, Standardize data: true |
Boosted Tree | Preset: Boosted tree, Ensemble method: AdaBoost, Learner type: Decision tree, Maximum number of splits: 20, Number of learners: 30 Learning rate: 0.1 |
MLPNN | Type: Feed Forward Learning rate: 0.1 |
4.1 Segmentation accuracy
In this step, we present the segmentation accuracy of proposed using relation:
WhereTPrepresents true positive values, TN denotes true negative values, FP denotes false positive values, and FN denotes the false negative values. These values are calculated by comparing each segmented image and their corresponding given ground truth image. The proposed CBCSF method is tested on all selected images of type ulcer and their few accuracy results are given in the Table 1. The maximum segmentation accuracy achieved is 97.46% and average accuracy of 87.9635%. A comparison is also being carried out with [14], which achieved maximum segmentation accuracy of 87.09%. The sample ulcer segmentation results with ground truth images are shown in Fig. 8, and bleeding segmentation results are shown in Fig. 9.
4.2 Classification accuracy
In this section, we present proposed classification results in terms of accuracy, sensitivity, FNR, FPR, and AUC. The classification results are computed into five distinct scenarios - given in Table 2. In the first scenario, all three classes such as ulcer, bleeding, and healthy are selected to extract their LBP features. For this purpose, 50:50 training and testing method is opted, The 50% samples from each class are selected for testing, and results are validated using 10-fold cross-validation. The maximum testing accuracy of scenario 1 achieved is 99.60%, sensitivity rate of 0.996, FPR 0.000, FNR 0.4, and AUC is 1.00 on MLPNN, given in Table 3. Moreover, the best accuracy on some other supervised learning methods such as Fine KNN, CSVM, QSVM, and WKNN using same features are 99.50%, 99.40%, 99.00%, and 99.00%, respectively. In Fig. 10, a confusion matrix is presented, which shows the authenticity of MLPNN. Moreover, the computation time of each classifier including MLPNN is also provided, having best time of 11.60 s (MLPNN), however, the worst testing computation time is 41.26 s (Fine tree), Table 3. The computational time of all classification methods are given in Fig. 11
In the second scenario, GLCM features are extracted to perform classification. For classification 10 fold cross validation is performed and achieved maximum classification accuracy in terms of accuracy 97.3%, FNR 2.7%, FPR 0.013, sensitivity 0.971, and AUC is 0.9866 as given in Table 4. The classification accuracy of MLPNN is proved by the confusion matrix presented in Fig. 12. A second highest accuracy, sensitivity, and AUC is 97.2%, 0.970, and 0.9866, which is achieved on QSVM. The computation time for classification method is also calculated and best-achieved computation time is 8.44 s for MLPNN, however, the worst execution time is 35.08 and classification accuracy is 82.2%, which is achieved on MGSVM. Moreover, the computation time of all classification methods is also plotted in Fig. 13.
In the third scenario, color features are extracted from tested images. The color features are extracted from RGB mapped images having dimension 1 × 128. The extracted color descriptors consist of 125 SVD and 3 mean features. Thereafter, perform 10 fold cross-validation and obtained maximum results upto 99.7% on MLPNN, which is presented in Table 5. In Fig. 14, a confusion matrix is shown, which confirms the performance of MLPNN. Tables 3, 4, and 5 show that color features performs better for classification of GI diseases. However, the best computation time for color features is 9.02 s as shown in Fig. 15, which was achieved on MLPNN. It is a little bit higher than GLCM features due to an increase in the number of features. Because the dimension of GLCM features is 1 × 42, however, color features are 1 × 128 in dimension. But, from Tables 3 and 4, it is clearly shown that color features perform better as compared to LBP and GLCM features.
After computation of classification results on individual feature vectors, fused features using the serial-based method in the fourth scenario. The major aim of feature fusion is to improve the classification accuracy and also reduce the computation time. The fused features are store into one matrix and performed 10-foldcross-validation. The fusion process produces maximum classification accuracy is 99.5% on MLPNN as presented in Table 6. The performance of MLPNN is confirmed by a confusion matrix shown in Fig. 16. However, we noticed that the classification accuracy of MLPNN on fused features is less than color features (99.70%) and LBP features (99.60%) but it improves the performance in terms of computation time, which is 8.116 s. The computation time of LBP features is 11.61, GLCM features are 8.44, and 9.02 s for color features, whereas the computation time of MLPNN on a fused vector is 8.116 as shown in Fig. 17, which is good as compared to previous scenarios.
Finally, the proposed feature selection method is applied to the fused feature vector and selects the best features to make the proposed method more reliable and efficient in terms classification accuracy and computational time. The LBP, GLCM, and color features are extracted from 50% testing images and fused in one matrix. Thereafter, select the best features and performed 10-fold cross-validation. The maximum achieved classification accuracy is 100% on MLPNN and FGSVM but theperformance of MLPNN is better as compared to FGSVM based on their computation time as given in Table 7. The performance of MLPNN is confirmed by confusion matrix given in Fig. 18. Moreover, the proposed selection results are proved through ROC plots as shown in Fig. 19. The ROC plots are shown for each class which reveals the maximum AUC and minimum FP rate.
The classification results of Table 7 presented that the other supervised learning classification methods perform well on best-selected features and achieved average classification accuracy is 99.03%. The best computation time on selected features is 3.624 s which is achieved on MLPNN, however, the second-best time is 7.007 s for FTree as shown in Fig. 20. The all above results and discussion, it is clearly shown that the proposed method performs well for probability based best-selected features and achieved improved performance in terms of accuracy, sensitivity, FPR, and computation time. In addition, we also performed three validation methods such as Hold-Out, Leave-out-one, and K-fold for computation of the classification results using proposed features selection method- results are presented in Tables 8 and 9. From the stats, it is quite cleared that the proposed method still performs exceptionally by achieving the classification accuracies of 99.7% and 99.6% (Table 10).
Moreover, a general comparison with existing methods is also presented in Table 8 in terms of accuracy and sensitivity rate. The time parameter is not added under the comparison table because in literature, most of the authors are not considering it and are only focused on the classification accuracy. From the analysis, it is concluded that the proposed method outperforms existing methods in terms of greater accuracy and sensitivity. Few labeled recognition results of proposed method are shown in Fig. 21.
5 Conclusion
A new method is proposed for detecting and classifying GI diseases from WCE images. Fusion of HSI active contour and a newly improved saliency method using MAP is the crux of this framework. We clearly verify from the statistical measures that all three selected classes are accurately classified with proposed by achieving a maximum accuracy of 100% and best computation time of 3.624 s. Moreover, we make use of multiple features including color, LBP, and GLCM to generate a robust feature set – comprising all good range of features.
In the above discussion, we conclude that color transformation plays a key role in ulcer segmentation from WCE images. It not only highlights primary infected regions but also the regions where RGB color space fails to reveal a difference. Moreover, fusion of set of features worked well. In the coming articles, we will be focusing on fusing more number of features as well as interested in adding additional steps of feature selection and dimensionality reduction. It will not only select most discriminant features but also computationally not very expensive in terms of time. Additionally, in the future, a deep CNN method (DenseNet, Inception V3) will be implemented by applying transfer learning using more than 20,000 images from a greater number of patients.
References
Akram T et al (2018) Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J Ambient Intell Humaniz Comput:1–20
Ali H et al (2018) Computer assisted gastric abnormalities detection using hybrid texture descriptors for chromoendoscopy images. Comput Methods Prog Biomed
Chan TF, Vese LA (2001) Active contours without edges. IEEE Trans Image Process 10(2):266–277
Charfi S, El Ansari M (2017) Computer-aided diagnosis system for ulcer detection in wireless capsule endoscopy videos. In: Advanced Technologies for Signal and Image Processing (ATSIP), 2017 International Conference on. IEEE
Charfi S, El Ansari M (2018) Computer-aided diagnosis system for colon abnormalities detection in wireless capsule endoscopy images. Multimed Tools Appl 77(3):4047–4064
Duan Q et al (2016) Visual saliency detection using information contents weighting. Optik 127(19):7418–7430
Fu Y et al (2014) Computer-aided bleeding detection in WCE video. IEEE J Biomed Health Inform 18(2):636–642
Iddan G et al (2000) Wireless capsule endoscopy. Nature 405(6785):417
Jeon G (2013) Contrast intensification in NTSC YIQ. International Journal of Control and Automation 6(4):157–166
Khan MA et al (2018) An implementation of optimized framework for action classification using multilayers neural network on selected fused features. Pattern Anal Applic: 1-21
Khan SA et al (2019) Lungs nodule detection framework from computed tomography images using support vector machine. Microsc Res Tech
Khan MA et al (2019) Brain tumor detection and classification: a framework of marker-based watershed algorithm and multilevel priority features selection. Microsc Res Tech
Khan MA et al (2019) Construction of saliency map and hybrid set of features for efficient segmentation and classification of skin lesion. Microsc Res Tech
Kundu A, Fattah S (2017) An asymmetric indexed image based technique for automatic ulcer detection in wireless capsule endoscopy images. In: Humanitarian Technology Conference (R10-HTC), 2017 IEEE Region 10. IEEE
Kundu A et al (2017) An automatic ulcer detection scheme using histogram in YIQ domain from wireless capsule endoscopy images. In: Region 10 Conference, TENCON 2017-2017 IEEE. IEEE
Li B, Meng MQ-H (2009) Computer-based detection of bleeding and ulcer in wireless capsule endoscopy images by chromaticity moments. Comput Biol Med 39(2):141–147
Li B, Meng MQ-H (2012) Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Trans Inf Technol Biomed 16(3):323–329
Liaqat A et al (2018) Automated ulcer and bleeding classification from wce images using multiple features fusion and selection. Journal of Mechanics in Medicine and Biology:1850038
Maghsoudi OH, Alizadeh M (2018) Feature based framework to detect diseases, tumor, and bleeding in wireless capsule endoscopy. arXiv preprint arXiv:1802.02232
Maghsoudi OH, Alizadeh M, Mirmomen M (2016) A computer aided method to detect bleeding, tumor, and disease regions in Wireless Capsule Endoscopy. in Signal Processing in Medicine and Biology Symposium (SPMB), 2016 IEEE. IEEE
Mergener K (2008) Update on the use of capsule endoscopy. Gastroenterol Hepatol 4(2):107
Nasir M et al (2018) An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microsc Res Tech
Pei S-C et al (2017) Compact LBP and WLBP descriptor with magnitude and direction difference for face recognition. In: Image Processing (ICIP), 2017 IEEE International Conference on. IEEE
Rashid M et al (2018) Object detection and classification: a joint selection and fusion strategy of deep convolutional neural network and SIFT point features. Multimed Tools Appl:1–27
Ribeiro MG et al (2019) Classification of colorectal cancer based on the association of multidimensional and multiresolution features. Expert Syst Appl 120:262–278
Sharif M et al (2017) A framework of human detection and action recognition based on uniform segmentation and combination of Euclidean distance and joint entropy-based features selection. EURASIP Journal on Image and Video Processing 2017(1):89
Sharif M et al (2018) Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J Ambient Intell Humaniz Comput:1-20
Sharif M et al (2018) Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Comput Electron Agric 150:220–234
Siegel R, DeSantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA Cancer J Clin 64(2):104–117
Siegel RL et al (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67(3):177–193
Sivakumar P, Kumar BM (2018) A novel method to detect bleeding frame and region in wireless capsule endoscopy video. Clust Comput: 1-7
Stewart GW (1993) On the early history of the singular value decomposition. SIAM Rev 35(4):551–566
Suman S et al (2017) Feature selection and classification of ulcerated lesions using statistical analysis for WCE images. Appl Sci 7(10):1097
Suman S et al (2017) Detection and classification of bleeding region in WCE images using color feature. In: Proceedings of the 15th International Workshop on Content-Based Multimedia Indexing. ACM
Xue Z, Li SZ, Teoh EK (2003) Bayesian shape model for facial feature extraction and recognition. Pattern Recogn 36(12):2819–2833
Yuan Y, Meng MQH (2017) Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 44(4):1379–1389
Yuan Y et al (2015) Saliency based ulcer detection for wireless capsule endoscopy diagnosis. IEEE Trans Med Imaging 34(10):2046–2057
Yuan Y et al (2017) Discriminative joint-feature topic model with dual constraints for WCE classification. IEEE Transactions on Cybernetics
Yuan Y, Li B, Meng MQ-H (2017) WCE abnormality detection based on saliency and adaptive locality-constrained linear coding. IEEE Trans Autom Sci Eng 14(1):149–159
Zhang S, Wang Z (2016) Cucumber disease recognition based on global-local singular value decomposition. Neurocomputing 205:341–348
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, M.A., Rashid, M., Sharif, M. et al. Classification of gastrointestinal diseases of stomach from WCE using improved saliency-based method and discriminant features selection. Multimed Tools Appl 78, 27743–27770 (2019). https://doi.org/10.1007/s11042-019-07875-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-07875-9